Prediction of Disordered Regions and Their Roles in the Anti-Pathogenic and Immunomodulatory Functions of Butyrophilins
Abstract
1. Introduction
2. Results
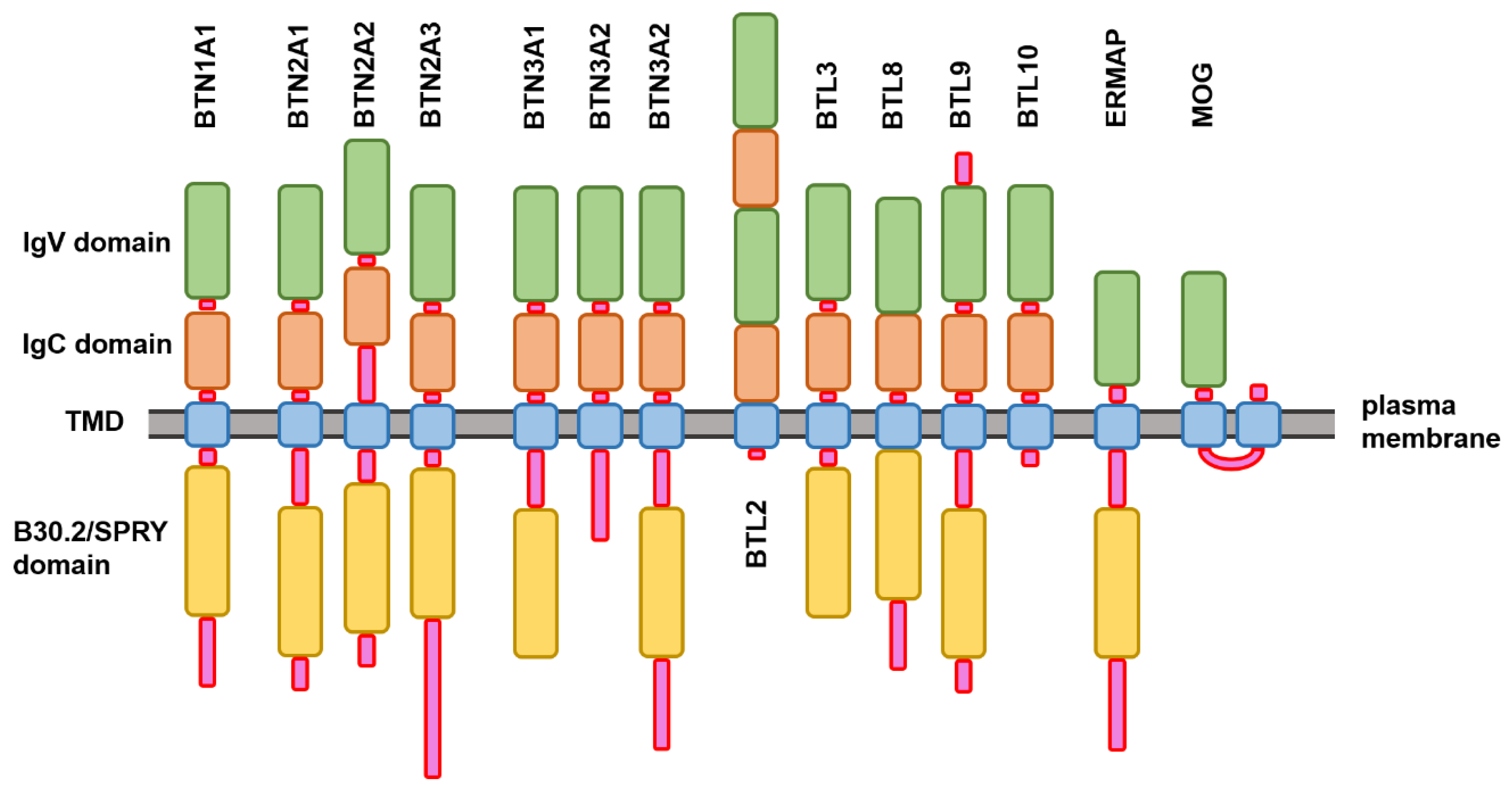
2.1. Structure and Functional Disorder of the Members of Human Butyrophilin Family
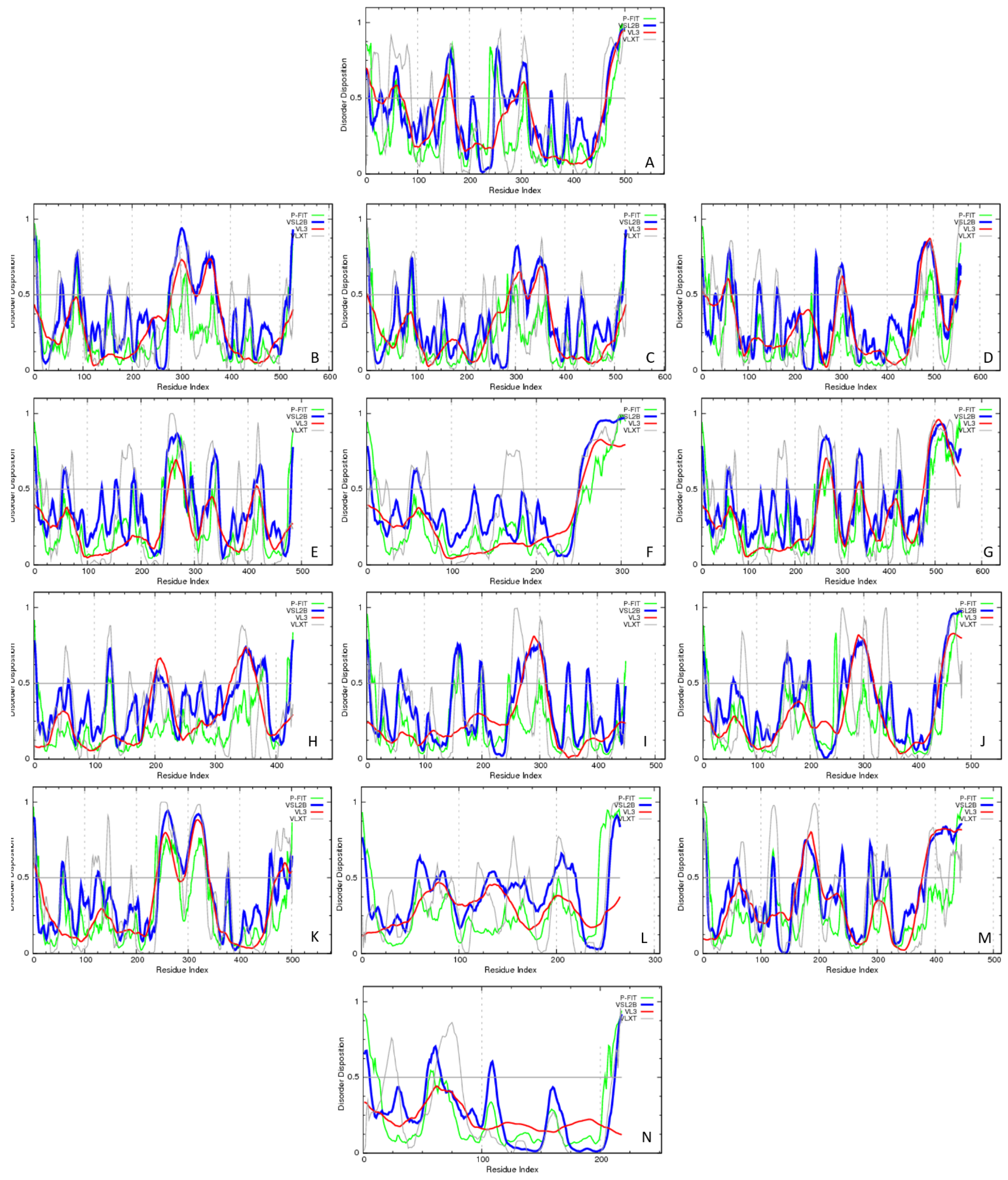
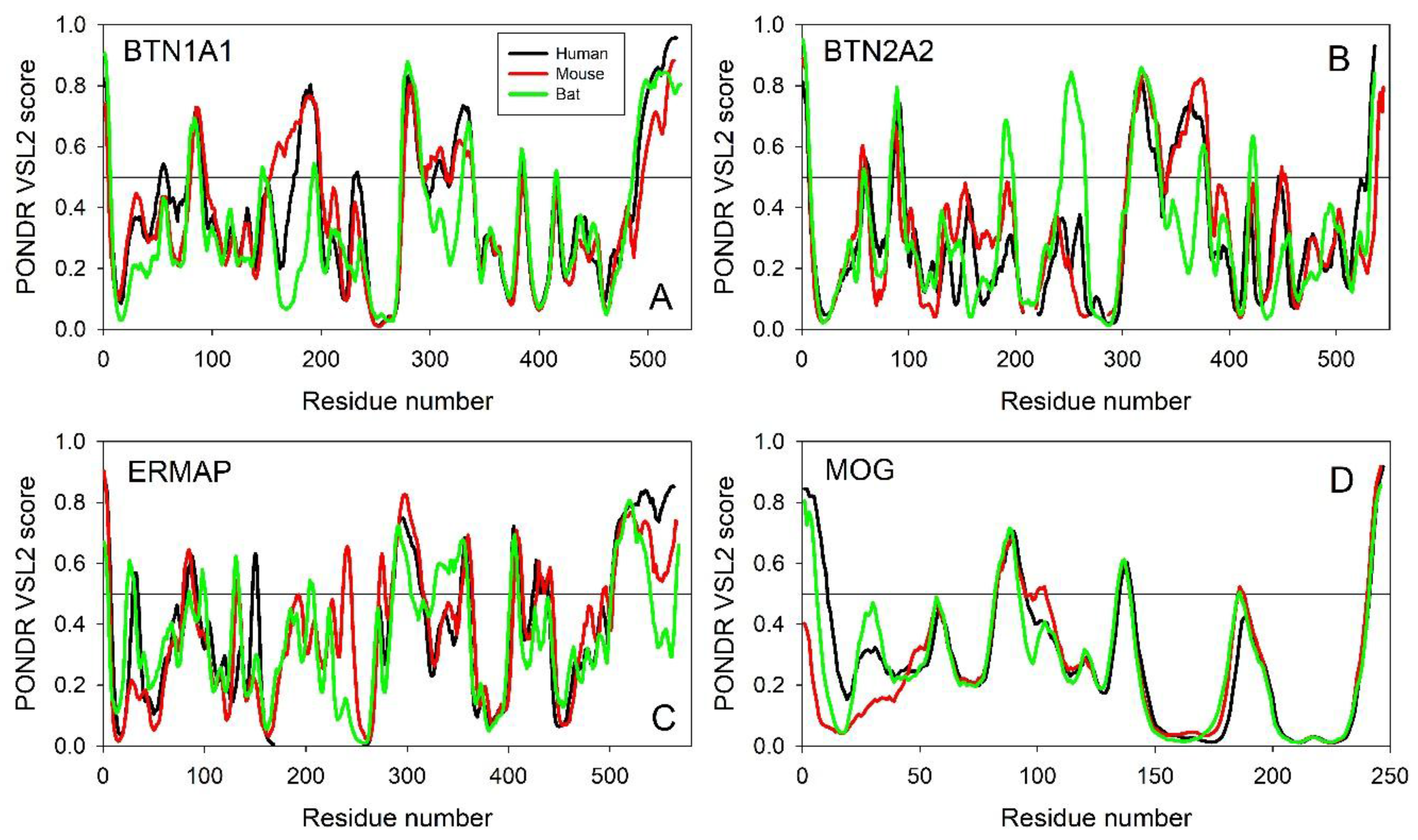
Evaluating the Overall Disorder Status of Human Butyrophilin Family Members
2.2. Predicted Intrinsic Disorder-Based Functionality of Human Butyrophilin Family Members
2.2.1. Finding Potential Disorder-Based Binding Sites, Molecular Recognition Features (MoRFs)
2.2.2. Predicted Intrinsic Disorder-Based Functionality of Human Butyrophilins
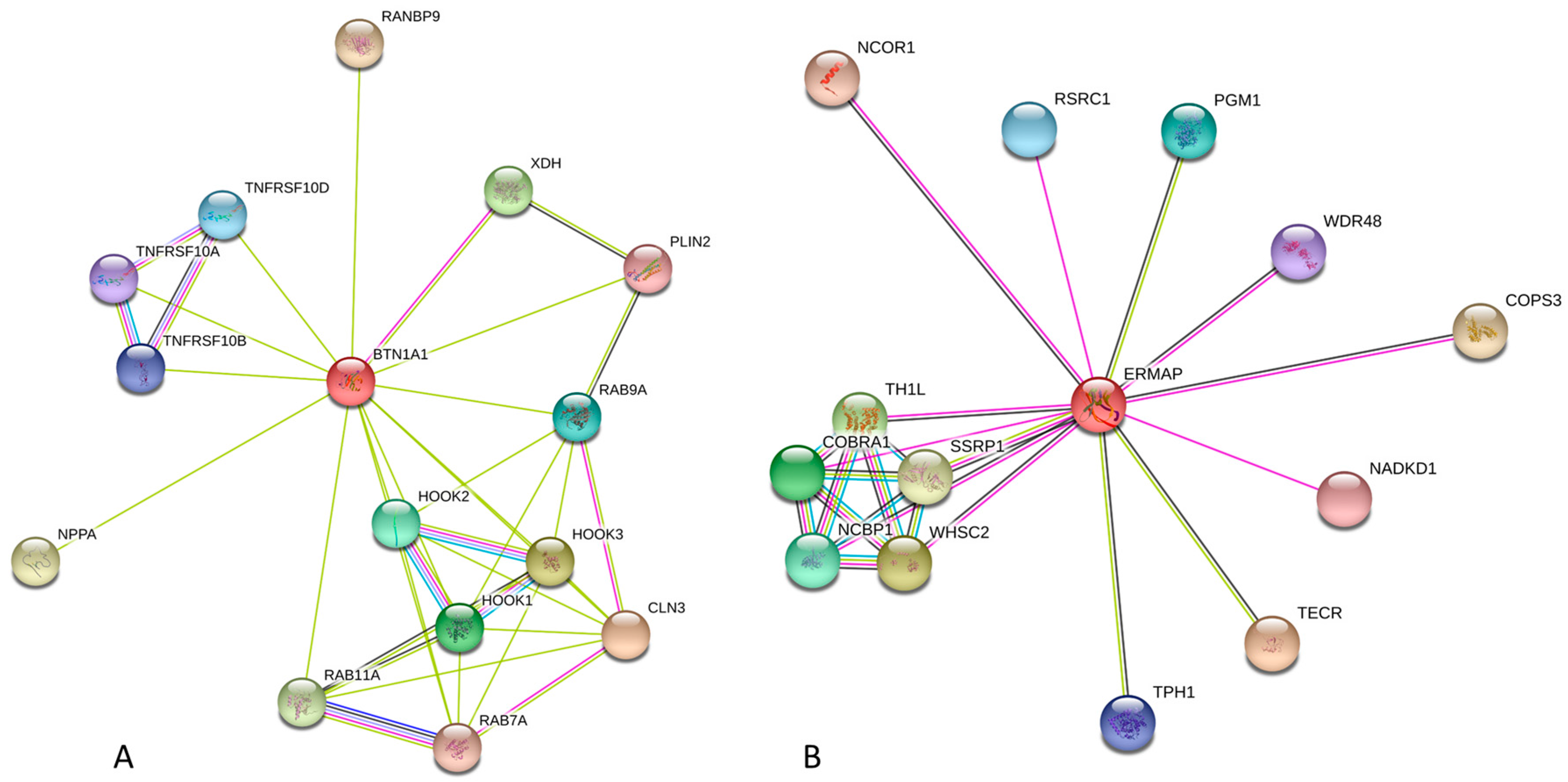
2.2.3. Protein-Protein Interactions of Human Butyrophilins
3. Discussion
3.1. Structural Organization of the Human Butyrophilin Family Members
3.2. Butyrophilins and Immune Response
3.3. Therapeutic Potentials of Butyrophilins
4. Experimental Section
4.1. Dataset
4.2. Methods
4.2.1. Multiple Sequence Alignment
4.2.2. Evaluation of Intrinsic Disorder Predisposition of Human Butyrophilins
4.2.3. Evaluation of the Functionality of Intrinsically Disordered Regions in Human Butyrophilins
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- O′Riordan, N.; Kane, M.; Joshi, L.; Hickey, R.M. Structural and functional characteristics of bovine milk protein glycosylation. Glycobiology 2014, 24, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.A.; Scallan, C.D.; Ceriani, R.L.; Hamosh, M. Structural and functional aspects of three major glycoproteins of the human milk fat globule membrane. Adv. Exp. Med. Biol. 2001, 501, 179–187. [Google Scholar] [PubMed]
- Williams, A.F.; Barclay, A.N. The immunoglobulin superfamily—Domains for cell surface recognition. Annu. Rev. Immunol. 1988, 6, 381–405. [Google Scholar] [CrossRef] [PubMed]
- Abeler-Dorner, L.; Swamy, M.; Williams, G.; Hayday, A.C.; Bas, A. Butyrophilins: An emerging family of immune regulators. Trends Immunol. 2012, 33, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Afrache, H.; Gouret, P.; Ainouche, S.; Pontarotti, P.; Olive, D. The butyrophilin (BTN) gene family: From milk fat to the regulation of the immune response. Immunogenetics 2012, 64, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Esser, C. A fat story-antigen presentation by butyrophilin 3A1 to gammadelta T cells. Cell. Mol. Immunol. 2014, 11, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.A.; Knezevic, B.R.; Ammann, J.U.; Rhodes, D.A.; Aw, D.; Palmer, D.B.; Mather, I.H.; Trowsdale, J. BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J. Immunol. 2010, 184, 3514–3525. [Google Scholar] [CrossRef] [PubMed]
- Mather, I.H. A review and proposed nomenclature for major proteins of the milk-fat globule membrane. J. Dairy Sci. 2000, 83, 203–247. [Google Scholar] [CrossRef]
- Ogg, S.L.; Weldon, A.K.; Dobbie, L.; Smith, A.J.; Mather, I.H. Expression of butyrophilin (BTN1A1) in lactating mammary gland is essential for the regulated secretion of milk-lipid droplets. Proc. Natl. Acad. Sci. USA 2004, 101, 10084–10089. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W.; Heid, H.W.; Grund, C.; Winter, S.; Freudenstein, C.; Schmid, E.; Jarasch, E.D.; Keenan, T.W. Antibodies to the major insoluble milk fat globule membrane-associated protein: Specific location in apical regions of lactating epithelial cells. J. Cell Biol. 1981, 89, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Ogg, S.L.; Komaragiri, M.V.; Mather, I.H. Structural organization and mammary-specific expression of the butyrophilin gene. Mamm. Genome 1996, 7, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Stammers, M.; Rowen, L.; Rhodes, D.; Trowsdale, J.; Beck, S. Btl-ii: A polymorphic locus with homology to the butyrophilin gene family, located at the border of the major histocompatibility complex class ii and class iii regions in human and mouse. Immunogenetics 2000, 51, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.Z.; Gordon, C.T.; Lai, Y.H.; Fujiwara, Y.; Peters, L.L.; Perkins, A.C.; Chui, D.H. Ermap, a gene coding for a novel erythroid specific adhesion/receptor membrane protein. Gene 2000, 242, 337–345. [Google Scholar] [CrossRef]
- Rhodes, D.A.; Stammers, M.; Malcherek, G.; Beck, S.; Trowsdale, J. The cluster of btn genes in the extended major histocompatibility complex. Genomics 2001, 71, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Ruddy, D.A.; Kronmal, G.S.; Lee, V.K.; Mintier, G.A.; Quintana, L.; Domingo, R., Jr.; Meyer, N.C.; Irrinki, A.; McClelland, E.E.; Fullan, A.; et al. A 1.1-mb transcript map of the hereditary hemochromatosis locus. Genome Res. 1997, 7, 441–456. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Permyakov, E.A.; Permyakov, S.E.; Breydo, L.; Redwan, E.M.; Almehdar, H.A.; Uversky, V.N. Disorder in milk proteins: Alpha-lactalbumin. Part c. Peculiarities of metal binding. Curr. Protein Pept. Sci. 2016, 17, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, E.A.; Permyakov, S.E.; Breydo, L.; Redwan, E.M.; Almehdar, H.A.; Uversky, V.N. Disorder in milk proteins: Alpha -lactalbumin. Part a. Structural properties and conformational behavior. Curr. Protein Pept. Sci. 2016, 17, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Permyakov, S.E.; Breydo, L.; Redwan, E.M.; Almehdar, H.A.; Permyakov, E.A. Disorder in milk proteins: Alpha-lactalbumin. Part b. A multifunctional whey protein acting as an oligomeric molten globular “oil container” in the anti-tumorigenic drugs, liprotides. Curr. Protein Pept. Sci. 2016, 17, 612–628. [Google Scholar] [CrossRef] [PubMed]
