Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies
Abstract
:1. The Burden and Risks of Bacterial Infections
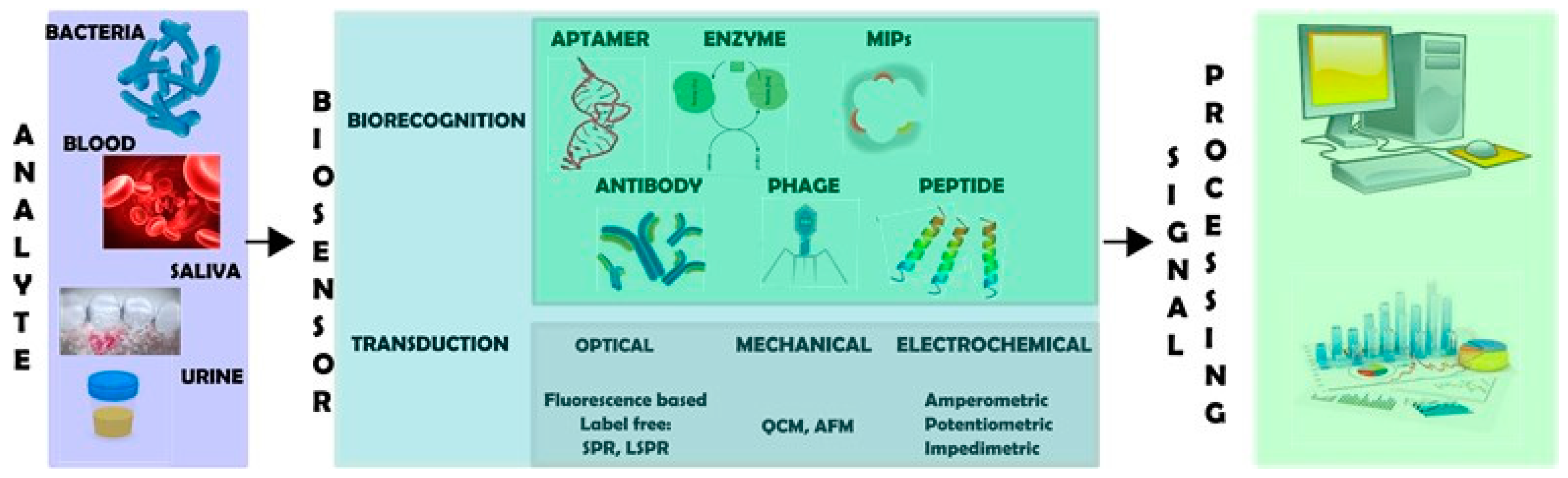
2. Biosensors for Detecting Bacterial Cells
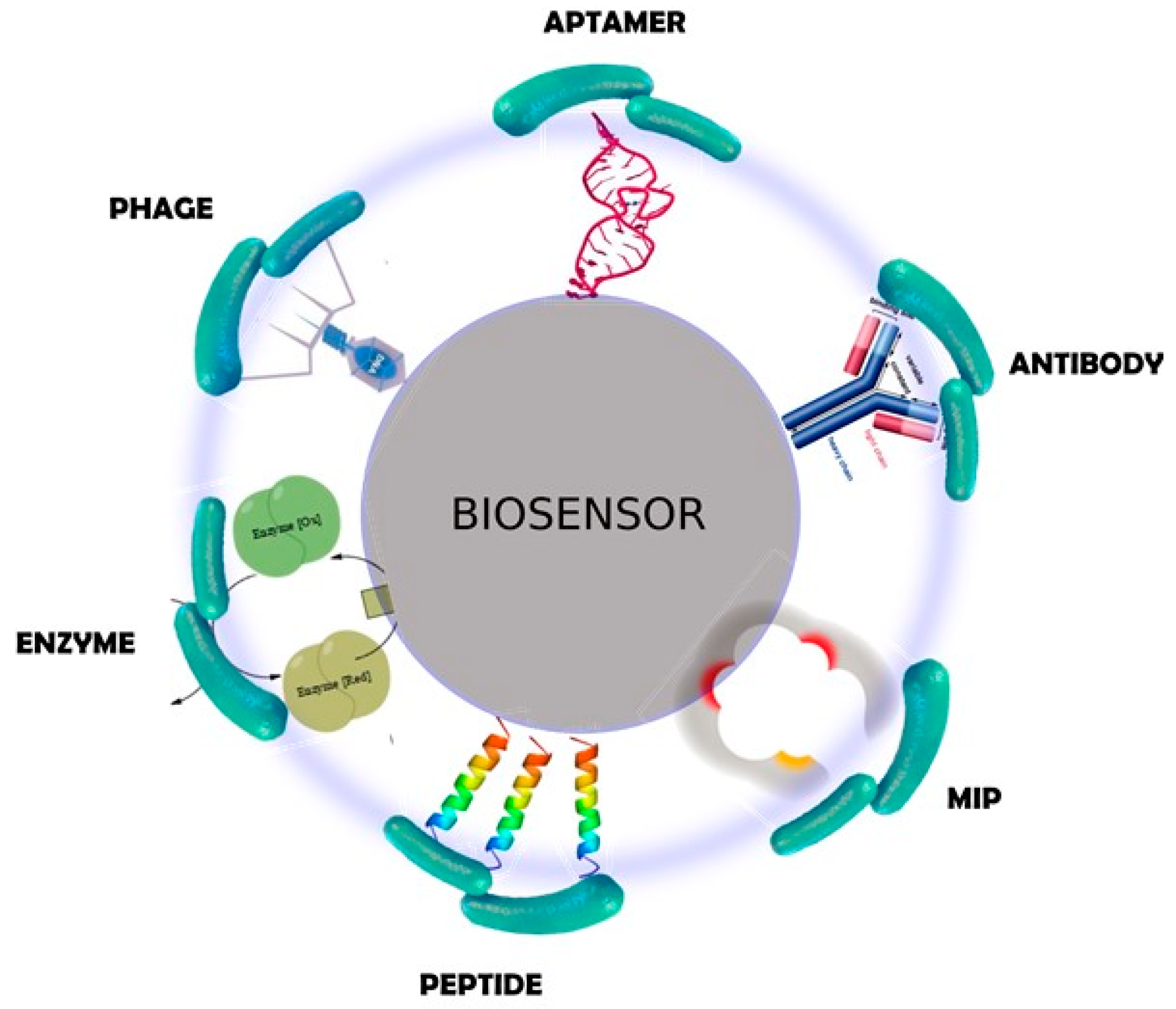
2.1. Biorecognition Elements
2.2. Transduction Systems
2.2.1. Mechanical Transducers
2.2.2. Optical Transducers
2.2.3. Electrochemical Transducers
2.3. Signal Amplifiers
3. AMP-Based Impedimetric Biosensors
3.1. AMPs as Biorecognition Elements
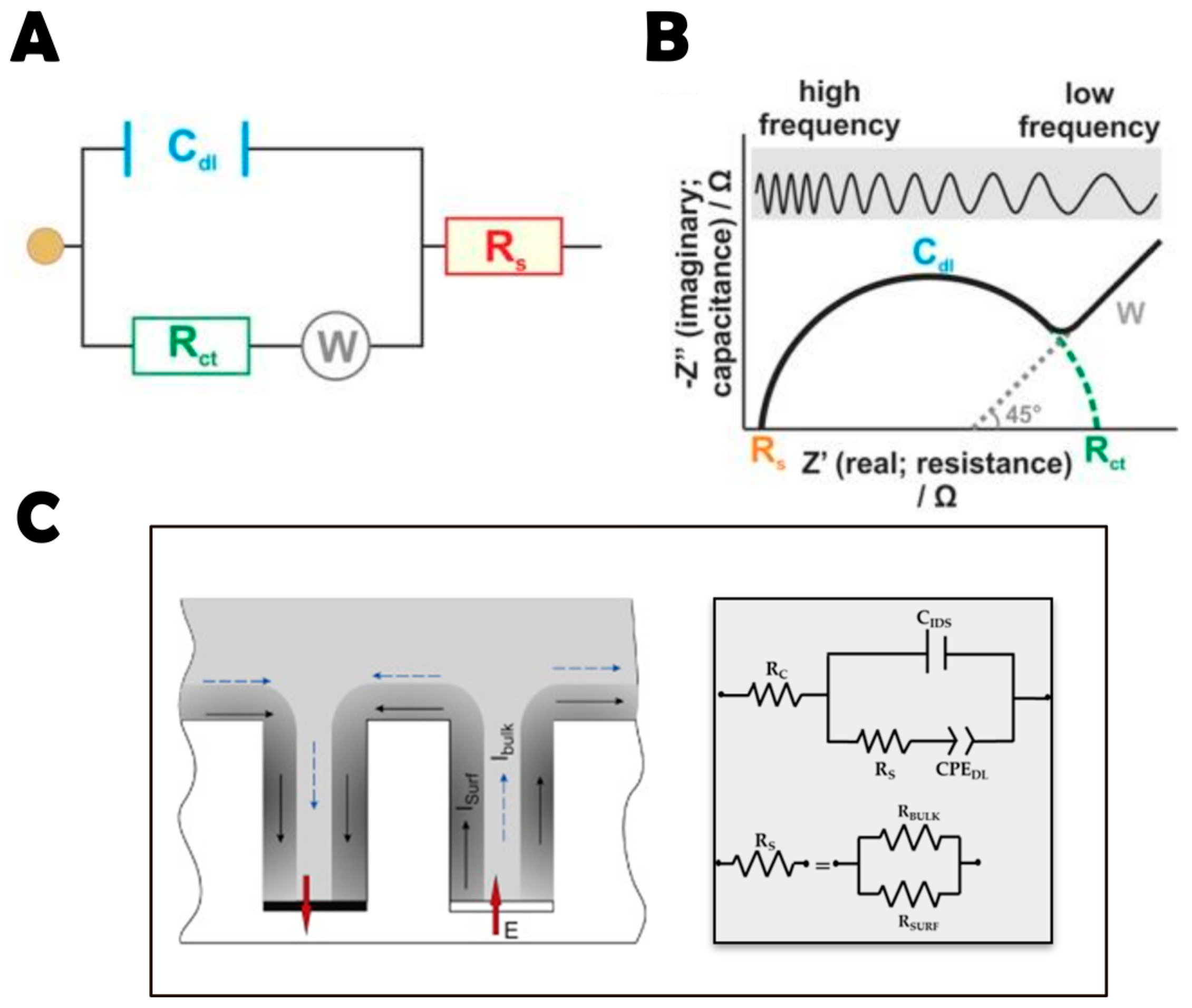
3.2. EIS Technique
3.3. Examples of AMP-Based Biosensors for Bacterial Detection
4. Conclusions
Funding
Conflicts of Interest
References
- De La Fuente-Núñez, C.; Cardoso, M.H.; De Souza Cândido, E.; Franco, O.L.; Hancock, R.E.W. Synthetic antibiofilm peptides. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- WHO 2018 Report. Available online: http://www.who.int/gho/mortality_burden_disease/causes_death/top_10/en (accessed on 15 May 2018).
- Riool, M.; de Breij, A.; Drijfhout, J.W.; Nibbering, P.H.; Zaat, S.A.J. Antimicrobial Peptides in Biomedical Device Manufacturing. Front. Chem. 2017, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Burlage, R.S.; Tillmann, J. Biosensors of bacterial cells. J. Microbiol. Methods 2017, 138, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Caygill, R.L.; Blair, G.E.; Millner, P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta 2010, 681, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Bahadır, E.B.; Sezgintürk, M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015, 478, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Pashazadeh, P.; Mokhtarzadeh, A.; Hasanzadeh, M.; Hejazi, M.; Hashemi, M.; de la Guardia, M. Nano-materials for use in sensing of salmonella infections: Recent advances. Biosens. Bioelectron. 2017, 87, 1050–1064. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Frank Cheng Biosensors for Bacterial Detection. Int. J. Biosens. Bioelectron. Biosens. 2017, 2, 7–9. [Google Scholar] [CrossRef]
- Jarocka, U.; Sawicka, R.; Góra-Sochacka, A.; Sirko, A.; Dehaen, W.; Radecki, J.; Radecka, H. An electrochemical immunosensor based on a 4,4′-thiobisbenzenethiol self-assembled monolayer for the detection of hemagglutinin from avian influenza virus H5N1. Sens. Actuators B Chem. 2016, 228, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Shi, S.; Ma, R.; Jia, W.; Wang, H. Highly sensitive electrochemical biosensor based on nonlinear hybridization chain reaction for DNA detection. Biosens. Bioelectron. 2016, 80, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Goldzstein, A.; Aamouche, A.; Homblé, F.; Voué, M.; Conti, J.; De Coninck, J.; Devouge, S.; Marchand-Brynaert, J.; Goormaghtigh, E. Ligand-receptor interactions in complex media: A new type of biosensors for the detection of coagulation factor VIII. Biosens. Bioelectron. 2009, 24, 831–836. [Google Scholar] [CrossRef]
- Chang, Y.F.; Fu, C.; Chen, Y.T.; Fang-Ju Jou, A.; Chen, C.C.; Chou, C.; Annie Ho, J.A. Use of liposomal amplifiers in total internal reflection fluorescence fiber-optic biosensors for protein detection. Biosens. Bioelectron. 2016, 77, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Wang, C.Y.; Huang, C.F.; Cheng, A.T. Interdigitated electrodes based on impedance biosensor for sensing peptide LL-37. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Boston, MA, USA, 30 August–3 September 2011; pp. 71–74. [Google Scholar] [CrossRef]
- Gs, S.; Cv, A.; Mathew, B.B. Biosensors: A Modern Day Achievement. J. Instrum. Technol. 2014, 2, 26–39. [Google Scholar] [CrossRef]
- Laura Estela, C.R.; Alejandro, R.P. Biofilms: A Survival and Resistance Mechanism of Microorganisms. In Antibiotic Resistant Bacteria—A Continuous Challenge in the New Millennium; InTech: Rijeka, Croatia, 2012; Volume 2, p. 64. ISBN 9789537619992. [Google Scholar]
