Marine Natural Peptides: Determination of Absolute Configuration Using Liquid Chromatography Methods and Evaluation of Bioactivities
Abstract
1. Introduction
2. Peptides from Marine Cyanobacteria and Other Bacteria
2.1. Cyclic Peptides
2.2. Cyclic Depsipeptides
2.3. Lipopeptides
3. Peptides from Marine-Derived Fungi
3.1. Cyclic Peptides
3.2. Cyclic Depsipeptides
4. Peptides from Marine Sponges
4.1. Cyclic Peptides
4.2. Cyclic Depsipeptides
4.3. Lipopeptides
5. Peptides from Other Marine Invertebrates and Algae
5.1. Cyclic Peptides
5.2. Cyclic Depsipeptides
5.3. Lipopeptides
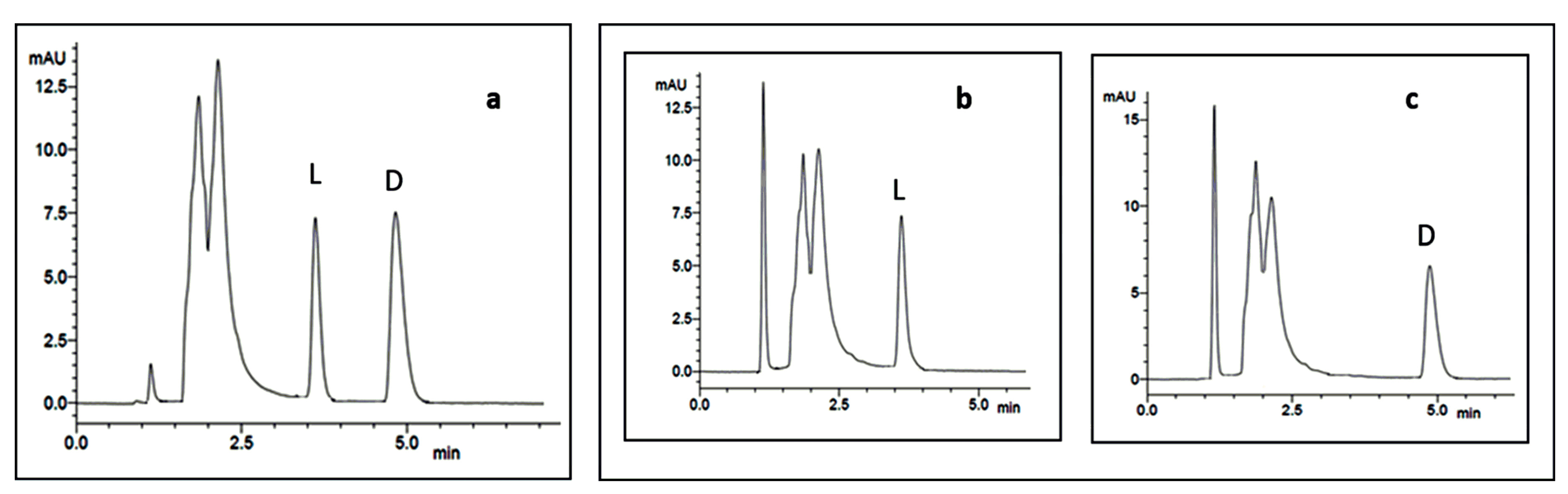
6. Case-Study: Chiral HPLC in the Analysis of the Stereochemistry of Cyclopeptides Isolated from Marine Sponge-Associated Fungi
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pinto, M.M.M.; Castanheiro, R.A.P.; Kijjoa, A. Xanthones from marine-derived microorganisms: Isolation, structure elucidation and biological activities. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Kijjoa, A.; Sawangwong, P. Drugs and cosmetics from the sea. Mar. Drugs 2004, 2, 73–82. [Google Scholar] [CrossRef]
- Saleem, M.; Ali, M.S.; Hussain, S.; Jabbar, A.; Ashraf, M.; Lee, Y.S. Marine natural products of fungal origin. Nat. Prod. Rep. 2007, 24, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Biomolecules produced by mangrove-associated microbes. Curr. Med. Chem. 2011, 18, 5224–5266. [Google Scholar] [CrossRef] [PubMed]
- Cheung, R.C.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M.; Rodriguez, A.D.; Berlinck, R.G.; Fusetani, N. Marine pharmacology in 2007–8: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp. Biochem. Physiol. C 2011, 153, 191–222. [Google Scholar]
- Aneiros, A.; Garateix, A. Bioactive peptides from marine sources: Pharmacological properties and isolation procedures. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 803, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.; Hamann, M.T. Marine natural products and their potential applications as anti-infective agents. Lancet Infect. Dis. 2003, 3, 338–348. [Google Scholar] [CrossRef]
- Harada, K.-I.; Fujii, K.; Mayumi, T.; Hibino, Y.; Suzuki, M.; Ikai, Y.; Oka, H. A method usingL/CMS for determination of absolute configuration of constituent amino acids in peptide—Advanced Marfey’s method. Tetrahedron Lett. 1995, 36, 1515–1518. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Hu, W.P.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2009, 26, 170–244. [Google Scholar] [CrossRef] [PubMed]
- Jimeno, J.; Faircloth, G.; Sousa-Faro, J.M.F.; Scheuer, P.; Rinehart, K. New marine derived anticancer therapeutics—A journey from the sea to clinical trials. Mar. Drugs 2004, 2, 14–29. [Google Scholar] [CrossRef]
- Baker, D.D.; Chu, M.; Oza, U.; Rajgarhia, V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–1244. [Google Scholar] [CrossRef] [PubMed]
- Glaser, K.B.; Mayer, A.M. A renaissance in marine pharmacology: From preclinical curiosity to clinical reality. Biochem. Pharmacol. 2009, 78, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Gogineni, V.; Hamann, M.T. Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology. Biochim. Biophys. Acta 2018, 1862, 81–196. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Abbas, S.Q.; Akhter, N.; Shagufta, B.I.; Shah, S.A.A.; Hassan, S.S.U. Emerging biopharmaceuticals from bioactive peptides derived from marine organisms. Chem. Biol. Drug Des. 2017, 90, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Jo, C.; Khan, F.F.; Khan, M.I.; Iqbal, J. Marine bioactive peptides: Types, structures, and physiological functions. Food Rev. Int. 2016, 33, 44–61. [Google Scholar] [CrossRef]
- He, Y.; Wang, B.; Dukor, R.K.; Nafie, L.A. Determination of absolute configuration of chiral molecules using vibrational optical activity: A review. Appl. Spectrosc. 2011, 65, 699–723. [Google Scholar] [CrossRef] [PubMed]
- Seco, J.M.; Quiñoá, E.; Riguera, R. The assignment of absolute configuration by NMR†. Chem. Rev. 2004, 104, 17–118. [Google Scholar] [CrossRef]
- Hoye, T.R.; Jeffrey, C.S.; Shao, F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007, 2, 2451–2458. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hopmann, K.H.; Hudecova, J.; Isaksson, J.; Novotna, J.; Stensen, W.; Andrushchenko, V.; Urbanova, M.; Svendsen, J.S.; Bour, P.; et al. Determination of absolute configuration and conformation of a cyclic dipeptide by NMR and chiral spectroscopic methods. J. Phys. Chem. A 2013, 117, 1721–1736. [Google Scholar] [CrossRef] [PubMed]