- Almehdar, H.A.; El-Fakharany, E.M.; Uversky, V.N.; Redwan, E.M. Disorder in milk proteins: Structure, functional disorder, and biocidal potentials of lactoperoxidase. Curr. Protein Pept. Sci. 2015, 16, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Redwan, E.M.; Xue, B.; Almehdar, H.A.; Uversky, V.N. Disorder in milk proteins: Caseins, intrinsically disordered colloids. Curr. Protein Pept. Sci. 2015, 16, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. Pondr-fit: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the embl-ebi. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.H.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Brown, C.J.; Obradovic, Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002, 62, 25–49. [Google Scholar] [PubMed]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Daughdrill, G.W.; Pielak, G.J.; Uversky, V.N.; Cortese, M.S.; Dunker, A.K. Natively disordered proteins. In Handbook of Protein Folding; Buchner, J., Kiefhaber, T., Eds.; Wiley-VCH, Verlag GmbH & Co.: Weinheim, Germany, 2005; pp. 271–353. [Google Scholar]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your id: Intrinsic disorder as an id for recognition, regulation and cell signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Uversky, V.N. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008, 4, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Multitude of binding modes attainable by intrinsically disordered proteins: A portrait gallery of disorder-based complexes. Chem. Soc. Rev. 2011, 40, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Disordered competitive recruiter: Fast and foldable. J. Mol. Biol. 2012, 418, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013, 19, 4191–4213. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002, 12, 54–60. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 2006, 362, 1043–1059. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Oldfield, C.J.; Mohan, A.; Radivojac, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Characterization of molecular recognition features, morfs, and their binding partners. J. Proteome Res. 2007, 6, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef] [PubMed]
- Disfani, F.M.; Hsu, W.L.; Mizianty, M.J.; Oldfield, C.J.; Xue, B.; Dunker, A.K.; Uversky, V.N.; Kurgan, L. Morfpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics 2012, 28, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Bayarjargal, M.; Tsunoda, T.; Patil, A.; Sharma, A. Morfpred-plus: Computational identification of morfs in protein sequences using physicochemical properties and hmm profiles. J. Theor. Biol. 2018, 437, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. Anchor: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. Iupred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Malhis, N.; Jacobson, M.; Gsponer, J. Morfchibi system: Software tools for the identification of morfs in protein sequences. Nucleic Acids Res. 2016, 44, W488–W493. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Cozzetto, D. Disopred3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015, 31, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Noguchi, T.; Tominaga, D.; Yamana, H. Mfspssmpred: Identifying short disorder-to-order binding regions in disordered proteins based on contextual local evolutionary conservation. BMC Bioinform. 2013, 14, 300. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)p(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005, 579, 3346–3354. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The string database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.; et al. The pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Mistry, J.; Schuster-Bockler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef] [PubMed]
- Han, J.D.; Bertin, N.; Hao, T.; Goldberg, D.S.; Berriz, G.F.; Zhang, L.V.; Dupuy, D.; Walhout, A.J.; Cusick, M.E.; Roth, F.P.; et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Hopfield, J.J.; Leibler, S.; Murray, A.W. From molecular to modular cell biology. Nature 1999, 402, C47–C52. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Nakamura, H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006, 580, 2041–2045. [Google Scholar] [CrossRef] [PubMed]
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006, 2, e100. [Google Scholar] [CrossRef] [PubMed]