- Ducret, A.; Valignat, M.-P.; Mouhamar, F.; Mignot, T.; Theodoly, O. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc. Natl. Acad. Sci. USA 2012, 109, 10036–10041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, P.K.; Janissen, R.; Monteiro, M.P.; Cavalli, A.; Murillo, D.M.; Merfa, M.V.; Cesar, C.L.; Carvalho, H.F.; De Souza, A.A.; Bakkers, E.P.A.M.; et al. Nanowire arrays as cell force sensors to investigate adhesin-enhanced holdfast of single cell bacteria and biofilm stability. Nano Lett. 2016, 16, 4656–4664. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Nogués, M.; Buxadera-Palomero, J.; Ginebra, M.P.; Manero, J.M.; Gil, F.J.; Mas-Moruno, C. All-in-one trifunctional strategy: A cell adhesive, bacteriostatic and bactericidal coating for titanium implants. Colloids Surf. B Biointerfaces 2018, 169, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Lu, Y.; Gau, V.; Liao, J.C.; Wong, P.K. Rapid Antimicrobial Susceptibility Testing with Electrokinetics Enhanced Biosensors for Diagnosis of Acute Bacterial Infections. Ann. Biomed. Eng. 2014, 42, 2314–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, P.; Bhunia, A.K. Cell-based biosensor for rapid screening of pathogens and toxins. Biosens. Bioelectron. 2010, 26, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Justino, C.I.L.; Freitas, A.C.; Pereira, R.; Duarte, A.C.; Rocha Santos, T.A.P. Recent developments in recognition elements for chemical sensors and biosensors. TrAC Trends Anal. Chem. 2015, 68, 2–17. [Google Scholar] [CrossRef]
- Templier, V.; Roux, A.; Roupioz, Y.; Livache, T. Ligands for label-free detection of whole bacteria on biosensors: A review. TrAC Trends Anal. Chem. 2016, 79, 71–79. [Google Scholar] [CrossRef]
- Arugula, M.A; Simonian, A. Novel trends in affinity biosensors: Current challenges and perspectives. Meas. Sci. Technol. 2014, 25, 032001. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Abramova, N.; Bratov, A.; Vigués, N.; Mas, J.; Muñoz, F.-X. Sensitivity and Response Time of Polyethyleneimine Modified Impedimetric Transducer for Bacteria Detection. Electroanalysis 2015, 27, 656–662. [Google Scholar] [CrossRef]
- Banerjee, P.; Bhunia, A.K. Mammalian cell-based biosensors for pathogens and toxins. Trends Biotechnol. 2009, 27, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-M.; Jeon, J.-W.; Kim, T.-Y.; Paek, S.-H. An innate immune system-mimicking, real-time biosensing of infectious bacteria. Analyst 2015, 140, 6061–6070. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Richter, Ł.; Janczuk-Richter, M.; Niedziółka-Jönsson, J.; Paczesny, J.; Hołyst, R. Recent advances in bacteriophage-based methods for bacteria detection. Drug Discov. Today 2017. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.W.; Morin, K.M.; Mello, C.M. Antimicrobial Peptides for Use in Biosensing Applications; US Army Research; Army Natick Research Development and Engineering Center: Natick, MA, USA, 2004; pp. 1–7. [Google Scholar]
- Pavan, S.; Berti, F. Short peptides as biosensor transducers. Anal. Bioanal. Chem. 2012, 402, 3055–3070. [Google Scholar] [CrossRef] [PubMed]
- Uzun, L.; Turner, A.P.F. Molecularly-imprinted polymer sensors: Realising their potential. Biosens. Bioelectron. 2016, 76, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Latif, U.; Qian, J.; Can, S.; Dickert, F.L. Biomimetic receptors for bioanalyte detection by quartz crystal microbalances—From molecules to cells. Sensors 2014, 14, 23419–23438. [Google Scholar] [CrossRef] [PubMed]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T.A.P. Analytical applications of affibodies. TrAC Trends Anal. Chem. 2015, 65, 73–82. [Google Scholar] [CrossRef]
- Ye, M.; Hu, J.; Peng, M.; Liu, J.; Liu, J.; Liu, H.; Zhao, X.; Tan, W. Generating aptamers by cell-SELEX for applications in molecular medicine. Int. J. Mol. Sci. 2012, 13, 3341–3353. [Google Scholar] [CrossRef] [PubMed]
- Šmuc, T.; Ahn, I.Y.; Ulrich, H. Nucleic acid aptamers as high affinity ligands in biotechnology and biosensorics. J. Pharm. Biomed. Anal. 2013, 81–82, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.M.; Lee, S.Y. Optical Biosensors for the Detection of Pathogenic Microorganisms. Trends Biotechnol. 2016, 34, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Miranda, O.R.; Li, X.; Garcia-Gonzalez, L.; Zhu, Z.J.; Yan, B.; Bunz, U.H.F.; Rotello, V.M. Colorimetric bacteria sensing using a supramolecular enzyme-nanoparticle biosensor. J. Am. Chem. Soc. 2011, 133, 9650–9653. [Google Scholar] [CrossRef] [PubMed]