- Berova, N.; Di Bari, L.; Pescitelli, G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 2007, 36, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Toome, V.; De Bernardo, S.; Weigele, M. A simple method for determining the absolute configuration of α-amino acids. Tetrahedron 1975, 31, 2625–2627. [Google Scholar] [CrossRef]
- Urriolabeitia, E.P.; Díaz-de-Villegas, M.D. A simple method for determining the absolute configuration of alpha-amino acids. J. Chem. Ed. 1999, 76, 77. [Google Scholar] [CrossRef]
- Gomez, E.D.; Duddeck, H. A new method proposed for the determination of absolute configurations of alpha-amino acids. Magn. Reson. Chem. 2009, 47, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Calcaterra, A.; D’Acquarica, I. The market of chiral drugs: Chiral switches versus de novo enantiomerically pure compounds. J. Pharm. Biomed. Anal. 2018, 147, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Dolowy, M.; Pyka, A. Application of TLC, HPLC and GC methods to the study of amino acid and peptide enantiomers: A review. Biomed. Chromatogr. 2014, 28, 84–101. [Google Scholar] [CrossRef] [PubMed]
- Marfey, P. Determination ofd-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Vijayasarathy, S.; Prasad, P.; Fremlin, L.J.; Ratnayake, R.; Salim, A.A.; Khalil, Z.; Capon, R.J. C3 and 2D C3 Marfey’s methods for amino acid analysis in natural products. J. Nat. Prod. 2016, 79, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, R.; Bruckner, H. Use of Marfey’s reagent and analogs for chiral amino acid analysis: Assessment and applications to natural products and biological systems. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 3148–3161. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, R.; Bruckner, H. Marfey’s reagent for chiral amino acid analysis: A review. Amino Acids 2004, 27, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Tiritan, M.E.; Ribeiro, A.R.; Fernandes, C.; Pinto, M. Chiral pharmaceuticals. In Kirk-Othmer Encyclopedia of Chemicl Technology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016. [Google Scholar]
- Okamoto, Y.; Ikai, T. Chiral HPLC for efficient resolution of enantiomers. Chem. Soc. Rev. 2008, 37, 2593–2608. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.; Phyo, Y.Z.; Silva, A.S.; Tiritan, M.E.; Kijjoa, A.; Pinto, M.M.M. Chiral stationary phases based on small molecules: An update of the last 17 years. Sep. Purif. Methods 2017, 33, 1–35. [Google Scholar] [CrossRef]
- Ribeiro, J.; Tiritan, M.; Pinto, M.; Fernandes, C. Chiral stationary phases for liquid chromatography based on chitin- and chitosan-derived marine polysaccharides. Symmetry 2017, 9, 190. [Google Scholar] [CrossRef]
- Fernandes, C.; Tiritan, M.E.; Pinto, M. Small molecules as chromatographic tools for hplc enantiomeric resolution: Pirkle-type chiral stationary phases evolution. Chromatographia 2013, 76, 871–897. [Google Scholar] [CrossRef]
- Ribeiro, A.R.; Castro, P.M.L.; Tiritan, M.E. Chiral pharmaceuticals in the environment. Environ. Chem. Lett. 2012, 10, 239–253. [Google Scholar] [CrossRef]
- Li, B.; T Haynie, D. Chiral Drug Separation. In Encyclopedia of Chemical Processing; Taylor & Francis: Boca Raton, FL, USA, 2006. [Google Scholar]
- Auckloo, B.N.; Wu, B. Structure, biological properties and applications of marine-derived polysaccharides. Curr. Org. Chem. 2016, 20, 2002–2012. [Google Scholar] [CrossRef]
- Bennur, T.; Ravi Kumar, A.; Zinjarde, S.S.; Javdekar, V. Nocardiopsis species: A potential source of bioactive compounds. J. Appl. Microbiol. 2016, 120, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ciavatta, M.L.; Lefranc, F.; Carbone, M.; Mollo, E.; Gavagnin, M.; Betancourt, T.; Dasari, R.; Kornienko, A.; Kiss, R. Marine mollusk-derived agents with antiproliferative activity as promising anticancer agents to overcome chemotherapy resistance. Med. Res. Rev. 2017, 37, 702–801. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.Y.; Dahiya, R.; Qin, H.L.; Mourya, R.; Maharaj, S. Natural proline-rich cyclopolypeptides from marine organisms: Chemistry, synthetic methodologies and biological status. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Fernando, I.P.S.; Nah, J.W.; Jeon, Y.J. Potential anti-inflammatory natural products from marine algae. Environ. Toxicol. Pharmacol. 2016, 48, 22–30. [Google Scholar] [CrossRef] [PubMed]
- França, P.H.B.; da Silva-Júnior, E.F.; Santos, B.V.O.; Alexandre-Moreira, M.S.; Quintans-Júnior, L.J.; de Aquino, T.M.; de Araújo-Júnior, J.X. Antileishmanial marine compounds: A review. Rec. Nat. Prod. 2017, 11, 92–113. [Google Scholar]