- Ekman, D.; Light, S.; Bjorklund, A.K.; Elofsson, A. What properties characterize the hub proteins of the protein-protein interaction network of saccharomyces cerevisiae? Genome Biol. 2006, 7, R45. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Chen, J.; Dunker, A.K.; Simon, I.; Tompa, P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 2006, 5, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.P.; Ganapathi, M.; Sandhu, K.S.; Dash, D. Intrinsic unstructuredness and abundance of pest motifs in eukaryotic proteomes. Proteins 2006, 62, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Garner, E.; Guilliot, S.; Romero, P.; Albrecht, K.; Hart, J.; Obradovic, Z.; Kissinger, C.; Villafranca, J.E. Protein disorder and the evolution of molecular recognition: Theory, predictions and observations. Pac. Symp. Biocomput. 1998, 3, 473–484. [Google Scholar]
- Sepulveda, M.; Armangue, T.; Martinez-Hernandez, E.; Arrambide, G.; Sola-Valls, N.; Sabater, L.; Tellez, N.; Midaglia, L.; Arino, H.; Peschl, P.; et al. Clinical spectrum associated with mog autoimmunity in adults: Significance of sharing rodent mog epitopes. J. Neurol. 2016, 263, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Orozco, G.; Eerligh, P.; Sanchez, E.; Zhernakova, S.; Roep, B.O.; Gonzalez-Gay, M.A.; Lopez-Nevot, M.A.; Callejas, J.L.; Hidalgo, C.; Pascual-Salcedo, D.; et al. Analysis of a functional btnl2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum. Immunol. 2005, 66, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, L.; Chen, S.; He, L.; Yang, X.; Shi, Y.; Cheng, J.; Zhang, L.; Gu, C.C.; Huang, J.; et al. Genome-wide association study in han chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 2012, 44, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Spitsberg, V.L. Invited review: Bovine milk fat globule membrane as a potential nutraceutical. J. Dairy Sci. 2005, 88, 2289–2294. [Google Scholar] [CrossRef]
- Meyer, T.; Lauschke, J.; Ruppert, V.; Richter, A.; Pankuweit, S.; Maisch, B. Isolated cardiac sarcoidosis associated with the expression of a splice variant coding for a truncated BTNL2 protein. Cardiology 2008, 109, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, K.C.; Lin, Y.J.; Chang, J.S.; Wan, L.; Tsai, F.J. BTNL2 gene polymorphisms may be associated with susceptibility to kawasaki disease and formation of coronary artery lesions in taiwanese children. Eur. J. Pediatr. 2010, 169, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, M.; Oguri, M.; Kato, K.; Yoshida, T.; Fujimaki, T.; Horibe, H.; Yokoi, K.; Watanabe, S.; Satoh, K.; Aoyagi, Y.; et al. Association of a polymorphism of BTN2A1 with type 2 diabetes mellitus in japanese individuals. Diabet. Med. 2011, 28, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, P.; du Bois, R.M. Genetics of sarcoidosis. Clin. Dermatol. 2007, 25, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, J. Genetics of sarcoidosis. Curr. Opin. Pulm. Med. 2008, 14, 434–439. [Google Scholar] [CrossRef] [PubMed]
- Chatham, W. Rheumatic manifestations of systemic disease: Sarcoidosis. Curr. Opin Rheumatol. 2010, 22, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.; Reindl, M. Immunopathogenic and clinical relevance of antibodies against myelin oligodendrocyte glycoprotein (MOG) in multiple sclerosis. J. Neural Transm. Suppl. 2000, 351–360. [Google Scholar]
- Stefferl, A.; Brehm, U.; Linington, C. The myelin oligodendrocyte glycoprotein (MOG): A model for antibody-mediated demyelination in experimental autoimmune encephalomyelitis and multiple sclerosis. J. Neural Transm. Suppl. 2000, 58, 123–133. [Google Scholar]
- Lebrero-Fernandez, C.; Wenzel, U.A.; Akeus, P.; Wang, Y.; Strid, H.; Simren, M.; Gustavsson, B.; Borjesson, L.G.; Cardell, S.L.; Ohman, L.; et al. Altered expression of butyrophilin (BTN) and btn-like (BTNL) genes in intestinal inflammation and colon cancer. Immun. Inflamm. Dis. 2016, 4, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.M.; Witzmann, F.A.; Cummings, O.W.; Schmidt, C.M. A pilot study of proteomic profiles of human hepatocellular carcinoma in the united states. J. Surg. Res. 2009, 155, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Le Page, C.; Marineau, A.; Bonza, P.K.; Rahimi, K.; Cyr, L.; Labouba, I.; Madore, J.; Delvoye, N.; Mes-Masson, A.M.; Provencher, D.M.; et al. BTN3A2 expression in epithelial ovarian cancer is associated with higher tumor infiltrating T cells and a better prognosis. PLoS ONE 2012, 7, e38541. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, L.M.; Kumar, A.; Boyle, E.A.; Zhang, Y.; McIntosh, L.M.; Kolb, S.; Stott-Miller, M.; Smith, T.; Karyadi, D.M.; Ostrander, E.A.; et al. Germline missense variants in the BTNL2 gene are associated with prostate cancer susceptibility. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.; Parodi, F.; Diedrich, K.; Angelini, G.; Gotz, C.; Viaggi, S.; Maric, I.; Coviello, D.; Pistillo, M.P.; Morabito, A.; et al. Analysis of the expression and single-nucleotide variant frequencies of the butyrophilin-like 2 gene in patients with uveal melanoma. JAMA Ophthalmol. 2016, 134, 1125–1133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cubillos-Ruiz, J.R.; Martinez, D.; Scarlett, U.K.; Rutkowski, M.R.; Nesbeth, Y.C.; Camposeco-Jacobs, A.L.; Conejo-Garcia, J.R. Cd277 is a negative co-stimulatory molecule universally expressed by ovarian cancer microenvironmental cells. Oncotarget 2010, 1, 329–338. [Google Scholar] [PubMed]