- Jokerst, J.C.; Adkins, J.A.; Bisha, B.; Mentele, M.M.; Goodridge, L.D.; Henry, C.S. Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens. Anal. Chem. 2012, 84, 2900–2907. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, Z.; Ackerman, J.D.; Yazdani, M.; Hou, S.; Nugen, S.R.; Rotello, V.M. Electrochemical nanoparticle-enzyme sensors for screening bacterial contamination in drinking water. Analyst 2015, 140, 4991–4996. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Knoll, W.; Dostalek, J. Bacterial pathogen surface plasmon resonance biosensor advanced by long range surface plasmons and magnetic nanoparticle assays. Anal. Chem. 2012, 84, 8345–8350. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Mouawad, F.; Lévesque, S.; Sacher, E.; Mandeville, R.; Meunier, M. The differential detection of methicillin-resistant, methicillin-susceptible and borderline oxacillin-resistant Staphylococcus aureus by surface plasmon resonance. Biosens. Bioelectron. 2013, 49, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; Wang, D.; Zhang, X.; Zuo, G.; Wei, H.; Yang, R.; Zhang, Z.; Cheng, Z.; Guo, Y.; Cui, Z.; et al. Rapid detection of Bacillus anthracis using monoclonal antibody functionalized QCM sensor. Biosens. Bioelectron. 2009, 24, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, R.; Wang, Y.; Su, X.; Ying, Y.; Wang, J.; Li, Y. Evaluation of different micro/nanobeads used as amplifiers in QCM immunosensor for more sensitive detection of E. coli O157:H7. Biosens. Bioelectron. 2011, 29, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Lin, C.S.; Chen, S.H.; Ye, R.; Wu, V.C.H. A piezoelectric immunosensor for specific capture and enrichment of viable pathogens by quartz crystal microbalance sensor, followed by detection with antibody-functionalized gold nanoparticles. Biosens. Bioelectron. 2012, 38, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Salam, F.; Uludag, Y.; Tothill, I.E. Real-time and sensitive detection of Salmonella typhimurium using an automated quartz crystal microbalance (QCM) instrument with nanoparticles amplification. Talanta 2013, 115, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Sungkanak, U.; Sappat, A.; Wisitsoraat, A.; Promptmas, C.; Tuantranont, A. Ultrasensitive detection of Vibrio cholerae O1 using microcantilever-based biosensor with dynamic force microscopy. Biosens. Bioelectron. 2010, 26, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Mutharasan, R. Rapid and sensitive immunodetection of Listeria monocytogenes in milk using a novel piezoelectric cantilever sensor. Biosens. Bioelectron. 2013, 45, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Laczka, O.; Maesa, J.M.; Godino, N.; del Campo, J.; Fougt-Hansen, M.; Kutter, J.P.; Snakenborg, D.; Muñoz-Pascual, F.X.; Baldrich, E. Improved bacteria detection by coupling magneto-immunocapture and amperometry at flow-channel microband electrodes. Biosens. Bioelectron. 2011, 26, 3633–3640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Ávila, B.E.F.; Pedrero, M.; Campuzano, S.; Escamilla-Gómez, V.; Pingarrón, J.M. Sensitive and rapid amperometric magnetoimmunosensor for the determination of Staphylococcus aureus. Anal. Bioanal. Chem. 2012, 403, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Varshney, M.; Li, Y. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle-antibody conjugates for detection of Escherichia coli O157:H7 in food samples. Biosens. Bioelectron. 2007, 22, 2408–2414. [Google Scholar] [CrossRef] [PubMed]
- Mejri, M.B.; Baccar, H.; Baldrich, E.; Del Campo, F.J.; Helali, S.; Ktari, T.; Simonian, A.; Aouni, M.; Abdelghani, A. Impedance biosensing using phages for bacteria detection: Generation of dual signals as the clue for in-chip assay confirmation. Biosens. Bioelectron. 2010, 26, 1261–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dweik, M.; Stringer, R.C.; Dastider, S.G.; Wu, Y.; Almasri, M.; Barizuddin, S. Specific and targeted detection of viable Escherichia coli O157:H7 using a sensitive and reusable impedance biosensor with dose and time response studies. Talanta 2012, 94, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Barreiros dos Santos, M.; Agusil, J.P.; Prieto-Simón, B.; Sporer, C.; Teixeira, V.; Samitier, J. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2013, 45, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Ye, W.W.; Zhang, Y.; Xiao, L.D.; Leung, P.H.M.; Li, Y.; Yang, M. Ultrasensitive detection of E. coli O157:H7 with biofunctional magnetic bead concentration via nanoporous membrane based electrochemical immunosensor. Biosens. Bioelectron. 2013, 41, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Alocilja, E.C. Gold nanoparticle-labeled biosensor for rapid and sensitive detection of bacterial pathogens. J. Biol. Eng. 2015, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Dou, W.; Zhao, G. Amperometric immunoassay for the detection of Salmonella pullorum using a screen—Printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Microchim. Acta 2016, 183, 757–764. [Google Scholar] [CrossRef]
- Bruno, J. Application of DNA Aptamers and Quantum Dots to Lateral Flow Test Strips for Detection of Foodborne Pathogens with Improved Sensitivity versus Colloidal Gold. Pathogens 2014, 3, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.Y.; Jeong, H.Y.; Kim, M.I.; Park, T.J. Colorimetric Detection System for Salmonella typhimurium Based on Peroxidase-Like Activity of Magnetic Nanoparticles with DNA Aptamers. J. Nanomater. 2015, 2015, 2. [Google Scholar] [CrossRef]
- Yoo, S.M.; Kim, D.-K.; Lee, S.Y. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta 2015, 132, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Tram, K.; Kanda, P.; Salena, B.J.; Huan, S.; Li, Y. Translating bacterial detection by DNAzymes into a litmus test. Angew. Chem. 2014, 53, 12799–12802. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.Z.; Song, H. Bin; Zuo, G.M.; Yang, R.F.; Wei, H.P.; Wang, D.B.; Cui, Z.Q.; Zhang, Z.P.; Cheng, Z.X.; Zhang, X.E. DNA probe functionalized QCM biosensor based on gold nanoparticle amplification for Bacillus anthracis detection. Biosens. Bioelectron. 2011, 26, 3398–3404. [Google Scholar] [CrossRef] [PubMed]
- Khemthongcharoen, N.; Wonglumsom, W.; Suppat, A.; Jaruwongrungsee, K.; Tuantranont, A.; Promptmas, C. Piezoresistive microcantilever-based DNA sensor for sensitive detection of pathogenic Vibrio cholerae O1 in food sample. Biosens. Bioelectron. 2015, 63, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Zelada-Guillen, G.A.; Riu, J.; Düzgün, A.; Rius, F.X. Immediate detection of living bacteria at ultralow concentrations using a carbon nanotube based potentiometrie aptasensor. Angew. Chem. 2009, 48, 7334–7337. [Google Scholar] [CrossRef] [PubMed]
- Zelada-Guillén, G.A.; Sebastián-Avila, J.L.; Blondeau, P.; Riu, J.; Rius, F.X. Label-free detection of Staphylococcus aureus in skin using real-time potentiometric biosensors based on carbon nanotubes and aptamers. Biosens. Bioelectron. 2012, 31, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.; Stoltenburg, R.; Strehlitz, B.; Frense, D.; Beckmann, D. Development of an impedimetric aptasensor for the detection of Staphylococcus aureus. Int. J. Mol. Sci. 2017, 18, 2484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, Z.; Si, C.; Ying, Y. Monitoring of Escherichia coli O157:H7 in food samples using lectin based surface plasmon resonance biosensor. Food Chem. 2013, 136, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Serra, B.; Gamella, M.; Reviejo, A.J.; Pingarron, J.M. Lectin-modified piezoelectric biosensors for bacteria recognition and quantification. Anal. Bioanal. Chem. 2008, 391, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Yakovleva, M.E.; Moran, A.P.; Safina, G.R.; Wadström, T.; Danielsson, B. Lectin typing of Campylobacter jejuni using a novel quartz crystal microbalance technique. Anal. Chim. Acta 2011, 694, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gamella, M.; Campuzano, S.; Parrado, C.; Reviejo, A.J.; Pingarron, J.M. Microorganisms recognition and quantification by lectin adsorptive affinity impedance. Talanta 2009, 78, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.S.L.; Oliveira, M.D.L.; De Melo, C.P.; Andrade, C.A.S. Impedimetric sensor of bacterial toxins based on mixed (Concanavalin A)/polyaniline films. Colloids Surf. B Biointerfaces 2014, 117, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Brosel-Oliu, S.; Galyamin, D.; Abramova, N.; Muñoz-Pascual, F.X.; Bratov, A. Impedimetric label-free sensor for specific bacteria endotoxin detection by surface charge registration. Electrochim. Acta 2017, 243, 142–151. [Google Scholar] [CrossRef]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.M.; Bock, W.J.; Mikulic, P.; Chinnappan, R.; Ng, A.; Tolba, M.; Zourob, M. Long period grating based biosensor for the detection of Escherichia coli bacteria. Biosens. Bioelectron. 