Comparative Transcriptome Analysis Identifies Putative Genes Involved in Dioscin Biosynthesis in Dioscorea zingiberensis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Generation of D. Zingiberensis Transcriptomes from its Leaf and Rhizome Tissues
2.2. CD-HIT and BlastP Analysis
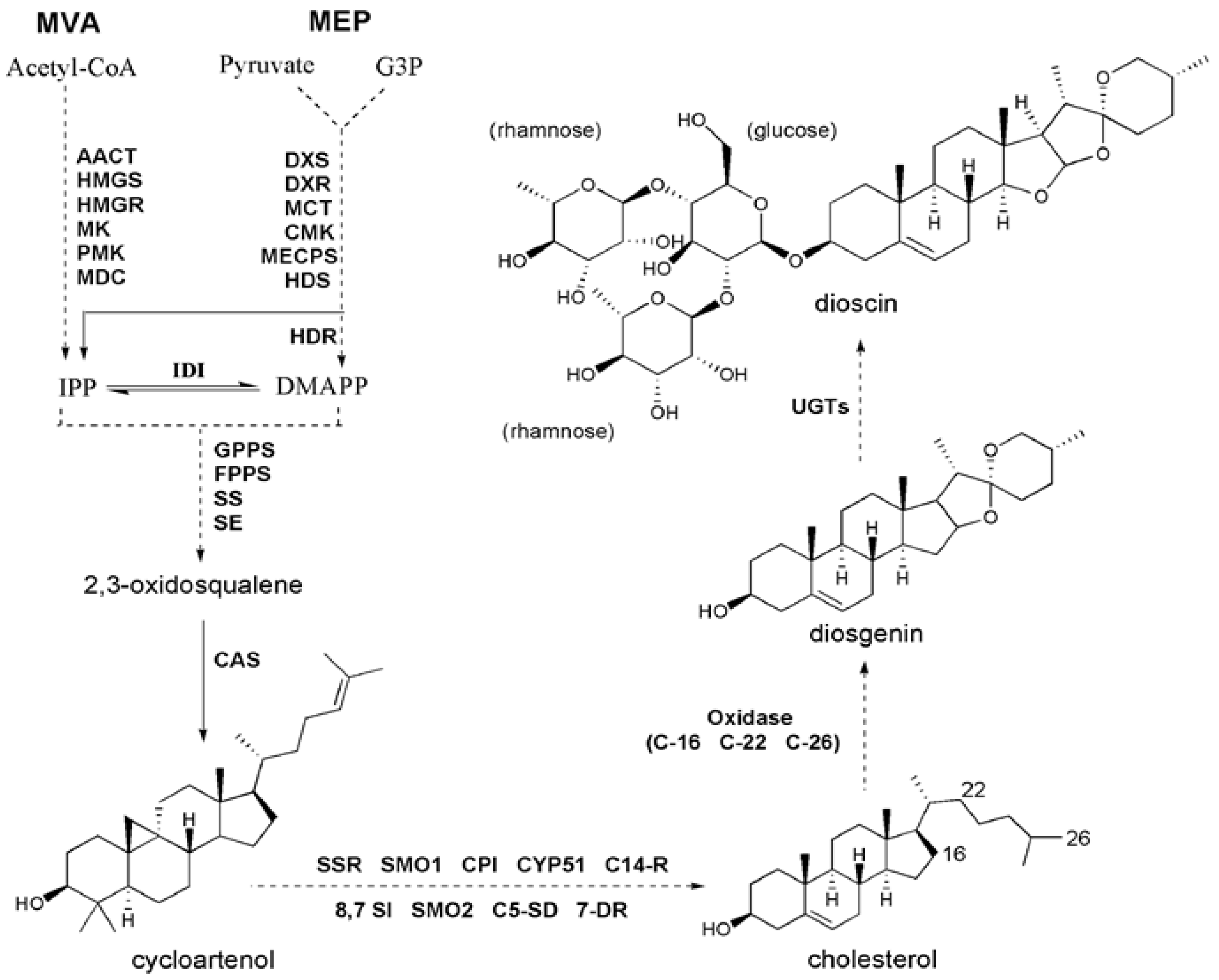
2.3. Identification of Genes Related to Steroidal Backbone Biosynthesis
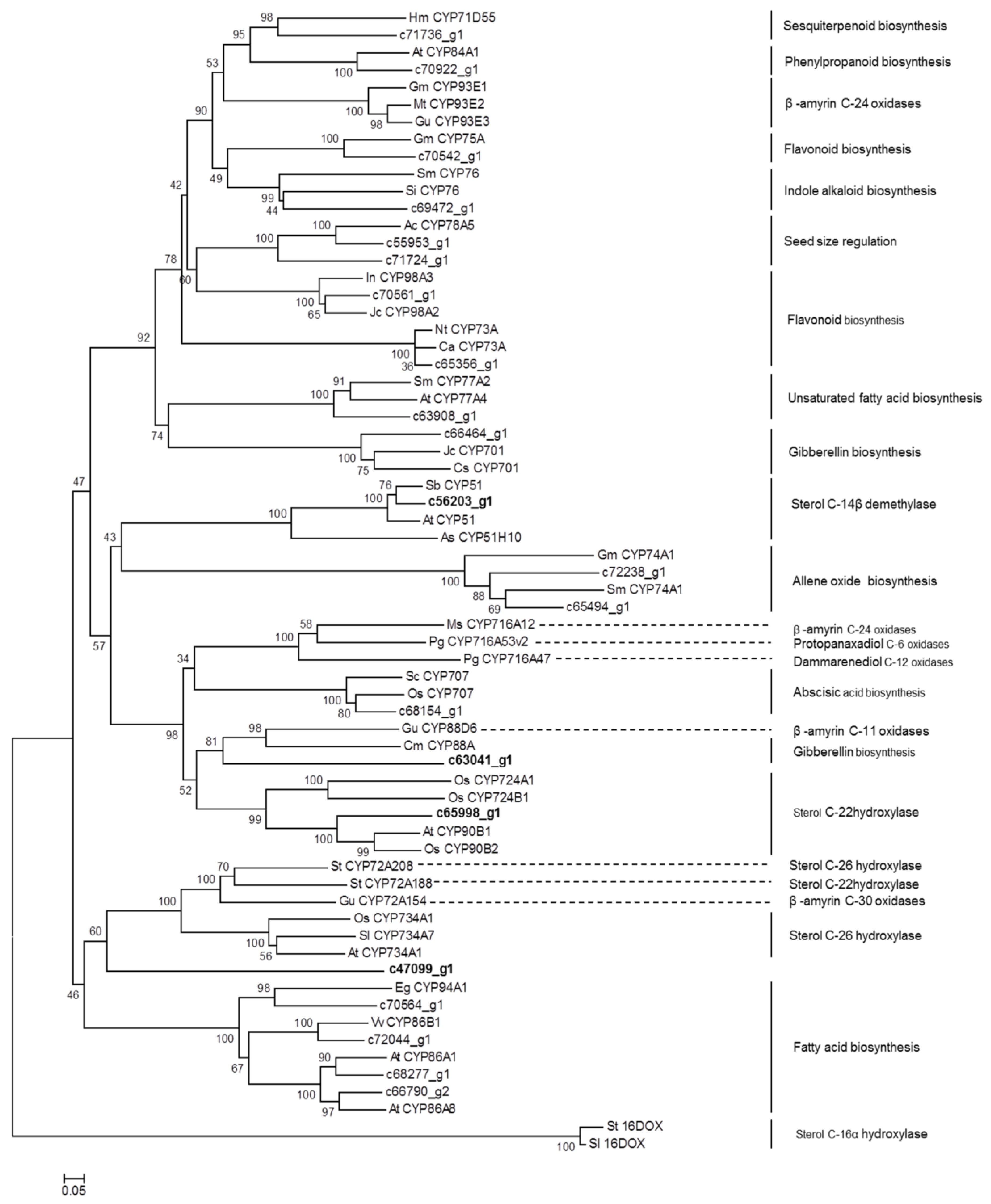
2.4. Identification of CYP450 Unigenes Putatively Involved in Diosgenin Biosynthesis
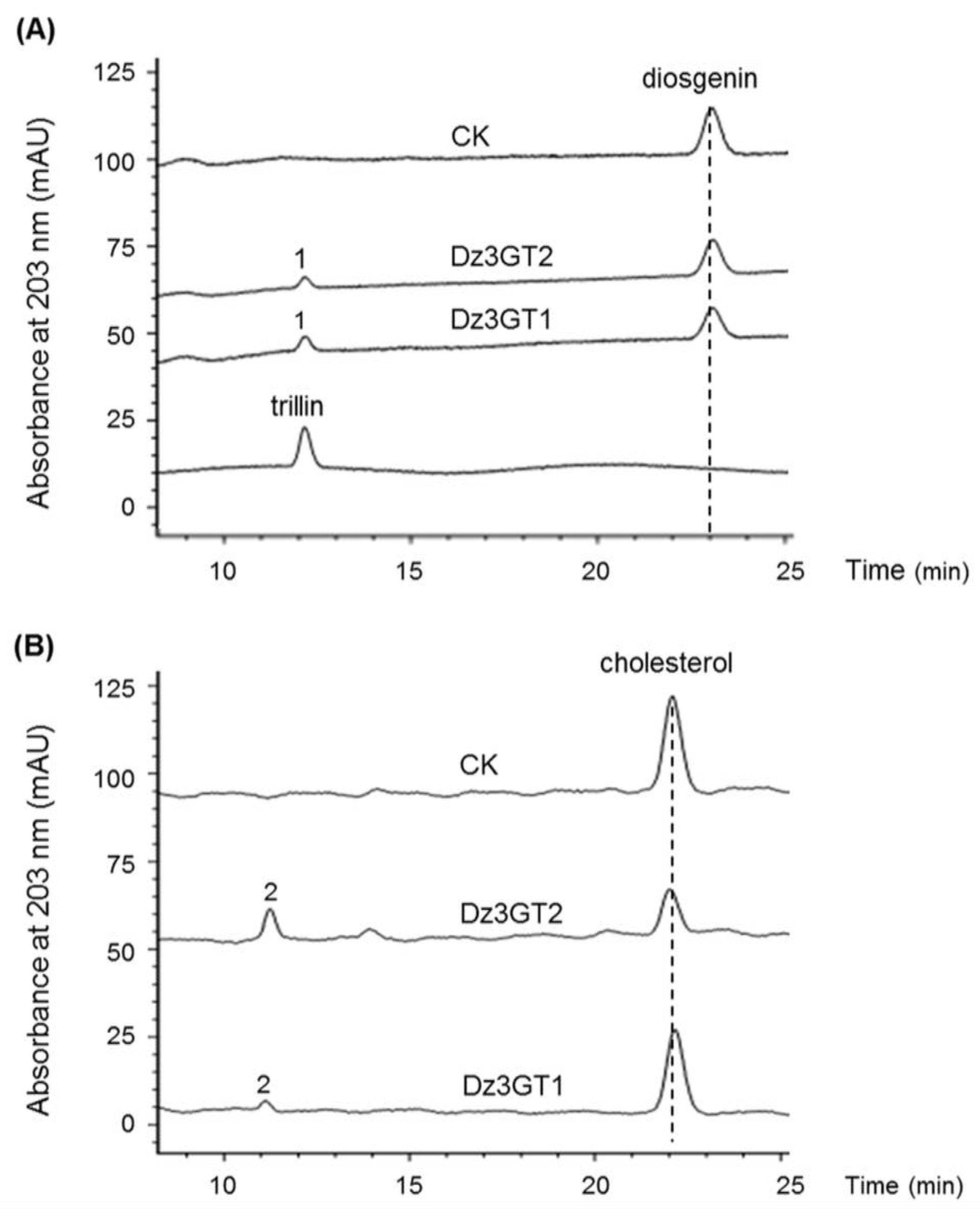
2.5. Identification of UGT Candidates Involved in Dioscin Biosynthesis
3. Materials and Methods
3.1. Plant Material
3.2. Transcriptome Sequencing, De Novo Assembly and Unigene Functional Annotation
3.3. CD-HIT Analysis
3.4. Blastp Analysis
3.5. Phylogenetic Analysis
3.6. Plant Metabolite Extraction
3.7. Recombinant Protein Expression in E. Coli and Purification
3.8. Enzyme Assays
3.9. HPLC (High Performance Liquid Chromatography) Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yi, D. Natural steroidal saponins, an exploitable field in new drug development. Chin. New Drugs J. 2000, 9, 521–524. [Google Scholar]
- Zhu, J.; Guo, X.; Fu, S.; Zhang, X.; Liang, X. Characterization of steroidal saponins in crude extracts from Dioscorea zingiberensis C. H. Wright by ultra-performance liquid chromatography/electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J. Pharm. Biomed. Anal. 2010, 53, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.G.; Jiang, W.; Nitin, M.; Bao, X.Q.; Chen, S.L.; Tao, Z.M. Characterizing diversity based on nutritional and bioactive compositions of yam germplasm (Dioscorea spp.) commonly cultivated in China. J. Food Drug Anal. 2016, 24, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, Y.; Han, X.; Qi, Y.; Xu, L.; Xu, Y.; Yin, L.; Sun, H.; Liu, K.; Peng, J. Anti-cancer effects of dioscin on three kinds of human lung cancer cell lines through inducing DNA damage and activating mitochondrial signal pathway. Food Chem. Toxicol. 