- Liu, B.; Newburg, D.S. Human milk glycoproteins protect infants against human pathogens. Breastfeed. Med. 2013, 8, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Szasz, C.; Buday, L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005, 30, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins from a to z. Int. J. Biochem. Cell Biol. 2011, 43, 1090–1103. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. The most important thing is the tail: Multitudinous functionalities of intrinsically disordered protein termini. FEBS Lett. 2013, 587, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [PubMed]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- DeForte, S.; Uversky, V.N. Order, disorder, and everything in between. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Dancing protein clouds: The strange biology and chaotic physics of intrinsically disordered proteins. J. Biol. Chem. 2016, 291, 6681–6688. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. P53 proteoforms and intrinsic disorder: An illustration of the protein structure-function continuum concept. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Malaney, P.; Uversky, V.N.; Dave, V. Pten proteoforms in biology and disease. Cell. Mol. Life Sci. 2017, 74, 2783–2794. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Looking at the recent advances in understanding alpha-synuclein and its aggregation through the proteoform prism. F1000Res 2017, 6, 525. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J. Adenovirus amazes at cold spring harbor. Nature 1977, 268, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Black, D.L. Mechanisms of alternative pre-messenger rna splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef] [PubMed]
- Stamm, S.; Ben-Ari, S.; Rafalska, I.; Tang, Y.; Zhang, Z.; Toiber, D.; Thanaraj, T.A.; Soreq, H. Function of alternative splicing. Gene 2005, 344, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Brett, D.; Hanke, J.; Lehmann, G.; Haase, S.; Delbruck, S.; Krueger, S.; Reich, J.; Bork, P. Est comparison indicates 38% of human mrnas contain possible alternative splice forms. FEBS Lett. 2000, 474, 83–86. [Google Scholar] [CrossRef]
- Johnson, J.M.; Castle, J.; Garrett-Engele, P.; Kan, Z.; Loerch, P.M.; Armour, C.D.; Santos, R.; Schadt, E.E.; Stoughton, R.; Shoemaker, D.D. Genome-wide survey of human alternative pre-mrna splicing with exon junction microarrays. Science 2003, 302, 2141–2144. [Google Scholar] [CrossRef] [PubMed]
- Graveley, B.R. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001, 17, 100–107. [Google Scholar] [CrossRef]
- Minneman, K.P. Splice variants of g protein-coupled receptors. Mol. Interv. 2001, 1, 108–116. [Google Scholar] [PubMed]
- Thai, T.H.; Kearney, J.F. Distinct and opposite activities of human terminal deoxynucleotidyltransferase splice variants. J. Immunol. 2004, 173, 4009–4019. [Google Scholar] [CrossRef] [PubMed]
- Scheper, W.; Zwart, R.; Baas, F. Alternative splicing in the n-terminus of alzheimer’s presenilin 1. Neurogenetics 2004, 5, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Chalancon, G.; Eustermann, S.; Wagner, G.P.; Fuxreiter, M.; Bateman, A.; Babu, M.M. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell. 2012, 46, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Chalancon, G.; Dunker, A.K.; Bateman, A.; Balaji, S.; Fuxreiter, M.; Babu, M.M. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Palakodeti, A.; Sandstrom, A.; Sundaresan, L.; Harly, C.; Nedellec, S.; Olive, D.; Scotet, E.; Bonneville, M.; Adams, E.J. The molecular basis for modulation of human vgamma9vdelta2 t cell responses by cd277/butyrophilin-3 (btn3a)-specific antibodies. J. Biol. Chem. 2012, 287, 32780–32790. [Google Scholar] [CrossRef] [PubMed]
- Shatsky, M.; Nussinov, R.; Wolfson, H.J. A method for simultaneous alignment of multiple protein structures. Proteins 2004, 56, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Morita, C.T. Sensor function for butyrophilin 3a1 in prenyl pyrophosphate stimulation of human vgamma2vdelta2 t cells. J. Immunol. 2015, 195, 4583–4594. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Henry, O.; Distefano, M.D.; Wang, Y.C.; Raikkonen, J.; Monkkonen, J.; Tanaka, Y.; Morita, C.T. Butyrophilin 3a1 plays an essential role in prenyl pyrophosphate stimulation of human vgamma2vdelta2 t cells. J. Immunol. 2013, 191, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Nerdal, P.T.; Peters, C.; Oberg, H.H.; Zlatev, H.; Lettau, M.; Quabius, E.S.; Sousa, S.; Gonnermann, D.; Auriola, S.; Olive, D.; et al. Butyrophilin 3a/cd277-dependent activation of human gammadelta t cells: Accessory cell capacity of distinct leukocyte populations. J. Immunol. 2016, 197, 3059–3068. [Google Scholar] [CrossRef] [PubMed]