2012, 35, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chai, Y. Blocking Non-Specific Binding for Phage-Based Magnetoelastic Biosensors. Biosens. J. 2015, 4, 1–2. [Google Scholar] [CrossRef]
- Chen, J.; Alcaine, S.D.; Jiang, Z.; Rotello, V.M.; Nugen, S.R. Detection of Escherichia coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015, 87, 8977–8984. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.V.; Sorokulova, I.B.; Petrenko, V.A.; Chen, I.H.; Barbaree, J.M.; Vodyanoy, V.J. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens. Bioelectron. 2006, 21, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Chen, H.; Horikawa, S.; Shen, W.; Simonian, A.; Chin, B.A. Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens. Bioelectron. 2010, 26, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Li, S.; Chin, B.A. Detection of Salmonella typhimurium Grown Directly on Tomato Surface Using Phage-Based Magnetoelastic Biosensors. Food Bioprocess Technol. 2013, 6, 682–689. [Google Scholar] [CrossRef]
- Chai, Y.; Li, S.; Horikawa, S.; Park, M.-K.; Vodyanoy, V.; Chin, B.A. Rapid and Sensitive Detection of Salmonella typhimurium on Eggshells by Using Wireless Biosensors. J. Food Prot. 2012, 75, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Park, J.W.; Wikle, H.C.; Chin, B.A. Evaluation of phage-based magnetoelastic biosensors for direct detection of Salmonella typhimurium on spinach leaves. Sens. Actuators B Chem. 2012, 176, 1134–1140. [Google Scholar] [CrossRef]
- Yemini, M.; Levi, Y.; Yagil, E.; Rishpon, J. Specific electrochemical phage sensing for Bacillus cereus and Mycobacterium smegmatis. Bioelectrochemistry 2007, 70, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Tlili, C.; Sokullu, E.; Safavieh, M.; Tolba, M.; Ahmed, M.U.; Zourob, M. Bacteria screening, viability, and confirmation assays using bacteriophage-impedimetric/loop-mediated isothermal amplification dual-response biosensors. Anal. Chem. 2013, 85, 4893–4901. [Google Scholar] [CrossRef] [PubMed]
- Tolba, M.; Ahmed, M.U.; Tlili, C.; Eichenseher, F.; Loessner, M.J.; Zourob, M. A bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of Listeria cells. Analyst 2012, 137, 5749–5756. [Google Scholar] [CrossRef] [PubMed]
- Shabani, A.; Marquette, C.A.; Mandeville, R.; Lawrence, M.F. Carbon microarrays for the direct impedimetric detection of Bacillus anthracis using Gamma phages as probes. Analyst 2013, 138, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Kulagina, N.V.; Shaffer, K.M.; Anderson, G.P.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptide-based array for Escherichia coli and Salmonella screening. Anal. Chim. Acta 2006, 575, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Kulagina, N.V.; Shaffer, K.M.; Ligler, F.; Taitt, C.R. Antimicrobial peptides as new recognition molecules for screening challenging species. Sens. Actuators B Chem. 2007, 121, 150–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, J.H.; Woo, D.H.; Chun, M.S.; Chang, M.S. Microfluidic based biosensing for Escherichia coli detection by embedding antimicrobial peptide-labeled beads. Sens. Actuators B Chem. 2014, 191, 211–218. [Google Scholar] [CrossRef]
