Anthocidins A–D, New 5-Hydroxyanthranilic Acid Related Metabolites from the Sea Urchin-Associated Actinobacterium, Streptomyces sp. HDa1
Abstract
1. Introduction
2. Results
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain Isolation and Cultivation
3.3. Extraction and Purification
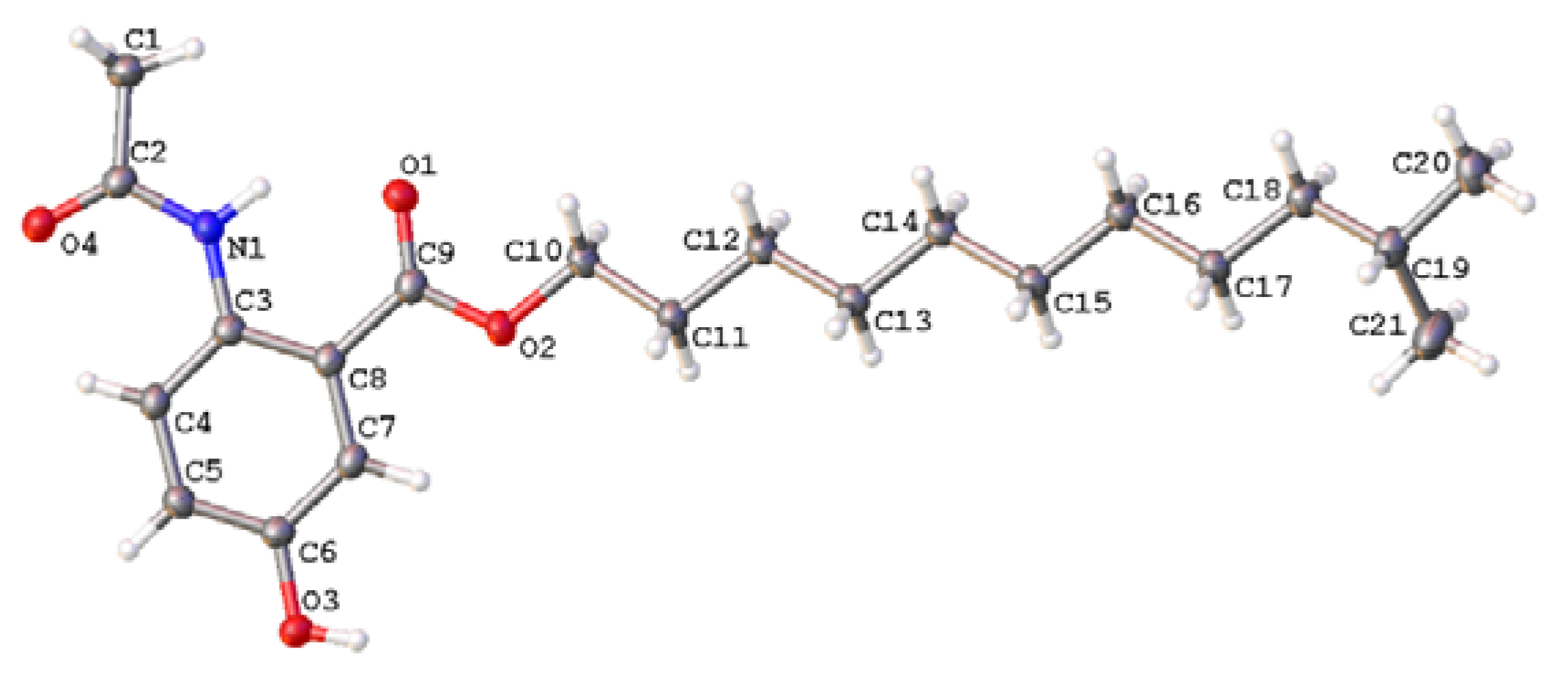
3.4. X-ray Single-Crystal Data of 1
3.5. Biological Assays
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Yang, C.; Horn, H.; Hajjar, D.; Ravasi, T.; Hentschel, U. Actinomycetes from red sea sponges: sources for chemical and phylogenetic diversity. Mar. Drugs 2014, 12, 2771–2789. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Zhen, X.; Li, X.L.; Chen, J.J.; Chen, T.J.; Yang, J.L.; Zhu, P. Tetrocarcin Q, a new spirotetronate with a unique glycosyl group from a marine-derived actinomycete Micromonospora carbonacea LS276. Mar. Drugs 2018, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Yang, X.Y.; Liu, T.; Xiao, H.; Wang, G.Y.; Zhou, M.J; Liu, F.W.; Zhang, Y.T.; Liu, D.; Chen, M.H.; et al. Fluostatins M–Q featuring a 6-5–6-6 ring skeleton and high oxidized A-rings from marine Streptomyces sp. PKU-MA00045. Mar. Drugs 2018, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Park, S.H.; Kwon, Y.; Lee, S.K.; Shin, J.; Nam, J.W.; Oh, D.C. QM-HiFSA-aided structure determination of succinilenes A–D, new triene polyols from a marine-derived Streptomyces sp. Mar. Drugs 2017, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Wang, Y.S.; Liu, C.L.; Zeng, Y.J.; Cheng, P.; Jiao, R.H.; Bao, S.X.; Huang, H.Q.; Tan, R.X.; Ge, H.M. Strepchazolins A and B: Two new alkaloids from a marine Streptomyces chartreusis NA02069. Mar. Drugs 2017, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Lacret, R.; Pérez-Victoria, I.; Oves-Costales, D.; De la Cruz, M.; Domingo, E.; Martín, J.; Díaz, C.; Vicente, F.; Genilloud, O.; Reyes, F. MDN-0170, a new napyradiomycin from Streptomyces sp. Strain CA-271078. Mar. Drugs 2016, 14, 188. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Eltamany, E.E.; Abdelmohsen, U.R.; Ibrahim, A.K.; Hassanean, H.A.; Hentschel, U.; Ahmed, S.A. New antibacterial xanthone from the marine sponge-derived Micrococcus sp. EG45. Bioorg. Med. Chem. Lett. 2014, 24, 4939–4942. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Cheng, C.; Viegelmann, C.; Zhang, T.; Grkovic, T.; Ahmed, S.; Quinn, R.J.; Hentschel, U.; Edrada-Ebel, R. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora. sp. EG49. Mar. Drugs 2014, 12, 1220–1244. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.K.; Zhou, Y.Q.; Han, H.; Wang, W.; Xiang, L.; Deng, X.Z.; Ge, H.M.; Jiao, R.H. New antibacterial phenone derivatives asperphenone A–C from mangrove-derived fungus Aspergillus sp. YHZ-1. Mar. Drugs 2018, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.K.; Gai, C.J.; Cai, C.H.; Chen, L.L.; Liu, S.B.; Zeng, Y.B.; Yuan, J.Z.; Mei, W.L.; Dai, H.F. Metabolites with insecticidal activity from Aspergillus fumigatus JRJ111048 isolated from mangrove plant Acrostichum. specioum endemic to Hainan island. Mar. Drugs 2017, 15, 381. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, T.M.; Shen, M.H.; Yang, M.Q.; Feng, Q.Y.; Tang, X.M.; Li, X.M. Spiculisporic acids B–D, three new γ-butenolide derivatives from a sea urchin-derived fungus Aspergillus sp. HDf2. Molecules 2012, 17, 13175–13182. [Google Scholar] [CrossRef] [PubMed]
- Ohkuma, H.; Tomita, K.; Hoshino, Y.; Suzuki, K.; Hasegawa, M.; Sawada, Y.; Konishi, M.; Hook, D.J.; Oki, T. 5-Hydroxyanthranilic acid derivatives as potent 5-lipoxygenase inhibitors. J. Antibiot. 1993, 46, 705–711. [Google Scholar] [CrossRef] [PubMed]
- De Kowalewski, D.G.; Kowalewski, V.J.; Botek, E.; Contreras, R.H.; Facelli, J.C. Experimental and theoretical study of the ethoxy group conformational effect on 13C chemical shifts in ortho-substituted phenetols. Magn. Reson. Chem. 1997, 35, 351–356. [Google Scholar] [CrossRef]
- Linder, J.; Moody, C.J. The total synthesis of siphonazole, a structurally unusual bis-oxazole natural product. Chem. Commun. 2007, 15, 1508–1509. [Google Scholar] [CrossRef] [PubMed]