- Shippy, R.R.; Lin, X.; Agabiti, S.S.; Li, J.; Zangari, B.M.; Foust, B.J.; Poe, M.M.; Hsiao, C.C.; Vinogradova, O.; Wiemer, D.F.; et al. Phosphinophosphonates and their tris-pivaloyloxymethyl prodrugs reveal a negatively cooperative butyrophilin activation mechanism. J. Med. Chem. 2017, 60, 2373–2382. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, C.H.; Lin, X.; Barney, R.J.; Shippy, R.R.; Li, J.; Vinogradova, O.; Wiemer, D.F.; Wiemer, A.J. Synthesis of a phosphoantigen prodrug that potently activates vgamma9vdelta2 t-lymphocytes. Chem. Biol. 2014, 21, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Morita, C.T.; Jin, C.; Sarikonda, G.; Wang, H. Nonpeptide antigens, presentation mechanisms, and immunological memory of human vgamma2vdelta2 t cells: Discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol. Rev. 2007, 215, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Gao, Y.; Scully, E.; Davis, C.T.; Anderson, J.F.; Welte, T.; Ledizet, M.; Koski, R.; Madri, J.A.; Barrett, A.; et al. Gamma delta t cells facilitate adaptive immunity against west nile virus infection in mice. J. Immunol. 2006, 177, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, W.; Pan, M.; Scully, E.; Girardi, M.; Augenlicht, L.H.; Craft, J.; Yin, Z. Gamma delta t cells provide an early source of interferon gamma in tumor immunity. J. Exp. Med. 2003, 198, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Vavassori, S.; Kumar, A.; Wan, G.S.; Ramanjaneyulu, G.S.; Cavallari, M.; El Daker, S.; Beddoe, T.; Theodossis, A.; Williams, N.K.; Gostick, E.; et al. Butyrophilin 3a1 binds phosphorylated antigens and stimulates human gammadelta t cells. Nat. Immunol. 2013, 14, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Li, J.; Puthenveetil, R.; Lin, X.; Poe, M.M.; Hsiao, C.C.; Vinogradova, O.; Wiemer, A.J. The butyrophilin 3a1 intracellular domain undergoes a conformational change involving the juxtamembrane region. FASEB J. 2017, 31, 4697–4706. [Google Scholar] [CrossRef] [PubMed]
- Harly, C.; Guillaume, Y.; Nedellec, S.; Peigne, C.M.; Monkkonen, H.; Monkkonen, J.; Li, J.; Kuball, J.; Adams, E.J.; Netzer, S.; et al. Key implication of cd277/butyrophilin-3 (btn3a) in cellular stress sensing by a major human gammadelta t-cell subset. Blood 2012, 120, 2269–2279. [Google Scholar] [CrossRef] [PubMed]
- Compte, E.; Pontarotti, P.; Collette, Y.; Lopez, M.; Olive, D. Frontline: Characterization of bt3 molecules belonging to the b7 family expressed on immune cells. Eur. J. Immunol. 2004, 34, 2089–2099. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, A.Y. Novel immune check-point regulators in tolerance maintenance. Front. Immunol. 2015, 6, 421. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, H.; Yoshizaki, S.; Tadaki, T.; Egawa, K.; Seo, N. Stimulation of human butyrophilin 3 molecules results in negative regulation of cellular immunity. J. Leukoc. Biol. 2010, 88, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Guenot, M.; Loizon, S.; Howard, J.; Costa, G.; Baker, D.A.; Mohabeer, S.Y.; Troye-Blomberg, M.; Moreau, J.F.; Dechanet-Merville, J.; Mercereau-Puijalon, O.; et al. Phosphoantigen burst upon plasmodium falciparum schizont rupture can distantly activate vgamma9vdelta2 t cells. Infect. Immun. 2015, 83, 3816–3824. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, P.; Kim, C.C.; Greenhouse, B.; Nankya, F.; Bowen, K.; Eccles-James, I.; Muhindo, M.K.; Arinaitwe, E.; Tappero, J.W.; Kamya, M.R.; et al. Loss and dysfunction of vdelta2(+) gammadelta t cells are associated with clinical tolerance to malaria. Sci. Transl. Med. 2014, 6, 251ra117. [Google Scholar] [CrossRef] [PubMed]
- Stanisic, D.I.; Cutts, J.; Eriksson, E.; Fowkes, F.J.; Rosanas-Urgell, A.; Siba, P.; Laman, M.; Davis, T.M.; Manning, L.; Mueller, I.; et al. Gammadelta t cells and cd14+ monocytes are predominant cellular sources of cytokines and chemokines associated with severe malaria. J. Infect. Dis. 2014, 210, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.; Loizon, S.; Guenot, M.; Mocan, I.; Halary, F.; de Saint-Basile, G.; Pitard, V.; Dechanet-Merville, J.; Moreau, J.F.; Troye-Blomberg, M.; et al. Control of plasmodium falciparum erythrocytic cycle: Gammadelta t cells target the red blood cell-invasive merozoites. Blood 2011, 118, 6952–6962. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.; Tongtawe, P.; Kriangkum, J.; Wimonwattrawatee, T.; Pattanapanyasat, K.; Bryant, L.; Shafiq, J.; Suntharsamai, P.; Looareesuwan, S.; Webster, H.K.; et al. Polyclonal expansion of peripheral gamma delta t cells in human plasmodium falciparum malaria. Infect. Immun. 1994, 62, 855–862. [Google Scholar] [PubMed]
- Roussilhon, C.; Agrapart, M.; Guglielmi, P.; Bensussan, A.; Brasseur, P.; Ballet, J.J. Human tcr gamma delta+ lymphocyte response on primary exposure to plasmodium falciparum. Clin. Exp. Immunol. 1994, 95, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.W. Multifunctional immune responses of hmbpp-specific vgamma2vdelta2 t cells in m. Tuberculosis and other infections. Cell. Mol. Immunol. 2013, 10, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.W. Protective immune responses of major vgamma2vdelta2 t-cell subset in m. Tuberculosis infection. Curr. Opin. Immunol. 2016, 42, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Correia, D.V.; Fogli, M.; Hudspeth, K.; da Silva, M.G.; Mavilio, D.; Silva-Santos, B. Differentiation of human peripheral blood vdelta1+ t cells expressing the natural cytotoxicity receptor nkp30 for recognition of lymphoid leukemia cells. Blood 2011, 118, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Kunzmann, V.; Wilhelm, M. Anti-lymphoma effect of gammadelta t cells. Leuk. Lymphoma 2005, 46, 671–680. [Google Scholar] [CrossRef] [PubMed]