2013, 59, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xu, H.; Peng, J.; Wang, C.; Jin, Y.; Liu, K.; Sun, H.; Qin, J. Potent anti-inflammatory effect of dioscin mediated by suppression of TNF-α-induced VCAM-1, ICAM-1and EL expression via the NF-κB pathway. Biochimie 2015, 110, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Yan, Q.; Xu, L.; Yin, L.; Xu, H.; Xu, Y.; Wang, C.; Sun, H.; Peng, J. Dioscin reduces ovariectomy-induced bone loss by enhancing osteoblastogenesis and inhibiting osteoclastogenesis. Pharmacol. Res. 2016, 108, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Jesus, M.; Martins, A.P.; Gallardo, E.; Silvestre, S. Diosgenin: Recent Highlights on Pharmacology and Analytical Methodology. J. Anal. Methods Chem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Joly, R.A.; Bonner, J.; Bennett, R.D.; Heftmann, E. The biosynthesis of steroidal sapogenins in Dioscorea floribunda from doubly labelled cholesterol. Phytochemistry 1969, 8, 1709–1711. [Google Scholar] [CrossRef]
- Stohs, S.J.; Kaul, B.; Staba, E.J. The metabolism of 14C-cholesterol by Dioscorea deltoidea suspension cultures. Phytochemistry 1969, 8, 1679–1686. [Google Scholar] [CrossRef]
- Bennett, R.D.; Heftmann, E.; Joly, R.A. Biosynthesis of diosgenin from 26-hydroxycholesterol in dioscorea floribunda. Phytochemistry 1970, 9, 349–353. [Google Scholar] [CrossRef]
- Sonawane, P.D.; Pollier, J.; Panda, S.; Szymanski, J.; Massalha, H.; Yona, M.; Unger, T.; Malitsky, S.; Arendt, P.; Pauwels, L. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 2016. [Google Scholar] [CrossRef] [PubMed]