- Bobylev, M.M.; Bobyleva, L.I.; Strobel, G.A. Synthesis and bioactivity of analogs of maculosin, a host-specific phytotoxin produced by Alternaria alternata on spotted knapweed (Centaurea maculosa). J. Agric. Food Chem. 1996, 44, 3960–3964. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–9 are available from the authors. |
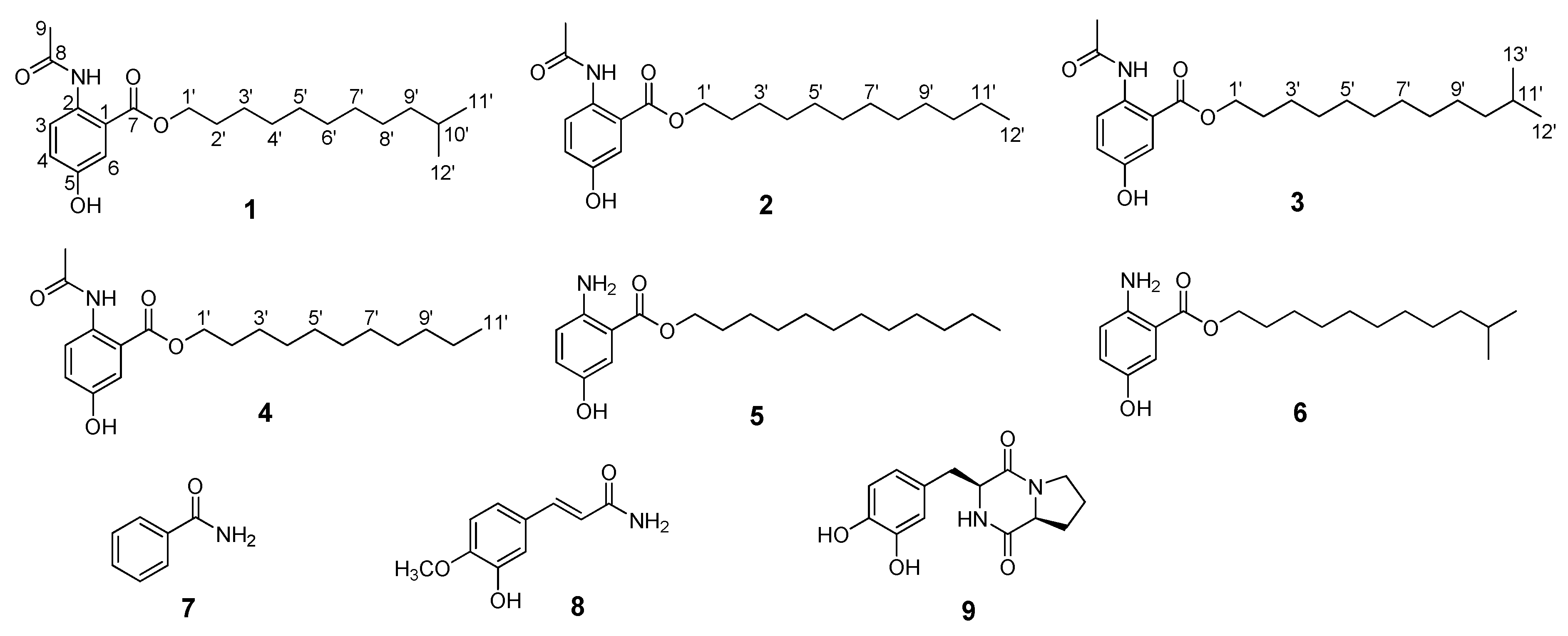

| Position | Anthocidin A (1) | Anthocidin B (2) | Anthocidin C (3) | ||||
|---|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | ||
| 1 | 116.8, C | 116.8, C | 116.7, C | ||||
| 2 | 134.1, C | 134.3, C | 135.1, C | ||||
| 2-NH | 10.93, s | 10.88, s | 10.84, s | ||||
| 3 | 122.1, CH | 8.53, d (9.1) | 122.1, CH | 8.50, d (9.0) | 122.2, CH | 8.53, d (8.9) | |
| 4 | 121.8, CH | 7.15, dd (9.1, 3.0) | 121.8, CH | 7.10, dd (9.0, 2.4) | 121.9, CH | 7.07, dd (8.9, 3.0) | |
| 5 | 151.7, C | 151.7, C | 150.9, C | ||||
| 6 | 116.9, CH | 7.55, d (3.0) | 116.9, CH | 7.52, d (2.4) | 116.8, CH | 7.50, d (3.0) | |
| 7 | 168.1, C | 168.1, C | 168.1, C | ||||
| 8 | 169.4, C | 169.3, C | 169.1, C | ||||
| 9 | 25.2, CH3 | 2.25, s | 25.3, CH3 | 2.22, s | 26.2, CH3 | 2.21, s | |
| 1′ | 65.7, CH2 | 4.31, t (6.7) | 65.7, CH2 | 4.28, t (6.6) | 65.8, CH2 | 4.30, t (6.7) | |
| 2′ | 28.6, CH2 | 1.77, m | 28.6, CH2 | 1.75, m | 28.7, CH2 | 1.76, m | |
| 3′ | 26.0, CH2 | 1.45, m | 26.0, CH2 | 1.42, m | 26.2, CH2 | 1.43, m | |
| 4′ | 27.4, CH2 | 1.20–1.38, m | 29.3 c, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 5′ | 29.3 b, CH2 | 1.20–1.38, m | 29.4 c, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 6′ | 29.5 b, CH2 | 1.20–1.38, m | 29.5 c, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 7′ | 29.7 b, CH2 | 1.20–1.38, m | 29.6 c, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 8′ | 29.9 b, CH2 | 1.20–1.38, m | 29.6 c, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 9′ | 39.1, CH2 | 1.16, m | 29.7 c, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 10′ | 27.9, CH | 1.53, m | 39.1, CH2 | 1.24–1.36, m | 22.0–39.0 d, CH2 | 1.20–1.40, m | |
| 11′ | 22.7, CH3 | 0.87, d (6.6) | 22.8, CH2 | 1.24–1.36, m | 28.1, CH | 1.51, m | |