- Kulagina, N.V.; Lassman, M.E.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptides for detection of bacteria in biosensor assays. Anal. Chem. 2005, 77, 6504–6508. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.-M.; Zhao, G.-C. Label-free detection of pathogenic bacteria via immobilized antimicrobial peptides. Talanta 2015, 137, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, X.; Yao, Q.; He, F. A novel method for the rapid detection of microbes in blood using pleurocidin antimicrobial peptide functionalized piezoelectric sensor. J. Microbiol. Methods 2017, 133, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, M.S.; Zhang, S.; Link, A.J.; McAlpine, M.C. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2010, 107, 19207–19212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
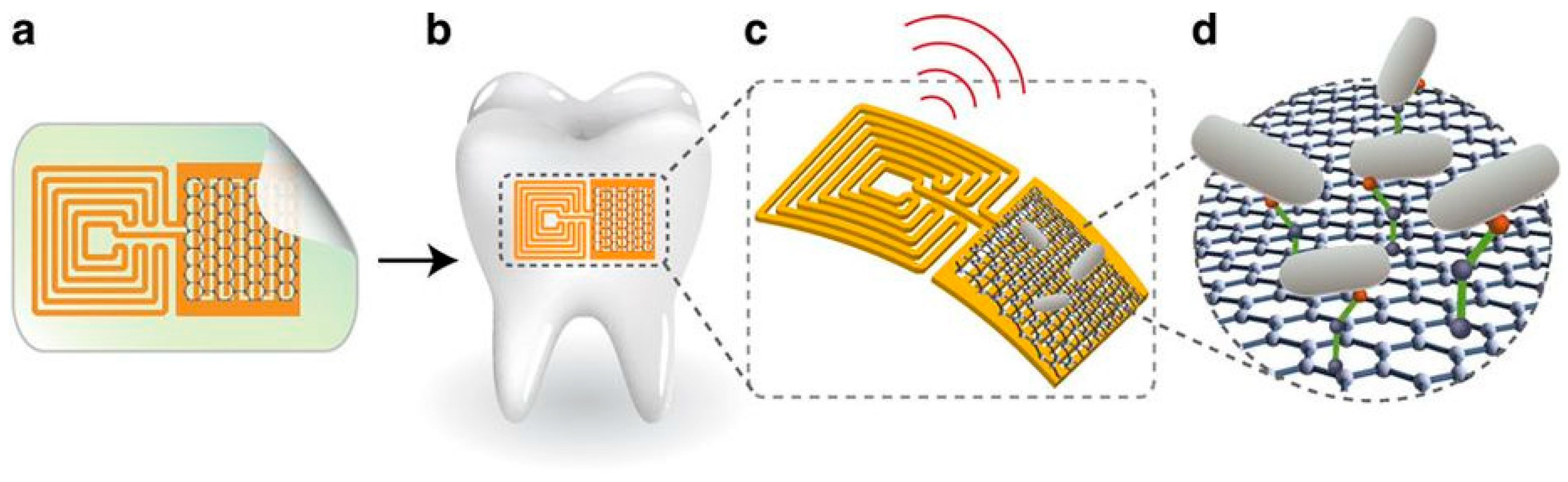
- Mannoor, M.S.; Tao, H.; Clayton, J.D.; Sengupta, A.; Kaplan, D.L.; Naik, R.R.; Verma, N.; Omenetto, F.G.; McAlpine, M.C. Graphene-based wireless bacteria detection on tooth enamel. Nat. Commun. 2012, 3, 763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lillehoj, P.B.; Kaplan, C.W.; He, J.; Shi, W.; Ho, C.-M. Rapid, Electrical Impedance Detection of Bacterial Pathogens Using Immobilized Antimicrobial Peptides. J. Lab. Autom. 2014, 19, 42–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etayash, H.; Jiang, K.; Thundat, T.; Kaur, K. Impedimetric detection of pathogenic gram-positive bacteria using an antimicrobial peptide from class IIa bacteriocins. Anal. Chem. 2014, 86, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Afrasiabi, R.; Fathi, F.; Wang, N.; Xiang, C.; Love, R.; She, Z.; Kraatz, H.B. Impedance based detection of pathogenic E. coli O157:H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens. Bioelectron. 2014, 58, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.A.S.; Nascimento, J.M.; Oliveira, I.S.; de Oliveira, C.V.J.; de Melo, C.P.; Franco, O.L.; Oliveira, M.D.L. Nanostructured sensor based on carbon nanotubes and clavanin A for bacterial detection. Colloids Surf. B Biointerfaces 2015, 135, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Marrakchi, M.; Xu, D.; Dong, H.; Andreescu, S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016, 80, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Nogués, M.; Brosel-Oliu, S.; Abramova, N.; Muñoz, F.X.; Bratov, A.; Mas-Moruno, C.; Gil, F.J. Impedimetric antimicrobial peptide-based sensor for the early detection of periodontopathogenic bacteria. Biosens. Bioelectron. 2016, 86, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Miranda, J.L.; Oliveira, M.D.L.; Oliveira, I.S.; Frias, I.A.M.; Franco, O.L.; Andrade, C.A.S. A simple nanostructured biosensor based on clavanin A antimicrobial peptide for gram-negative bacteria detection. Biochem. Eng. J. 2017, 124, 108–114. [Google Scholar] [CrossRef]
- Law, J.W.F.; Mutalib, N.S.A.; Chan, K.G.; Lee, L.H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2014, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Arlett, J.L.; Myers, E.B.; Roukes, M.L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 2011, 6, 203–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramgir, N.S.; Yang, Y.; Zacharias, M. Nanowire-based sensors. Small 2010, 6, 1705–1722. [Google Scholar] [CrossRef] [PubMed]
- Duque, T.; Chaves Ribeiro, A.C.; de Camargo, H.S.; da Costa Filho, P.A.; Mesquita Cavalcante, H.P.; Lopes, D. New Insights on Optical Biosensors: Techniques, Construction and Application. In State of the Art in Biosensors—General Aspects; InTech: Rijeka, Croatia, 2013; ISBN 978-953-51-1004-0. [Google Scholar] [Green Version]
- Lafleur, J.P.; Jönsson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Hamo, H.B.; Kushmaro, A.; Marks, R.S.; Grüner, C.; Rauschenbach, B.; Abdulhalim, I. Highly sensitive and specific detection of E. coli by a SERS nanobiosensor chip utilizing metallic nanosculptured thin films. Analyst 2015, 140, 3201–3209. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Niessner, R.; Haisch, C. SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal. Chem. 2014, 86, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Guilbault, G.G.; Lubrano, G.J. An enzyme electrode for the amperometric determination of glucose. Anal. Chim. Acta 1973, 64, 439–455. [Google Scholar] [CrossRef]