- McClanahan, J.; Fukushima, P.I.; Stetler-Stevenson, M. Increased peripheral blood gamma delta t-cells in patients with lymphoid neoplasia: A diagnostic dilemma in flow cytometry. Cytometry 1999, 38, 280–285. [Google Scholar] [CrossRef]
- Decaup, E.; Duault, C.; Bezombes, C.; Poupot, M.; Savina, A.; Olive, D.; Fournie, J.J. Phosphoantigens and butyrophilin 3a1 induce similar intracellular activation signaling in human tcrvgamma9+ gammadelta t lymphocytes. Immunol. Lett. 2014, 161, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Riano, F.; Karunakaran, M.M.; Starick, L.; Li, J.; Scholz, C.J.; Kunzmann, V.; Olive, D.; Amslinger, S.; Herrmann, T. Vgamma9vdelta2 tcr-activation by phosphorylated antigens requires butyrophilin 3 a1 (BTN3A1) and additional genes on human chromosome 6. Eur. J. Immunol. 2014, 44, 2571–2576. [Google Scholar] [CrossRef] [PubMed]
- Chapoval, A.I.; Smithson, G.; Brunick, L.; Mesri, M.; Boldog, F.L.; Andrew, D.; Khramtsov, N.V.; Feshchenko, E.A.; Starling, G.C.; Mezes, P.S. Btnl8, a butyrophilin-like molecule that costimulates the primary immune response. Mol. Immunol. 2013, 56, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Lebrero-Fernandez, C.; Bergstrom, J.H.; Pelaseyed, T.; Bas-Forsberg, A. Murine butyrophilin-like 1 and btnl6 form heteromeric complexes in small intestinal epithelial cells and promote proliferation of local T lymphocytes. Front Immunol. 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Malcherek, G.; Mayr, L.; Roda-Navarro, P.; Rhodes, D.; Miller, N.; Trowsdale, J. The b7 homolog butyrophilin btn2a1 is a novel ligand for dc-sign. J. Immunol. 2007, 179, 3804–3811. [Google Scholar] [CrossRef] [PubMed]
- Sarter, K.; Leimgruber, E.; Gobet, F.; Agrawal, V.; Dunand-Sauthier, I.; Barras, E.; Mastelic-Gavillet, B.; Kamath, A.; Fontannaz, P.; Guery, L.; et al. Btn2a2, a T cell immunomodulatory molecule coregulated with mhc class II genes. J. Exp. Med. 2016, 213, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, P.; Charavaryamath, C.; Moshynskyy, I.; Lockerbie, B.; Kaushik, R.S.; Loewen, M.E.; Kidney, B.A.; Stuart, C.; Simko, E. Evaluation of inhibition of f4ac positive escherichia coli attachment with xanthine dehydrogenase, butyrophilin, lactadherin and fatty acid binding protein. BMC Vet. Res. 2015, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, P.; Huang, Y.Y.; Lockerbie, B.; Shahriar, F.; Kelly, J.; Gordon, J.R.; Middleton, D.M.; Loewen, M.E.; Kidney, B.A.; Simko, E. Identification of escherichia coli f4ac-binding proteins in porcine milk fat globule membrane. Can. J. Vet. Res. 2015, 79, 120–128. [Google Scholar] [PubMed]
- Vojdani, A.; Campbell, A.W.; Anyanwu, E.; Kashanian, A.; Bock, K.; Vojdani, E. Antibodies to neuron-specific antigens in children with autism: Possible cross-reaction with encephalitogenic proteins from milk, chlamydia pneumoniae and streptococcus group a. J. Neuroimmunol. 2002, 129, 168–177. [Google Scholar] [CrossRef]
- Stefferl, A.; Schubart, A.; Storch, M.; Amini, A.; Mather, I.; Lassmann, H.; Linington, C. Butyrophilin, a milk protein, modulates the encephalitogenic t cell response to myelin oligodendrocyte glycoprotein in experimental autoimmune encephalomyelitis. J. Immunol. 2000, 165, 2859–2865. [Google Scholar] [CrossRef] [PubMed]
- Mana, P.; Goodyear, M.; Bernard, C.; Tomioka, R.; Freire-Garabal, M.; Linares, D. Tolerance induction by molecular mimicry: Prevention and suppression of experimental autoimmune encephalomyelitis with the milk protein butyrophilin. Int. Immunol. 2004, 16, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, P.; Shojaei, H.; Schittek, B.; Gieseler, F.; Wollenberg, B.; Kalthoff, H.; Kabelitz, D.; Wesch, D. Lysis of a broad range of epithelial tumour cells by human gamma delta t cells: Involvement of nkg2d ligands and t-cell receptor- versus nkg2d-dependent recognition. Scand. J. Immunol. 2007, 66, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Bryant, N.L.; Gillespie, G.Y.; Lopez, R.D.; Markert, J.M.; Cloud, G.A.; Langford, C.P.; Arnouk, H.; Su, Y.; Haines, H.L.; Suarez-Cuervo, C.; et al. Preclinical evaluation of ex vivo expanded/activated gammadelta t cells for immunotherapy of glioblastoma multiforme. J. Neurooncol. 2011, 101, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Chitadze, G.; Lettau, M.; Luecke, S.; Wang, T.; Janssen, O.; Furst, D.; Mytilineos, J.; Wesch, D.; Oberg, H.H.; Held-Feindt, J.; et al. Nkg2d- and t-cell receptor-dependent lysis of malignant glioma cell lines by human gammadelta t cells: Modulation by temozolomide and a disintegrin and metalloproteases 10 and 17 inhibitors. Oncoimmunology 2016, 5, e1093276. [Google Scholar] [CrossRef] [PubMed]