| 12′ | 22.7, CH3 | 0.87, d (6.6) | 14.1, CH3 | 0.87, t (6.8) | 19.4, CH3 | 0.85, d (6.4) | |
| 13′ | 11.5, CH3 | 0.83, d (6.4) | |||||
| Position | Anthocidin D (4) | ||||
|---|---|---|---|---|---|
| δC | δH (J in Hz) | Position | δC | δH (J in Hz) | |
| 1 | 116.4, C | 1′ | 65.7, CH2 | 4.30, t (6.7) | |
| 2 | 135.2, C | 2′ | 28.6, CH2 | 1.77, m | |
| 2-NH | 10.81, s | 3′ | 26.0, CH2 | 1.43, m | |
| 3 | 122.1, CH | 8.56, d (9.0) | 4′ | 29.5 b, CH2 | 1.20–1.40, m |
| 4 | 121.7, CH | 7.04, dd (9.0, 3.0) | 5′ | 29.6 b, CH2 | 1.20–1.40, m |
| 5 | 150.4, C | 6′ | 29.6 b, CH2 | 1.20–1.40, m | |
| 5-OH | 7′ | 29.3 b, CH2 | 1.20–1.40, m | ||
| 6 | 116.5, CH | 7.48, d (3.0) | 8′ | 29.2 b, CH2 | 1.20–1.40, m |
| 7 | 167.9, C | 9′ | 31.9, CH2 | 1.20–1.40, m | |
| 8 | 168.8, C | 10′ | 22.7, CH2 | 1.20–1.40, m | |
| 9 | 25.3, CH3 | 2.21, s | 11′ | 14.1, CH3 | 0.88, t (7.0) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Z.-K.; Wang, R.; Chen, S.-Q.; Chen, F.-X.; Liu, T.-M.; Yang, M.-Q. Anthocidins A–D, New 5-Hydroxyanthranilic Acid Related Metabolites from the Sea Urchin-Associated Actinobacterium, Streptomyces sp. HDa1. Molecules 2018, 23, 1032. https://doi.org/10.3390/molecules23051032
Guo Z-K, Wang R, Chen S-Q, Chen F-X, Liu T-M, Yang M-Q. Anthocidins A–D, New 5-Hydroxyanthranilic Acid Related Metabolites from the Sea Urchin-Associated Actinobacterium, Streptomyces sp. HDa1. Molecules. 2018; 23(5):1032. https://doi.org/10.3390/molecules23051032
Chicago/Turabian StyleGuo, Zhi-Kai, Rong Wang, Shi-Quan Chen, Fu-Xiao Chen, Tian-Mi Liu, and Ming-Qiu Yang. 2018. "Anthocidins A–D, New 5-Hydroxyanthranilic Acid Related Metabolites from the Sea Urchin-Associated Actinobacterium, Streptomyces sp. HDa1" Molecules 23, no. 5: 1032. https://doi.org/10.3390/molecules23051032
APA StyleGuo, Z.-K., Wang, R., Chen, S.-Q., Chen, F.-X., Liu, T.-M., & Yang, M.-Q. (2018). Anthocidins A–D, New 5-Hydroxyanthranilic Acid Related Metabolites from the Sea Urchin-Associated Actinobacterium, Streptomyces sp. HDa1. Molecules, 23(5), 1032. https://doi.org/10.3390/molecules23051032