- Mehrafarin, A.; Qaderi, A.; Rezazadeh, S.; Naghdi Badi, H.; Noormohammadi, G.; Zand, E. Bioengineering of important secondary metabolites and metabolic pathways in fenugreek (Trigonella foenum-graecum L.). J. Med. Plants 2010, 9, 1–18. [Google Scholar]
- Xiao, M.; Zhang, Y.; Chen, X.; Lee, E.J.; Barber, C.J.; Chakrabarty, R.; Desgagnépenix, I.; Haslam, T.M.; Kim, Y.B.; Liu, E. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 2013, 166, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Teoh, K.H.; Reed, D.W.; Maes, L.; Goossens, A.; Olson, D.J.H.; Ross, A.R.S.; Covello, P.S. The Molecular Cloning of Artemisinic Aldehyde Δ11(13) Reductase and Its Role in Glandular Trichome-dependent Biosynthesis of Artemisinin in Artemisia annua. J. Biol. Chem. 2008, 283, 21501–21508. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhou, Y.J.; Hillwig, M.L.; Shen, Y.; Yang, L.; Wang, Y.; Zhang, X.; Liu, W.; Peters, R.J.; Chen, X. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. USA 2013, 110, 12108–12113. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Zhou, C.; Li, J.; Zhang, Y. Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata. Plant J. Cell Mol. Biol. 2017, 90, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Ciura, J.; Szeliga, M.; Grzesik, M.; Tyrka, M. Next-generation sequencing of representational difference analysis products for identification of genes involved in diosgenin biosynthesis in fenugreek (Trigonella foenum-graecum). Planta 2017, 245, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, K.; Ghosh, A.; Kumar, V.; Chaudhary, S.; Srivastava, N.; Katudia, K.; Tiwari, T.; Chikara, S.K. De Novo Transcriptome Sequencing in L. to Identify Genes Involved in the Biosynthesis of Diosgenin. Plant Genome 2013, 6, 1–11. [Google Scholar] [CrossRef]