- Strid, J.; Roberts, S.J.; Filler, R.B.; Lewis, J.M.; Kwong, B.Y.; Schpero, W.; Kaplan, D.H.; Hayday, A.C.; Girardi, M. Acute upregulation of an nkg2d ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nat. Immunol. 2008, 9, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Groh, V.; Wu, J.; Steinle, A.; Phillips, J.H.; Lanier, L.L.; Spies, T. Activation of nk cells and t cells by nkg2d, a receptor for stress-inducible mica. Science 1999, 285, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Guerra, N.; Tan, Y.X.; Joncker, N.T.; Choy, A.; Gallardo, F.; Xiong, N.; Knoblaugh, S.; Cado, D.; Greenberg, N.M.; Raulet, D.H. Nkg2d-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 2008, 28, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Raulet, D.H.; Guerra, N. Oncogenic stress sensed by the immune system: Role of natural killer cell receptors. Nat. Rev. Immunol. 2009, 9, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Benyamine, A.; Le Roy, A.; Mamessier, E.; Gertner-Dardenne, J.; Castanier, C.; Orlanducci, F.; Pouyet, L.; Goubard, A.; Collette, Y.; Vey, N.; et al. Btn3a molecules considerably improve vgamma9vdelta2t cells-based immunotherapy in acute myeloid leukemia. Oncoimmunology 2016, 5, e1146843. [Google Scholar] [CrossRef] [PubMed]
- Werter, I.M.; Schneiders, F.L.; Scotet, E.; Verheul, H.M.; de Gruijl, T.D.; van der Vliet, H.J. Vgamma9vdelta2-t cells as antigen presenting cells for inkt cell based cancer immunotherapy. Oncoimmunology 2014, 3, e955343. [Google Scholar] [CrossRef] [PubMed]
- Correia, D.V.; Lopes, A.; Silva-Santos, B. Tumor cell recognition by gammadelta t lymphocytes: T-cell receptor vs. Nk-cell receptors. Oncoimmunology 2013, 2, e22892. [Google Scholar] [CrossRef] [PubMed]
- Hannani, D.; Ma, Y.; Yamazaki, T.; Dechanet-Merville, J.; Kroemer, G.; Zitvogel, L. Harnessing gammadelta t cells in anticancer immunotherapy. Trends Immunol. 2012, 33, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.Q.; Correia, D.V.; Grosso, A.R.; Lanca, T.; Ferreira, C.; Lacerda, J.F.; Barata, J.T.; Silva, M.G.; Silva-Santos, B. Identification of a panel of ten cell surface protein antigens associated with immunotargeting of leukemias and lymphomas by peripheral blood gammadelta t cells. Haematologica 2010, 95, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, M.; O’Brien, R.L.; Born, W.K. Gammadelta t cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010, 10, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Peedicayil, A.; Vierkant, R.A.; Hartmann, L.C.; Fridley, B.L.; Fredericksen, Z.S.; White, K.L.; Elliott, E.A.; Phelan, C.M.; Tsai, Y.Y.; Berchuck, A.; et al. Risk of ovarian cancer and inherited variants in relapse-associated genes. PLoS ONE 2010, 5, e8884. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Kato, K.; Oguri, M.; Horibe, H.; Kawamiya, T.; Yokoi, K.; Fujimaki, T.; Watanabe, S.; Satoh, K.; Aoyagi, Y.; et al. Association of polymorphisms of btn2a1 and ilf3 with myocardial infarction in japanese individuals with or without hypertension, diabetes mellitus or chronic kidney disease. Int. J. Mol. Med. 2011, 27, 745–752. [Google Scholar] [PubMed]
- Yamada, Y.; Nishida, T.; Ichihara, S.; Sawabe, M.; Fuku, N.; Nishigaki, Y.; Aoyagi, Y.; Tanaka, M.; Fujiwara, Y.; Yoshida, H.; et al. Association of a polymorphism of btn2a1 with myocardial infarction in east asian populations. Atherosclerosis 2011, 215, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Fujimaki, T.; Kato, K.; Oguri, M.; Yohida, T.; Horibe, H.; Yokoi, K.; Watanabe, S.; Satoh, K.; Aoyagi, Y.; Tanaka, M.; et al. Association of a polymorphism of btn2a1 with dyslipidemia in east asian populations. Exp. Ther. Med. 2011, 2, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Matsui, K.; Takeuchi, I.; Fujimaki, T. Association of genetic variants with dyslipidemia and chronic kidney disease in a longitudinal population-based genetic epidemiological study. Int. J. Mol. Med. 2015, 35, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Horibe, H.; Ueyama, C.; Fujimaki, T.; Oguri, M.; Kato, K.; Ichihara, S.; Yamada, Y. Association of a polymorphism of btn2a1 with dyslipidemia in community-dwelling individuals. Mol. Med. Rep. 2014, 9, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Kato, K.; Oguri, M.; Horibe, H.; Kawamiya, T.; Yokoi, K.; Fujimaki, T.; Watanabe, S.; Satoh, K.; Aoyagi, Y.; et al. Association of a polymorphism of btn2a1 with chronic kidney disease in individuals with or without hypertension or diabetes mellitus. Exp. Ther. Med. 2011, 2, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Oguri, M.; Fujimaki, T.; Horibe, H.; Kato, K.; Ichihara, S.; Yamada, Y. Association of a polymorphism of btn2a1 with chronic kidney disease in community-dwelling individuals. Biomed. Rep. 2013, 1, 868–872. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoshida, T.; Kato, K.; Horibe, H.; Oguri, M.; Fukuda, M.; Satoh, K.; Aoyagi, Y.; Shinkai, S.; Nozawa, Y.; Yamada, Y. Association of a genetic variant of btn2a1 with chronic kidney disease in japanese individuals. Nephrology (Carlton) 2011, 16, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Horibe, H.; Kato, K.; Oguri, M.; Yoshida, T.; Fujimaki, T.; Kawamiya, T.; Yokoi, K.; Watanabe, S.; Satoh, K.; Aoyagi, Y.; et al. Association of a polymorphism of btn2a1 with hypertension in japanese individuals. Am. J. Hypertens. 2011, 24, 924–929. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murakata, Y.; Fujimaki, T.; Yamada, Y. Association of a butyrophilin, subfamily 2, member a1 gene polymorphism with hypertension. Biomed. Rep. 2014, 2, 818–822. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamada, Y.; Matsui, K.; Takeuchi, I.; Oguri, M.; Fujimaki, T. Association of genetic variants with hypertension in a longitudinal population-based genetic epidemiological study. Int. J. Mol. Med. 2015, 35, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Oguri, M.; Kato, K.; Yoshida, T.; Fujimaki, T.; Horibe, H.; Yokoi, K.; Watanabe, S.; Satoh, K.; Aoyagi, Y.; Tanaka, M.; et al. Association of a genetic variant of btn2a1 with metabolic syndrome in east asian populations. J. Med. Genet. 2011, 48, 787–792. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. Uniprot: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. Espritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein disorder prediction: Implications for structural proteomics. Structure 2003, 11, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Linding, R.; Russell, R.B.; Neduva, V.; Gibson, T.J. Globplot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003, 31, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 2005, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, T.; Walsh, I.; Martin, A.J.; Tosatto, S.C. Mobidb: A comprehensive database of intrinsic protein disorder annotations. Bioinformatics 2012, 28, 2080–2081. [Google Scholar] [CrossRef] [PubMed]
- Potenza, E.; Domenico, T.D.; Walsh, I.; Tosatto, S.C. Mobidb 2.0: An improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Silman, I.; Sussman, J.L. Foldindex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef] [PubMed]
- Campen, A.; Williams, R.M.; Brown, C.J.; Meng, J.; Uversky, V.N.; Dunker, A.K. Top-idp-scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 2008, 15, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.L.; Kurgan, L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012, 13, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Kurgan, L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J. Biomol. Struct. Dyn. 2014, 32, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. Prdos: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Chatr-Aryamontri, A.; Boucher, L.; Oughtred, R.; Livstone, M.S.; Nixon, J.; Van Auken, K.; Wang, X.; Shi, X.; et al. The biogrid interaction database: 2011 update. Nucleic Acids Res. 2011, 39, D698–D704. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not Available. |



| Name/UniProt ID (Protein Length) a | PPIDRmean b | PPIDRFIT c | Long IDPRs d | AIBS e | MoRFChibi_Web f | Number of Binding Partners g |
|---|---|---|---|---|---|---|
| BTN1A1/Q13410 (500 a.a.r.) | 21.4 ± 4.1 | 17.6 | 44–87 456–500 | 124–125 137–140 162–166 459–462 515–517 | 481–493 | 14 (3) |
| BTN2A1/Q7KYR7 (499 a.a.r.) | 12.3 ± 4.8 | 9.0 | 303–343 | 22–23 260–267 | N.P. | 3 (38) |
| BTN2A2/Q8WVV5 (491 a.a.r.) | 10.2 ± 3.3 | 8.1 | 255–333 | 277–285 | N.P. | 8 (34) |
| BTN2A3/Q96KV6 (559 a.a.r.) | 13.3 ± 2.9 | 11.3 | 461–509 | 18–23 123–124 136–139 551–562 | 523–558 | Unknown |
| BTN3A1/O00481 (484 a.a.r.) | 14.5 ± 3.8 | 16.5 | 248–290 | 125–128 337–338 380–391 | N.P. | 12 (14) |
| BTN3A2/P78410 (305 a.a.r.) | 13.5 ± 2.5 | 18.0 | 150–305 | 125–128 | 278–304 | 6 (Unknown) |
| BTN3A3/O00478 (555 a.a.r.) | 19.7 ± 2.1 | 20.4 | 247–289 481–545 | 495–513 549–563 575–584 125–128 381–388 464–467 537–540 | 524–531 545–555 | 4 (9) |
| BTNL2/Q9UIR0 (428 a.a.r.) | 11.2 ± 2.4 | 7.5 | 324–369 | 354–361 265–269 341–244 | 36–41 359–362 | 14 (Unknown) |
| BTNL3/Q6UXE8 (449 a.a.r.) | 11.5 ± 3.3 | 8.0 | 268–310 | 254–258 353–355 | N.P. | 49 (10) |
| BTNL8/Q6UX41 (483 a.a.r.) | 18.8 ± 3.5 | 10.6 | 272–318 441–483 | 241–258 | 454–464 473–482 | 26 (42) |
| BTNL9/Q6UXG8 (501 a.a.r.) | 19.8 ± 2.5 | 20.0 | 243–339 | 324–332 533–535 | 485–501 | 38 (Unknown) |
| BTNL10/A8MVZ5 (265 a.a.r.) | 12.1 ± 3.9 | 12.8 | 197–219 | 126–131 265–273 | 1–6 40–44 | Unknown |
| ERMAP/Q96PL5 (446 a.a.r.) | 17.6 ± 4.1 | 11.0 | 378–446 | 168–175 364–365 455–459 | 34–38 422–429 | 13 (14) |
| MOG/Q16653 (218 a.a.r.) | 12.1 ± 3.3 | 13.8 | 61–82 | 14–20 | 1–9 19–25 35–56 64–72 | 85 (1) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Redwan, E.M.; Al-Hejin, A.M.; Almehdar, H.A.; Elsaway, A.M.; Uversky, V.N. Prediction of Disordered Regions and Their Roles in the Anti-Pathogenic and Immunomodulatory Functions of Butyrophilins. Molecules 2018, 23, 328. https://doi.org/10.3390/molecules23020328
Redwan EM, Al-Hejin AM, Almehdar HA, Elsaway AM, Uversky VN. Prediction of Disordered Regions and Their Roles in the Anti-Pathogenic and Immunomodulatory Functions of Butyrophilins. Molecules. 2018; 23(2):328. https://doi.org/10.3390/molecules23020328
Chicago/Turabian StyleRedwan, Elrashdy M., Ahmed M. Al-Hejin, Hussein A. Almehdar, Abdelrahman M. Elsaway, and Vladimir N. Uversky. 2018. "Prediction of Disordered Regions and Their Roles in the Anti-Pathogenic and Immunomodulatory Functions of Butyrophilins" Molecules 23, no. 2: 328. https://doi.org/10.3390/molecules23020328
APA StyleRedwan, E. M., Al-Hejin, A. M., Almehdar, H. A., Elsaway, A. M., & Uversky, V. N. (2018). Prediction of Disordered Regions and Their Roles in the Anti-Pathogenic and Immunomodulatory Functions of Butyrophilins. Molecules, 23(2), 328. https://doi.org/10.3390/molecules23020328