- Hou, X.M.; Xu, R.F.; Gu, Y.C.; Wang, C.Y.; Shao, C.L. Biological and chemical diversity of coral-derived microorganisms. Curr. Med. Chem. 2015, 22, 3707–3762. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.-Y.; Wang, B.-G. Mycochemistry of marine algicolous fungi. Fungal Divers. 2016, 80, 301–342. [Google Scholar] [CrossRef]
- Jin, Z. Muscarine, imidazole, oxazole and thiazole alkaloids. Nat. Prod. Rep. 2016, 33, 1268–1317. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Yu, W.; Sun, D. Advances in application of marine bioactive peptides in drug development. Chin. J. Org. Chem. 2017, 37, 1681. [Google Scholar] [CrossRef]
- Maciel, E.; Costa Leal, M.; Lillebo, A.I.; Domingues, P.; Domingues, M.R.; Calado, R. Bioprospecting of marine macrophytes using MS-based lipidomics as a new approach. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Oh, G.-W.; Ko, S.-C.; Lee, D.H.; Heo, S.-J.; Jung, W.-K. Biological activities and biomedical potential of sea cucumber (Stichopus japonicus): A review. Fish. Aquat. Sci. 2017, 20. [Google Scholar] [CrossRef]
- Pangestuti, R.; Kim, S.K. Bioactive peptide of marine origin for the prevention and treatment of non-communicable diseases. Mar. Drugs 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Rangel, M.; Santana, C.; Pinheiro, A.; Anjos, L.; Barth, T.; Júnior, O.; Fontes, W.; Castro, M. Marine depsipeptides as promising pharmacotherapeutic agents. Curr. Protein Pept. Sci. 2016, 18, 72–91. [Google Scholar] [CrossRef]
- Shannon, E.; Abu-Ghannam, N. Antibacterial derivatives of marine algae: An overview of pharmacological mechanisms and applications. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S. Therapeutic importance of peptides from marine source: A mini review. Indian J. Geo Mar. Sci. 2016, 45, 1422–1431. [Google Scholar]
- Sila, A.; Bougatef, A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- Suleria, H.A.R.; Gobe, G.; Masci, P.; Osborne, S.A. Marine bioactive compounds and health promoting perspectives; innovation pathways for drug discovery. Trends Food Sci. Technol. 2016, 50, 44–55. [Google Scholar] [CrossRef]
- Wang, L.; Dong, C.; Li, X.; Han, W.; Su, X. Anticancer potential of bioactive peptides from animal sources (Review). Oncol. Rep. 2017, 38, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Grant Burgess, J.; Wright, P.C. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2006, 23, 26–78. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Seo, Y.; Lee, H.S.; Rho, J.R.; Mo, S.J. A new cyclic peptide from a marine-derived bacterium of the genus Nocardiopsis. J. Nat. Prod. 2003, 66, 883–884. [Google Scholar] [CrossRef] [PubMed]
- Grach-Pogrebinsky, O.; Carmeli, S. Three novel anabaenopeptins from the cyanobacterium Anabaena sp. Tetrahedron 2008, 64, 10233–10238. [Google Scholar] [CrossRef]
- Han, B.; Gross, H.; Goeger, D.E.; Mooberry, S.L.; Gerwick, W.H. Aurilides B and C, cancer cell toxins from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2006, 69, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Kanoh, K.; Imagawa, H.; Adachi, K.; Nishizawa, M.; Shizuri, Y. Urukthapelstatin A, a novel cytotoxic substance from marine-derived Mechercharimyces asporophorigenens YM11–542. II. Physico-chemical properties and structural elucidation. J. Antibiot. 2007, 60, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Matthew, S.; Ross, C.; Paul, V.J.; Luesch, H. Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron 2008, 64, 4081–4089. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, H.; Chen, Y.; Tan, J.; Song, Y.; Zou, J.; Tian, X.; Hua, Y.; Ju, J. Marthiapeptide A, an anti-infective and cytotoxic polythiazole cyclopeptide from a 60 L scale fermentation of the deep sea-derived Marinactinospora thermotolerans SCSIO 00652. J. Nat. Prod. 2012, 75, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.C.; Li, S.; Nam, S.J.; Liu, Z.; Zhang, C. Nocardiamides A and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037. J. Nat. Prod. 2013, 76, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, Q.; Liu, X.; Chen, Y.; Zhang, Y.; Sun, A.; Zhang, W.; Zhang, J.; Ju, J. Cyclic Hexapeptides from the deep south china sea-derived streptomyces scopuliridis scsio zj46 active against pathogenic gram-positive bacteria. J. Nat. Prod. 2014, 77, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Iwasaki, A.; Sumimoto, S.; Kanamori, Y.; Ohno, O.; Iwatsuki, M.; Ishiyama, A.; Hokari, R.; Otoguro, K.; Omura, S.; et al. Janadolide, a cyclic polyketide-peptide hybrid possessing a tert-butyl group from an okeania sp. marine cyanobacterium. J. Nat. Prod. 2016, 79, 1862–1866. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.A.; Al-Lihaibi, S.S.; Alarif, W.M.; Abdel-Lateff, A.; Nogata, Y.; Washio, K.; Morikawa, M.; Okino, T. Wewakazole B, a cytotoxic cyanobactin from the cyanobacterium moorea producens collected in the red sea. J. Nat. Prod. 2016, 79, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Davyt, D.; Serra, G. Thiazole and oxazole alkaloids: Isolation and synthesis. Mar. Drugs 2010, 8, 2755–2780. [Google Scholar] [CrossRef] [PubMed]