- Nakayasu, M.; Umemoto, N.; Ohyama, K.; Fujimoto, Y.; Lee, H.J.; Watanabe, B.; Muranaka, T.; Saito, K.; Sugimoto, Y.; Mizutani, M. A Dioxygenase Catalyzes Steroid 16α-Hydroxylation in Steroidal Glycoalkaloid Biosynthesis. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Fujita, S.; Ohnishi, T.; Watanabe, B.; Yokota, T.; Takatsuto, S.; Fujioka, S.; Yoshida, S.; Sakata, K.; Mizutani, M. Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. Cell Mol. Biol. 2006, 45, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Umemoto, N.; Nakayasu, M.; Ohyama, K.; Yotsu-Yamashita, M.; Mizutani, M.; Seki, H.; Saito, K.; Muranaka, T. Two Cytochrome P450 Monooxygenases Catalyze Early Hydroxylation Steps in the Potato Steroid Glycoalkaloid Biosynthetic Pathway. Plant Physiol. 2016, 171, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Nomura, T.; Watanabe, B.; Ohta, D.; Yokota, T.; Miyagawa, H.; Sakata, K.; Mizutani, M. Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 2006, 67, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kawabe, A.; Tokidasegawa, A.; Shimizu, B.; Takatsuto, S.; Shimada, Y.; Fujioka, S.; Mizutani, M. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. Cell Mol. Biol 2011, 67, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Seki, H.; Ohyama, K.; Sawai, S.; Mizutani, M.; Ohnishi, T.; Sudo, H.; Akashi, T.; Aoki, T.; Saito, K.; Muranaka, T. Licorice beta-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. USA 2008, 105, 14204–14209. [Google Scholar] [CrossRef] [PubMed]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R. Biosynthesis of Antinutritional Alkaloids in Solanaceous Crops Is Mediated by Clustered Genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Amit, R.; Mami, Y.; Hiroki, T.; Michimi, N.; Mareshige, K.; Hideyuki, S.; Kazuki, S. RNA-seq Transcriptome Analysis of Panax japonicus, and Its Comparison with Other Panax Species to Identify Potential Genes Involved in the Saponins Biosynthesis. Front. Plant Sci. 2016, 7, e0144. [Google Scholar]