- Bratov, A.; Abramova, N. Response of a microcapillary impedimetric transducer to changes in surface conductance at liquid/solid interface. J. Colloid Interface Sci. 2013, 403, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Grossi, M.; Riccò, B. Electrical impedance spectroscopy (EIS) for biological analysis and food characterization: A review. J. Sens. Sens. Syst. 2017, 6, 303–325. [Google Scholar] [CrossRef]
- Basu, P.K.; Indukuri, D.; Keshavan, S.; Navratna, V.; Vanjari, S.R.K.; Raghavan, S.; Bhat, N. Graphene based E. coli sensor on flexible acetate sheet. Sens. Actuators B Chem. 2014, 190, 342–347. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Varshney, M.; Li, Y. Interdigitated array microelectrodes based impedance biosensors for detection of bacterial cells. Biosens. Bioelectron. 2009, 24, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Prodromidis, M.I. Impedimetric immunosensors—A review. Electrochim. Acta 2010, 55, 4227–4233. [Google Scholar] [CrossRef]
- Brosel-oliu, S.; Uria, N.; Abramova, N.; Bratov, A. Impedimetric Sensors for Bacteria Detection. In Biosensors—Micro and Nanoscale Applications; InTech: Rijeka, Croatia, 2015; Volume 9, pp. 258–287. ISBN 978-953-51-2173-2. [Google Scholar]
- Settu, K.; Chen, C.J.; Liu, J.T.; Chen, C.L.; Tsai, J.Z. Impedimetric method for measuring ultra-low E. coli concentrations in human urine. Biosens. Bioelectron. 2015, 66, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.T.; Settu, K.; Tsai, J.Z.; Chen, C.J. Impedance sensor for rapid enumeration of E. coli in milk samples. Electrochim. Acta 2015, 182, 89–95. [Google Scholar] [CrossRef]
- Hammond, J.L.; Formisano, N.; Estrela, P.; Carrara, S.; Tkac, J. Electrochemical biosensors and nanobiosensors. Essays Biochem. 2016, 60, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Bashir, R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol. Adv. 2008, 26, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Feng, A.; Huo, M.; Yuan, J. Ferrocene-based supramolecular structures and their applications in electrochemical responsive systems. Chem. Commun. 2014, 50, 13005–13014. [Google Scholar] [CrossRef] [PubMed]
- Bratov, A.; Abramova, N.; Marco, M.P.; Sanchez-Baeza, F. Three-Dimensional Interdigitated Electrode Array as a Tool for Surface Reactions Registration. Electroanalysis 2012, 24, 69–75. [Google Scholar] [CrossRef]
- Godoy-Gallardo, M.; Mas-Moruno, C.; Fernández-Calderón, M.C.; Pérez-Giraldo, C.; Manero, J.M.; Albericio, F.; Gil, F.J.; Rodríguez, D. Covalent immobilization of hLf1-11 peptide on a titanium surface reduces bacterial adhesion and biofilm formation. Acta Biomater. 2014, 10, 3522–3534. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Gallardo, M.; Mas-Moruno, C.; Yu, K.; Manero, J.M.; Gil, F.J.; Kizhakkedathu, J.N.; Rodriguez, D. Antibacterial properties of hLf1-11 peptide onto titanium surfaces: A comparison study between silanization and surface initiated polymerization. Biomacromolecules 2015, 16, 483–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyos-Nogués, M.; Velasco, F.; Ginebra, M.P.; Manero, J.M.; Gil, F.J.; Mas-Moruno, C. Regenerating bone via multifunctional coatings: The blending of cell integration and bacterial inhibition properties on the surface of biomaterials. ACS Appl. Mater. Interfaces 2017, 9, 21618–21630. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Kim, S.N.; Naik, R.R.; McAlpine, M.C. Biomimetic peptide nanosensors. Acc. Chem. Res. 2012, 45, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.; Nurunnabi, M.; Morshed, M.; Paul, A.; Polini, A.; Kuila, T.; Al Hariri, M.; Lee, Y.K.; Jaffa, A.A. Recent advances in application of biosensors in tissue engineering. BioMed Res. Int. 2014, 2014, 307519. [Google Scholar] [CrossRef] [PubMed]

| Transducer | ||||
|---|---|---|---|---|
| Optical | Mechanical | Electrochemical | ||
| Adhesin (Ad) or None | --- | Ad + Nanowires [18] | --- | |
| Bioreceptor | Enzyme (Ez) | (i) Ez NP + colorimetric [43] (ii) Bacteria-specific Ez + colorimetric [44] | --- | Ez NP + DPV [45] |
| Antibody (Ab) | (i) Ab + MNP + SPR [46] (ii) Ab + SPR [47] | (i) Ab + QCM [48] (ii) Ab + nanobeads + QCM [49] (iii) Au NP + Ab + QCM [50,51] (iv) Ab + cantilever [52] (v) Ab + PEMC [53] | (i) Ab + amperometric [54,55] (ii) Ab+ impedimetric [14,56,57,58,59,60] (iii) Ab MNP + DPV [61] (iv) Ab Mbeads + amperometric [54,55,62] | |