- Horgen, F.D.; Yoshida, W.Y.; Scheuer, P.J. Malevamides A–C, new depsipeptides from the marine cyanobacterium Symploca laete-viridis. J. Nat. Prod. 2000, 63, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Luesch, H.; Yoshida, W.Y.; Moore, R.E.; Paul, V.J. Isolation and structure of the cytotoxin lyngbyabellin b and absolute configuration of lyngbyapeptin a from the marine cyanobacteriumlyngbya majuscula. J. Nat. Prod. 2000, 63, 1437–1439. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Yoshida, W.Y.; Moore, R.E.; Paul, V.J. Tasipeptins A and B: New cytotoxic depsipeptides from the marine cyanobacterium Symploca sp. J. Nat. Prod. 2003, 66, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Gross, H.; McPhail, K.L.; Goeger, D.; Maier, C.S.; Gerwick, W.H. Wewakamide A and guineamide G, cyclic depsipeptides from the marine cyanobacteria Lyngbya semiplena and Lyngbya majuscula. J. Microbiol. Biotechnol. 2011, 21, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, S.P.; Owle, C.S.; Montaser, R.; Luesch, H.; Paul, V.J. Malyngamide 3 and cocosamides A and B from the marine cyanobacterium Lyngbya majuscula from Cocos Lagoon, Guam. J. Nat. Prod. 2011, 74, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Almaliti, J.; Malloy, K.L.; Glukhov, E.; Spadafora, C.; Gutierrez, M.; Gerwick, W.H. Dudawalamides A-D, antiparasitic cyclic depsipeptides from the marine cyanobacterium moorea producens. J. Nat. Prod. 2017, 80, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Luesch, H.; Pangilinan, R.; Yoshida, W.Y.; Moore, R.E.; Paul, V.J. Pitipeptolides A and B, new cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2001, 64, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Shiota, I.; Sumimoto, S.; Matsubara, T.; Sato, T.; Suenaga, K. Kohamamides A, B, and C, cyclic depsipeptides from an okeania sp. marine cyanobacterium. J. Nat. Prod. 2017, 80, 1948–1952. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Huang, H.; Li, J.; Song, Y.; Jiang, R.; Liu, J.; Zhang, S.; Hua, Y.; Ju, J. New anti-infective cycloheptadepsipeptide congeners and absolute stereochemistry from the deep sea-derived streptomyces drozdowiczii SCSIO 10141. Tetrahedron 2014, 70, 7795–7801. [Google Scholar] [CrossRef]
- Montaser, R.; Abboud, K.A.; Paul, V.J.; Luesch, H. Pitiprolamide, a proline-rich dolastatin 16 analogue from the marine cyanobacterium Lyngbya majuscula from Guam. J. Nat. Prod. 2011, 74, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Yoshida, W.Y.; Quon, M.K.; Moore, R.E.; Paul, V.J. The structure of Palau’amide, a potent cytotoxin from a species of the marine cyanobacterium Lyngbya. J. Nat. Prod. 2003, 66, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Montaser, R.; Paul, V.J.; Luesch, H. Pitipeptolides C-F, antimycobacterial cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula from Guam. Phytochemistry 2011, 72, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.G.; Yoshida, W.Y.; Quon, M.K.; Moore, R.E.; Paul, V.J. Ulongapeptin, a cytotoxic cyclic depsipeptide from a Palauan marine cyanobacterium Lyngbya sp. J. Nat. Prod. 2003, 66, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Plaza, A.; Bewley, C.A. Largamides A–H, unusual cyclic peptides from the marine cyanobacterium Oscillatoria sp. J. Org. Chem. 2006, 71, 6898–6907. [Google Scholar] [CrossRef] [PubMed]
- Bunyajetpong, S.; Yoshida, W.Y.; Sitachitta, N.; Kaya, K. Trungapeptins A–C, cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2006, 69, 1539–1542. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, S.P.; Ritson-Williams, R.; Paul, V.J. Carriebowmide, a new cyclodepsipeptide from the marine cyanobacterium Lyngbya polychroa. J. Nat. Prod. 2008, 71, 2060–2063. [Google Scholar] [CrossRef] [PubMed]