- Ye, T.; Song, W.; Zhang, J.J.; An, M.; Feng, S.; Yan, S.; Li, J. Identification and functional characterization of DzS3GT, a cytoplasmic glycosyltransferase catalyzing biosynthesis of diosgenin 3-O-glucoside in Dioscorea zingiberensis. Plant Cell Tissue Organ Culture 2017, 129, 399–410. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Xian, A.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S.; Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2014, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |

| Item | Sample | Number | Q20(%) | Q30(%) | GC(%) | N50(bp) |
|---|---|---|---|---|---|---|
| Raw read | Aug_L | 55,798,190 | ND | ND | ND | ND |
| Aug_R | 45,367,360 | ND | ND | ND | ND | |
| Nov_R | 42,564,054 | ND | ND | ND | ND | |
| Clean read | Aug_L | 53,330,426 | 97.68 | 94.42 | 49.52 | ND |
| Aug_R | 43,234,636 | 97.68 | 94.44 | 47.54 | ND | |
| Nov_R | 40,686,226 | 97.72 | 94.51 | 47.87 | ND | |
| Total | Unigenes | 143,245 | ND | ND | ND | 814 |
| Enzyme Name | Gene ID | FPKM (Aug_R) | FPKM (Aug_L) | FPKM (Nov_R) | log2ratio (Nov_RvsAug_R) | q-Value (Nov_RvsAug_R) | log2ratio (Aug_LvsAug_R) | q-Value (Aug_LvsAug_R) | |
|---|---|---|---|---|---|---|---|---|---|
| MVA | |||||||||
| AACT-1 | c46893_g1 | 4.86 | 0 | 0.17 | −5.1594 | 0.13758 | −3.6187 | 0.18281 | |
| AACT-2 | c72984_g1 | 38.7 | 32.07 | 67.57 | 0.59302 | 1.30E-05 | −0.50544 | 0.99456 | |
| AACT-3 | c102447_g1 | 3.08 | 0.05 | 0 | −0.40148 | 0.99421 | −0.67435 | 0.99456 | |
| HMGS | c61824_g1 | 4.19 | 0 | 0.71 | −2.9209 | 0.35955 | −3.6026 | 0.1887 | |
| HMGR | c66823_g1 | 560.62 | 515.37 | 476.23 | −0.58795 | 0.0030532 | −0.48117 | 0.2103 | |