| DNA/RNA/ Aptamer (Ap)/Aptazyme (Apz) | (i) Ap + colorimetric [63,64] (ii) Ap + LSPR [65] (iii) Apz + Mbeads + colorimetric [66] | (i) thssDNA + QCM [67] (ii) ssDNA + cantilever [68] | (i) Ap + potentiometric [69,70] (ii) Ap + impedimetric [71] | |
| Lectin (L) | L + SPR [72] | L + piezoelectric [73,74] | L + impedimetric [75,76,77] | |
| Phage (Ph) | (i) Ph + SPR [78,79] (ii) Ph + MES + colorimetric [80] (iii) Ph + Mbeads + colorimetric [81] | (i) Ph + QCM [82] (ii) Ph + MES [83,84,85,86] | (i) Ph + amperometric [87] (ii) Ph + impedimetric [57,88,89,90] | |
| AMP | AMP + fluorescence spectroscopy [91,92,93,94] | AMP + QCM [95,96] | AMP + impedimetric [95,97,98,99,100,101,102,103,104,105] | |
| AMP Sequence | Bacteria Detected | LoD (CFU/mL) | Application | Type of Transd. | Sensor Structure | Detection Time | Ref. |
|---|---|---|---|---|---|---|---|
| Magainin I | E. coli O157:H7; S. typhimurium | 1.6 × 105; 6.5 × 104 cells/mL | --- | FS | Silanized glass slide on array-based biosensor | 70 min | [94] |
| Cecropin A, parasin, magainin I, polymyxin B and E | E. coli O157:H7; S. typhimurium | See Table 3 | Detection of foodborne contaminants | FS | Glass slides-PDMS on a mixed “sandwich” assay (multi-AMP array) | --- | [91] |
| Cecropin (A, B, and P), parasin, magainin I, polymyxin (B and E), melittin, bactenecin | C. burnetti; B. melitensis; VEE; vaccinia virus | 5 × 105 cells/mL; 5 × 104; <5 × 105 cells/mL; <5 × 105 | Detection of inactivated targets of biodefense interest | FS | Glass slides-PDMS on array-based biosensor | --- | [92] |
| Magainin I | E. coli O157:H7; S. typhimurium | 103 | Detection of an infectious outbreak from a broad spectrum of pathogenic species | EIS | Gold surface-cysteine on IDEA | --- | [97] |
| GBP + OHP | E. coli; H. pylori; S. aureus | 103 in wireless operation mode | Duodenal ulcers and stomach cancers | EIS | IDEA with graphene resistive sensors in a silk support | --- | [98] |
| Magainin I | E. coli O157:H7; E. coli K12; B. subtilis; S. epidermis | 103 | Life-threatening gastrointestinal infections | EIS | Ferrocene-Magainin conjugate on a gold electrode | --- | [101] |
| Leucocin A | L. monocytogenes; S. aureus | 103 | --- | EIS | Gold surface- cysteamine on IDEA | 20 min | [100] |
| G10KHc, C16G2cys | P. aeruginosa; S. mutans | 105 | Infectious diseases | EIS | Gold surface-cysteine on microfluidic chip | 25 min | [99] |
| Clavanin A | K. pneumoniae; E. faecalis; E. coli; B. subtilis | 102; 102; <103; <103 | Detect pathogens with high resistance to conventional antibiotics | EIS | Nanostructured sensor based on carbon nanotubes on gold electrode | --- | [102] |
| GIGKFLHSAGKGKAFVGEIMKS | E. coli O157:H7 | 1.5 × 103 | Bacterial infections | EIS | Mixed self-assembled monolayer on a three electrode system | 30 min | [95] |
| WK3(QL)6K2G3C | E. coli; P. aeruginosa; S. aureus; S. epidermidis | 102 | Bacterial infections | EIS | Gold disk electrode | --- | [103] |
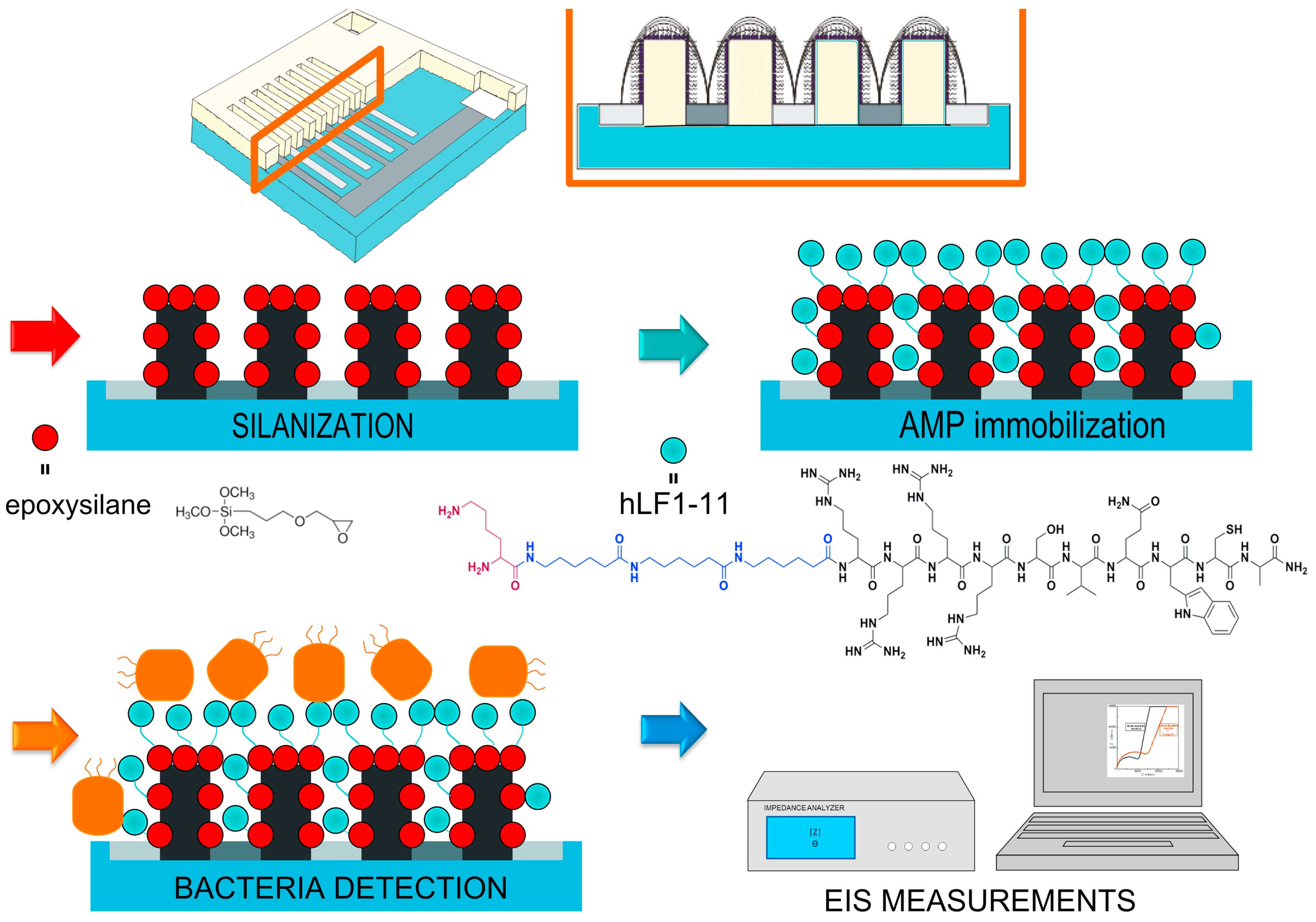
| hLF1-11 | S. sanguinis | KCl: 3.5 × 101; AS: 102 | Bacterial infections | EIS | 3D-IDEA based on silicon dioxide insulating substrate | 30 min | [104] |
| Clavanin A | E. coli; S. typhimurium; E. faecalis; S. aureus | ~ 10 | Dental infections | EIS | AuNPsCys | 70 min | [105] |
| AMP | E. coli | S. typhimurium |
|---|---|---|
| Polymyxin B | 1 × 105 | 5 × 105 |
| Polymyxin E | 5 × 105 | 5 × 106 |
| Magainin | 5 × 104 | 1 × 105 |
| Cecropin A | 1 × 105 | 5 × 105 |
| Parasin | 5 × 105 | 1 × 106 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. https://doi.org/10.3390/molecules23071683
Hoyos-Nogués M, Gil FJ, Mas-Moruno C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules. 2018; 23(7):1683. https://doi.org/10.3390/molecules23071683
Chicago/Turabian StyleHoyos-Nogués, Mireia, F. J. Gil, and Carlos Mas-Moruno. 2018. "Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies" Molecules 23, no. 7: 1683. https://doi.org/10.3390/molecules23071683
APA StyleHoyos-Nogués, M., Gil, F. J., & Mas-Moruno, C. (2018). Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules, 23(7), 1683. https://doi.org/10.3390/molecules23071683