- Linington, R.G.; Edwards, D.J.; Shuman, C.F.; McPhail, K.L.; Matainaho, T.; Gerwick, W.H. Symplocamide A, a potent cytotoxin and chymotrypsin inhibitor from the marine Cyanobacterium Symploca sp. J. Nat. Prod. 2008, 71, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Taori, K.; Paul, V.J.; Luesch, H. Kempopeptins A and B, serine protease inhibitors with different selectivity profiles from a marine cyanobacterium, Lyngbya sp. J. Nat. Prod. 2008, 71, 1625–1629. [Google Scholar] [CrossRef] [PubMed]
- Matthew, S.; Paul, V.J.; Luesch, H. Tiglicamides A–C, cyclodepsipeptides from the marine cyanobacterium Lyngbya confervoides. Phytochemistry 2009, 70, 2058–2063. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Puddick, J.; Prinsep, M.R.; Lee, P.P.; Tan, L.T. Hantupeptins B and C, cytotoxic cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. Phytochemistry 2010, 71, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Nunnery, J.K.; Engene, N.; Esquenazi, E.; Byrum, T.; Dorrestein, P.C.; Gerwick, W.H. Palmyramide A, a cyclic depsipeptide from a Palmyra Atoll collection of the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2010, 73, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Salvador, L.A.; Biggs, J.S.; Paul, V.J.; Luesch, H. Veraguamides A-G, cyclic hexadepsipeptides from a dolastatin 16-producing cyanobacterium Symploca cf. hydnoides from Guam. J. Nat. Prod. 2011, 74, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Meickle, T.; Gunasekera, S.P.; Liu, Y.; Luesch, H.; Paul, V.J. Porpoisamides A and B, two novel epimeric cyclic depsipeptides from a Florida Keys collection of Lyngbya sp. Bioorg. Med. Chem. 2011, 19, 6576–6580. [Google Scholar] [CrossRef] [PubMed]
- Vining, O.B.; Medina, R.A.; Mitchell, E.A.; Videau, P.; Li, D.; Serrill, J.D.; Kelly, J.X.; Gerwick, W.H.; Proteau, P.J.; Ishmael, J.E.; et al. Depsipeptide companeramides from a Panamanian marine cyanobacterium associated with the coibamide producer. J. Nat. Prod. 2015, 78, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.D.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Piperazimycins: Cytotoxic hexadepsipeptides from a marine-derived bacterium of the genus Streptomyces. J. Org. Chem. 2007, 72, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, C.C.; Thimmaiah, M.; Shaala, L.A.; Hau, A.M.; Malmo, J.M.; Ishmael, J.E.; Youssef, D.T.; McPhail, K.L. Cyclic depsipeptides, grassypeptolides D and E and Ibu-epidemethoxylyngbyastatin 3, from a Red Sea Leptolyngbya cyanobacterium. J. Nat. Prod. 2011, 74, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Maloney, K.N.; Nam, S.J.; Haste, N.M.; Raju, R.; Aalbersberg, W.; Jensen, P.R.; Nizet, V.; Hensler, M.E.; Fenical, W. Fijimycins A–C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 6557–6562. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, J.I.; Vansach, T.; Yoshida, W.Y.; Sakamoto, B.; Porzgen, P.; Horgen, F.D. Halogenated fatty acid amides and cyclic depsipeptides from an eastern Caribbean collection of the cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2009, 72, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, P.D.; Byrum, T.; Liu, W.T.; Dorrestein, P.C.; Gerwick, W.H. Viequeamide A, a cytotoxic member of the kulolide superfamily of cyclic depsipeptides from a marine button cyanobacterium. J. Nat. Prod. 2012, 75, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Kjaerulff, L.; Nielsen, A.; Mansson, M.; Gram, L.; Larsen, T.O.; Ingmer, H.; Gotfredsen, C.H. Identification of four new agr quorum sensing-interfering cyclodepsipeptides from a marine Photobacterium. Mar. Drugs 2013, 11, 5051–5062. [Google Scholar] [CrossRef] [PubMed]
- Nogle, L.M.; Okino, T.; Gerwick, W.H. Antillatoxin B, a neurotoxic lipopeptide from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2001, 64, 983–985. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, J.B.; Ernst-Russell, M.A.; de Ropp, J.S.; Molinski, T.F. Lobocyclamides A–C, lipopeptides from a cryptic cyanobacterial mat containing Lyngbya confervoides. J. Org. Chem. 2002, 67, 8210–8215. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290–344. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2014, 31, 160–258. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, J.; Luo, C.; Ding, W.; Cox, D.G. A new cyclopeptide with antifungal activity from the co-culture broth of two marine mangrove fungi. Nat. Prod. Res. 2014, 28, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.T.; Cheng, X.C.; Jensen, P.R.; Fenical, W. Scytalidamides A and B, new cytotoxic cyclic heptapeptides from a marine fungus of the genus Scytalidium. J. Org. Chem. 2003, 68, 8767–8773. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Kanoh, K.; Wisespongp, P.; Nishijima, M.; Shizuri, Y. Clonostachysins A and B, new anti-dinoflagellate cyclic peptides from a marine-derived fungus. J. Antibiot. 2005, 58, 145–150. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Bao, J.; Zhang, X.Y.; Tu, Z.C.; Shi, Y.M.; Qi, S.H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhu, H.; Hong, K.; Wang, Y.; Liu, P.; Wang, X.; Peng, X.; Zhu, W. Novel cyclic hexapeptides from marine-derived fungus, Aspergillus sclerotiorum PT06–1. Org. Lett. 2009, 11, 5262–5265. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Song, Y.; Chen, Y.; Huang, H.; Zhang, W.; Ju, J. Cyclic heptapeptides, cordyheptapeptides C-E, from the marine-derived fungus Acremonium persicinum SCSIO 115 and their cytotoxic activities. J. Nat. Prod. 2012, 75, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Prompanya, C.; Fernandes, C.; Cravo, S.; Pinto, M.M.; Dethoup, T.; Silva, A.M.; Kijjoa, A. A new cyclic hexapeptide and a new isocoumarin derivative from the marine sponge-associated fungus Aspergillus similanensis KUFA 0013. Mar. Drugs 2015, 13, 1432–1450. [Google Scholar] [CrossRef] [PubMed]
- May Zin, W.W.; Buttachon, S.; Dethoup, T.; Fernandes, C.; Cravo, S.; Pinto, M.M.; Gales, L.; Pereira, J.A.; Silva, A.M.; Sekeroglu, N.; et al. New cyclotetrapeptides and a new diketopiperzine derivative from the marine sponge-associated fungus neosartorya glabra KUFA 0702. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, K.M.; Renner, M.K.; Jensen, P.R.; Fenical, W. Exumolides A and B: Antimicroalgal cyclic depsipeptides produced by a marine fungus of the genus Scytalidium. Tetrahedron Lett. 1998, 39, 2463–2466. [Google Scholar] [CrossRef]
- Amagata, T.; Morinaka, B.I.; Amagata, A.; Tenney, K.; Valeriote, F.A.; Lobkovsky, E.; Clardy, J.; Crews, P. A chemical study of cyclic depsipeptides produced by a sponge-derived fungus. J. Nat. Prod. 2006, 69, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Lira, S.P.; Vita-Marques, A.M.; Seleghim, M.H.; Bugni, T.S.; LaBarbera, D.V.; Sette, L.D.; Sponchiado, S.R.; Ireland, C.M.; Berlinck, R.G. New destruxins from the marine-derived fungus Beauveria felina. J. Antibiot. 2006, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-C.; Jensen, P.R.; Fenical, W. Zygosporamide, a cytotoxic cyclic depsipeptide from the marine-derived fungus Zygosporium masonii. Tetrahedron Lett. 2006, 47, 8625–8628. [Google Scholar] [CrossRef]
- Aurelio, L.; Brownlee, R.T.C.; Dang, J.; Hughes, A.B.; Polya, G.M. Determination of the complete absolute configuration of petriellin A. Aust. J. Chem. 2006, 59, 407. [Google Scholar] [CrossRef]
- Kim, M.Y.; Sohn, J.H.; Ahn, J.S.; Oh, H. Alternaramide, a cyclic depsipeptide from the marine-derived fungus Alternaria sp. SF-5016. J. Nat. Prod. 2009, 72, 2065–2068. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Lang, G.; Steffens, S.; Schaumann, K. Petrosifungins A and B, novel cyclodepsipeptides from a sponge-derived strain of Penicillium brevicompactum. J. Nat. Prod. 2004, 67, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Kotoni, D.; Ciogli, A.; D’Acquarica, I.; Kocergin, J.; Szczerba, T.; Ritchie, H.; Villani, C.; Gasparrini, F. Enantioselective ultra-high and high performance liquid chromatography: A comparative study of columns based on the Whelk-O1 selector. J. Chromatogr. A 2012, 1269, 226–241. [Google Scholar] [CrossRef] [PubMed]
- Kotoni, D.; Ciogli, A.; Molinaro, C.; D’Acquarica, I.; Kocergin, J.; Szczerba, T.; Ritchie, H.; Villani, C.; Gasparrini, F. Introducing enantioselective ultrahigh-pressure liquid chromatography (eUHPLC): Theoretical inspections and ultrafast separations on a new sub-2-mum Whelk-O1 stationary phase. Anal. Chem. 2012, 84, 6805–6813. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.J.; Yuan, W.; Liao, X.J.; Han, B.N.; Wang, S.P.; Li, Z.Y.; Xu, S.H.; Zhang, W.; Lin, H.W. Oryzamides A-E, cyclodepsipeptides from the sponge-derived fungus nigrospora oryzae PF18. J. Nat. Prod. 2016, 79, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Kralj, A.; Kehraus, S.; Krick, A.; van Echten-Deckert, G.; Konig, G.M. Two new depsipeptides from the marine fungus Spicellum roseum. Planta Med. 2007, 73, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; She, Z.; Lin, Y.; Vrijmoed, L.L.; Lin, W. Cyclic peptides from an endophytic fungus obtained from a mangrove leaf (Kandelia candel). J. Nat. Prod. 2007, 70, 1696–1699. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.K.; Seo, C.H.; Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 2015, 13, 618–654. [Google Scholar] [CrossRef] [PubMed]