| MK-1 | c58223_g1 | 5.82 | 10.08 | 6.3 | −0.20776 | 0.99421 | 0.43378 | 0.75911 | |
| MK-2 | c76083_g1 | 2.81 | 0 | 0 | −3.3916 | 0.34457 | −3.3936 | 0.27635 | |
| PMK | c67242_g5 | 17.33 | 16.48 | 16.47 | −0.42636 | 0.99421 | −0.43222 | 0.99456 | |
| MVD | c56352_g1 | 32.56 | 27.55 | 71.17 | 0.77398 | 0.00014198 | −0.60355 | 0.99456 | |
| IDI-1 | c11491_g1 | 110.38 | 67.91 | 168.01 | −1.2525 | 0.99421 | −0.71196 | 0.99456 | |
| IDI-2 | c74391_g1 | 3.27 | 0 | 0 | 0.25038 | 0.0045152 | −1.0658 | 0.0053014 | |
| MEP | |||||||||
| DXS-1 | c72095_g4 | 7.95 | 29.55 | 13.16 | 0.44095 | 0.27058 | 1.4152 | 1.02E-06 | |
| DXS-2 | c54473_g1 | 1.48 | 16.01 | 2.96 | 0.64894 | 0.99421 | 3.0798 | 8.03E-07 | |
| DXR | c71143_g1 | 21.4 | 111.68 | 22.72 | −0.26747 | 0.99421 | 2.0223 | 8.93E-24 | |
| MCT | / | / | / | / | |||||
| CMK | c61103_g1 | 7.83 | 20.07 | 7.43 | −0.43182 | 0.99421 | 0.9928 | 0.072373 | |
| MECPS | c61331_g1 | 24.64 | 102.4 | 32.58 | 0.045319 | 0.99421 | 1.6868 | 6.12E-10 | |
| HDS | c71989_g1 | 12.68 | 81.59 | 15.17 | −0.093038 | 0.99421 | 2.3319 | 4.61E-48 | |
| HDR | c67492_g1 | 20.44 | 116.92 | 19.53 | −0.41845 | 0.99421 | 2.1561 | 2.70E-30 | |
| IPP to Cholesterol | |||||||||
| GPPS-1 | c97556_g1 | 9.45 | 45.39 | 5.51 | −1.1357 | 0.98019 | 1.8974 | 3.81E-06 | |
| GPPS-2 | c90599_g1 | 11.17 | 56.51 | 12.66 | −0.17417 | 0.99421 | 1.9746 | 3.10E-09 | |
| FPPS | c67887_g1 | 25.07 | 33.68 | 18.17 | −0.81943 | 0.87666 | 0.061922 | 0.76188 | |
| SS | c58461_g1 | 72.56 | 69.42 | 26.82 | −1.7907 | 4.01E-06 | −0.42756 | 0.99456 | |
| SE | c56806_g1 | 247.1 | 11.46 | 48.8 | −2.6935 | 6.27E-52 | −4.7909 | 1.01E-90 | |
| CAS | c73551_g1 | 66.58 | 244.71 | 39.38 | −1.1029 | 0.00062765 | −0.15504 | 0.70716 | |
| SSR-1 | c71749_g1 | 114.37 | 24.43 | 63.32 | −1.2053 | 9.8524E-07 | −2.5864 | 2.0831E-28 | |