- Kamalakkannan, P. Marine sponges a good source of bioactive compounds in anticancer agents. Int. J. Pharm. Sci. Rev. Res. 2015, 31, 132–135. [Google Scholar]
- Muller, W.E.; Schroder, H.C.; Wiens, M.; Perovic-Ottstadt, S.; Batel, R.; Muller, I.M. Traditional and modern biomedical prospecting: Part II-the Benefits: Approaches for a sustainable exploitation of biodiversity (secondary metabolites and biomaterials from sponges). Evid. Based Complement. Alternat. Med. 2004, 1, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Mehbub, M.F.; Lei, J.; Franco, C.; Zhang, W. Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 2014, 12, 4539–4577. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, A.; Debitus, C.; Gomez-Paloma, L. Haliclamide, a novel cyclic metabolite from the Vanuatu marine sponge Haliclona sp. Tetrahedron 2001, 57, 4443–4446. [Google Scholar] [CrossRef]
- Zhang, X.; Jacob, M.R.; Rao, R.R.; Wang, Y.H.; Agarwal, A.K.; Newman, D.J.; Khan, I.A.; Clark, A.M.; Li, X.C. Antifungal cyclic peptides from the marine sponge Microscleroderma herdmani. Res. Rep. Med. Chem. 2012, 2, 7–14. [Google Scholar] [PubMed]
- Aviles, E.; Rodriguez, A.D. Euryjanicins E-G, poly-phenylalanine and poly-proline cyclic heptapeptides from the Caribbean sponge Prosuberites laughlini. Tetrahedron 2013, 69, 10797–10804. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Jeon, J.E.; Won, T.H.; Sim, C.J.; Oh, D.C.; Oh, K.B.; Shin, J. New cyclic cystine bridged peptides from the sponge Suberites waedoensis. Mar. Drugs 2014, 12, 2760–2770. [Google Scholar] [CrossRef] [PubMed]
- Yeung, B.K.; Nakao, Y.; Kinnel, R.B.; Carney, J.R.; Yoshida, W.Y.; Scheuer, P.J.; Kelly-Borges, M. The kapakahines, cyclic peptides from the marine sponge cribrochalina olemda. J. Org. Chem. 1996, 61, 7168–7173. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Matsunaga, S.; Nakao, Y.; Furihata, K.; West, L.; Faulkner, D.J.; Fusetani, N. Koshikamide B, a cytotoxic peptide lactone from a marine sponge Theonella sp. J. Org. Chem. 2008, 73, 7889–7894. [Google Scholar] [CrossRef] [PubMed]
- Festa, C.; De Marino, S.; Sepe, V.; Monti, M.C.; Luciano, P.; D’Auria, M.V.; Débitus, C.; Bucci, M.; Vellecco, V.; Zampella, A. Perthamides C and D, two new potent anti-inflammatory cyclopeptides from a Solomon Lithistid sponge Theonella swinhoei. Tetrahedron 2009, 65, 10424–10429. [Google Scholar] [CrossRef]
- Festa, C.; De Marino, S.; Sepe, V.; D’Auria, M.V.; Bifulco, G.; Andrés, R.; Terencio, M.C.; Payá, M.; Debitus, C.; Zampella, A. Perthamides C–F, potent human antipsoriatic cyclopeptides. Tetrahedron 2011, 67, 7780–7786. [Google Scholar] [CrossRef]
- Mohammed, R.; Peng, J.; Kelly, M.; Hamann, M.T. Cyclic heptapeptides from the Jamaican sponge Stylissa caribica. J. Nat. Prod. 2006, 69, 1739–1744. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cheng, W.; de Voogd, N.J.; Proksch, P.; Lin, W. Stylissatins B–D, cycloheptapeptides from the marine sponge Stylissa massa. Tetrahedron Lett. 2016, 57, 4288–4292. [Google Scholar] [CrossRef]
- Afifi, A.H.; El-Desoky, A.H.; Kato, H.; Mangindaan, R.E.P.; de Voogd, N.J.; Ammar, N.M.; Hifnawy, M.S.; Tsukamoto, S. Carteritins A and B, cyclic heptapeptides from the marine sponge Stylissa carteri. Tetrahedron Lett. 2016, 57, 1285–1288. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Edrada-Ebel, R.; Mohamed, G.A.; Youssef, D.T.A.; Wray, V.; Proksch, P. Callyaerin G, a new cytotoxic cyclic peptide from the marine sponge Callyspongia aerizusa. Arkivoc 2008, 2008, 164. [Google Scholar] [CrossRef]
- Zhan, K.X.; Jiao, W.H.; Yang, F.; Li, J.; Wang, S.P.; Li, Y.S.; Han, B.N.; Lin, H.W. Reniochalistatins A-E, cyclic peptides from the marine sponge Reniochalina stalagmitis. J. Nat. Prod. 2014, 77, 2678–2684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.J.; Yi, Y.H.; Yang, G.J.; Hu, M.Y.; Cao, G.D.; Yang, F.; Lin, H.W. Proline-containing cyclopeptides from the marine sponge Phakellia fusca. J. Nat. Prod. 2010, 73, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Valeria D’Auria, M.; Zampella, A.; Paloma, L.G.; Minale, L.; Debitus, C.; Roussakis, C.; Le Bert, V. Callipeltins B and C; bioactive peptides from a marine Lithistida sponge Callipelta sp. Tetrahedron 1996, 52, 9589–9596. [Google Scholar] [CrossRef]