| SSR-2 | c70423_g1 | 333.58 | 118.07 | 110.38 | −1.9485 | 1.2352E-46 | −1.8588 | 4.3029E-43 | |
| SMO1 | c62143_g1 | 787.76 | 15.24 | 131 | −2.938 | 2.493E-172 | −6.0534 | 2.5658E-285 | |
| CPI | c57629_g1 | 16.77 | 11.25 | 42.91 | 1.0016 | 0.010324 | −0.9308 | 0.92087 | |
| CYP51 | c56203_g1 | 1248.79 | 77.57 | 445.7 | −1.8394 | 9.6484E-154 | −4.3694 | 0 | |
| C14-R | c69698_g1 | 62.54 | 21.13 | 43.66 | −0.87364 | 0.26089 | −1.9294 | 3.8533E-06 | |
| 8,7 SI | c69931_g1 | 57.39 | 36.66 | 70.93 | −0.046848 | 0.48601 | −1.0062 | 0.0167 | |
| SMO2 | c50487_g1 | 4.47 | 0 | 0.25 | −4.5004 | 0.3795 | NA | NA | |
| C5-SD | c67458_g1 | 580.81 | 245.51 | 146.88 | −2.3389 | 8.7667E-70 | −1.607 | 4.0749E-36 | |
| 7-DR | c60553_g1 | 74.97 | 111.84 | 64.56 | −0.56945 | 0.94196 | 0.21529 | 0.0091335 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liang, Q.; Li, C.; Liu, M.; Zhang, Y. Comparative Transcriptome Analysis Identifies Putative Genes Involved in Dioscin Biosynthesis in Dioscorea zingiberensis. Molecules 2018, 23, 454. https://doi.org/10.3390/molecules23020454
Li J, Liang Q, Li C, Liu M, Zhang Y. Comparative Transcriptome Analysis Identifies Putative Genes Involved in Dioscin Biosynthesis in Dioscorea zingiberensis. Molecules. 2018; 23(2):454. https://doi.org/10.3390/molecules23020454
Chicago/Turabian StyleLi, Jia, Qin Liang, Changfu Li, Mengdi Liu, and Yansheng Zhang. 2018. "Comparative Transcriptome Analysis Identifies Putative Genes Involved in Dioscin Biosynthesis in Dioscorea zingiberensis" Molecules 23, no. 2: 454. https://doi.org/10.3390/molecules23020454
APA StyleLi, J., Liang, Q., Li, C., Liu, M., & Zhang, Y. (2018). Comparative Transcriptome Analysis Identifies Putative Genes Involved in Dioscin Biosynthesis in Dioscorea zingiberensis. Molecules, 23(2), 454. https://doi.org/10.3390/molecules23020454