- Randazzo, A.; Bifulco, G.; Giannini, C.; Bucci, M.; Debitus, C.; Cirino, G.; Gomez-Paloma, L. Halipeptins A and B: Two novel potent anti-inflammatory cyclic depsipeptides from the Vanuatu marine sponge Haliclona species. J. Am. Chem. Soc. 2001, 123, 10870–10876. [Google Scholar] [CrossRef] [PubMed]
- Capon, R.J.; Ford, J.; Lacey, E.; Gill, J.H.; Heiland, K.; Friedel, T. Phoriospongin A and B: Two new nematocidal depsipeptides from the Australian marine sponges Phoriospongia sp. and Callyspongia bilamellata. J. Nat. Prod. 2002, 65, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Plaza, A.; Gustchina, E.; Baker, H.L.; Kelly, M.; Bewley, C.A. Mirabamides A-D, depsipeptides from the sponge Siliquariaspongia mirabilis that inhibit HIV-1 fusion. J. Nat. Prod. 2007, 70, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.D.; Pham, N.B.; Fechner, G.; Zencak, D.; Vu, H.T.; Hooper, J.N.; Quinn, R.J. Cytotoxic cyclic depsipeptides from the Australian marine sponge Neamphius huxleyi. J. Nat. Prod. 2012, 75, 2200–2208. [Google Scholar] [CrossRef] [PubMed]
- Coello, L.; Reyes, F.; Martin, M.J.; Cuevas, C.; Fernandez, R. Isolation and structures of pipecolidepsins A and B, cytotoxic cyclic depsipeptides from the Madagascan sponge Homophymia lamellosa. J. Nat. Prod. 2014, 77, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Rodriguez-Acebes, R.; Garcia-Ramos, Y.; Martinez, V.; Murcia, C.; Digon, I.; Marco, I.; Pelay-Gimeno, M.; Fernandez, R.; Reyes, F.; et al. Stellatolides, a new cyclodepsipeptide family from the sponge Ecionemia acervus: Isolation, solid-phase total synthesis, and full structural assignment of stellatolide A. J. Am. Chem. Soc. 2014, 136, 6754–6762. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.P.; Carroll, J.; Naylor, S.; Crews, P. An Antifungal Cyclodepsipeptide, Cyclolithistide A, from the SpongeTheonella swinhoei. J. Org. Chem. 1998, 63, 8757–8764. [Google Scholar] [CrossRef]
- Okada, Y.; Matsunaga, S.; van Soest, R.W.; Fusetani, N. Nagahamide A, an antibacterial depsipeptide from the marine sponge Theonella swinhoei. Org. Lett. 2002, 4, 3039–3042. [Google Scholar] [CrossRef] [PubMed]
- Plaza, A.; Bifulco, G.; Keffer, J.L.; Lloyd, J.R.; Baker, H.L.; Bewley, C.A. Celebesides A–C and theopapuamides B-D, depsipeptides from an Indonesian sponge that inhibit HIV-1 entry. J. Org. Chem. 2009, 74, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Ratnayake, A.S.; Bugni, T.S.; Feng, X.; Harper, M.K.; Skalicky, J.J.; Mohammed, K.A.; Andjelic, C.D.; Barrows, L.R.; Ireland, C.M. Theopapuamide, a cyclic depsipeptide from a Papua New Guinea lithistid sponge Theonella swinhoei. J. Nat. Prod. 2006, 69, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Plaza, A.; Bifulco, G.; Masullo, M.; Lloyd, J.R.; Keffer, J.L.; Colin, P.L.; Hooper, J.N.; Bell, L.J.; Bewley, C.A. Mutremdamide A and koshikamides C-H, peptide inhibitors of HIV-1 entry from different Theonella species. J. Org. Chem. 2010, 75, 4344–4355. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.C.; Wakimoto, T.; Abe, I. Sulfoureido lipopeptides from the marine sponge discodermia kiiensis. J. Nat. Prod. 2016, 79, 2418–2422. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, M.T.; Vale, C.; Rauter, A.P. Halogenated compounds from marine algae. Mar. Drugs 2010, 8, 2301–2317. [Google Scholar] [CrossRef] [PubMed]
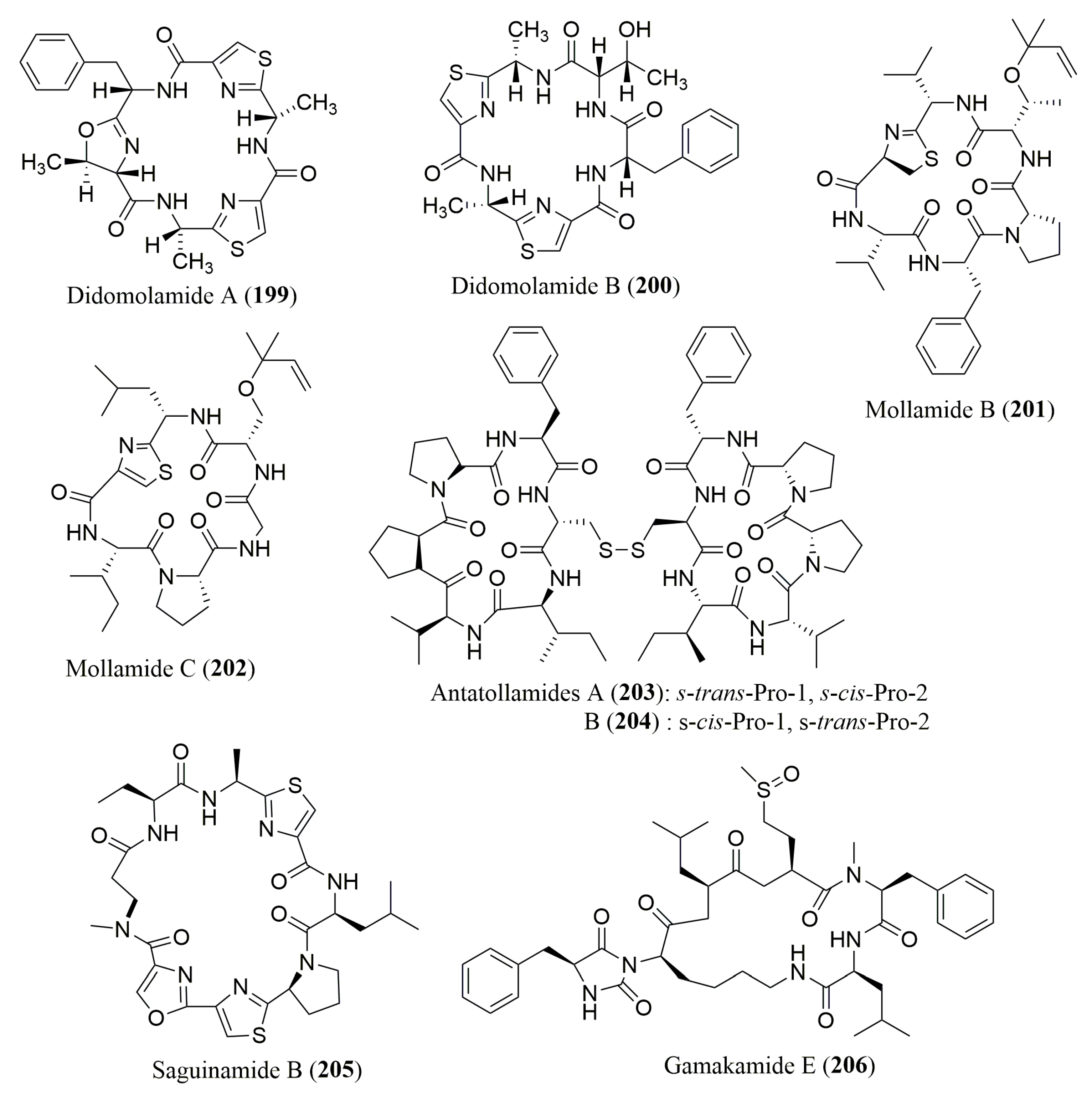
- Rudi, A.; Chill, L.; Aknin, M.; Kashman, Y. Didmolamide A and B, two new cyclic hexapeptides from the marine ascidian Didemnum molle. J. Nat. Prod. 2003, 66, 575–577. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.S.; Wang, B.; Dunbar, D.C.; Desai, P.V.; Patny, A.; Avery, M.; Hamann, M.T. Mollamides B and C, Cyclic hexapeptides from the indonesian tunicate Didemnum molle. J. Nat. Prod. 2008, 71, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Salib, M.N.; Molinski, T.F. Cyclic Hexapeptide Dimers, Antatollamides A and B, from the Ascidian Didemnum molle. A Tryptophan-Derived Auxiliary for l- and d-Amino Acid Assignments. J. Org. Chem. 2017, 82, 10181–10187. [Google Scholar] [CrossRef] [PubMed]
- Dalisay, D.S.; Rogers, E.W.; Edison, A.S.; Molinski, T.F. Structure elucidation at the nanomole scale. 1. Trisoxazole macrolides and thiazole-containing cyclic peptides from the nudibranch Hexabranchus sanguineus. J. Nat. Prod. 2009, 72, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Satake, M.; Horigome, Y.; Oshima, Y.; Yasumoto, T. Gamakamide-E, a strongly bitter tasting cyclic peptide with a hydantoin structure from cultured oysters crassostrea gigas. Fish. Aquatic. Sci. 2012, 15, 15–19. [Google Scholar] [CrossRef]
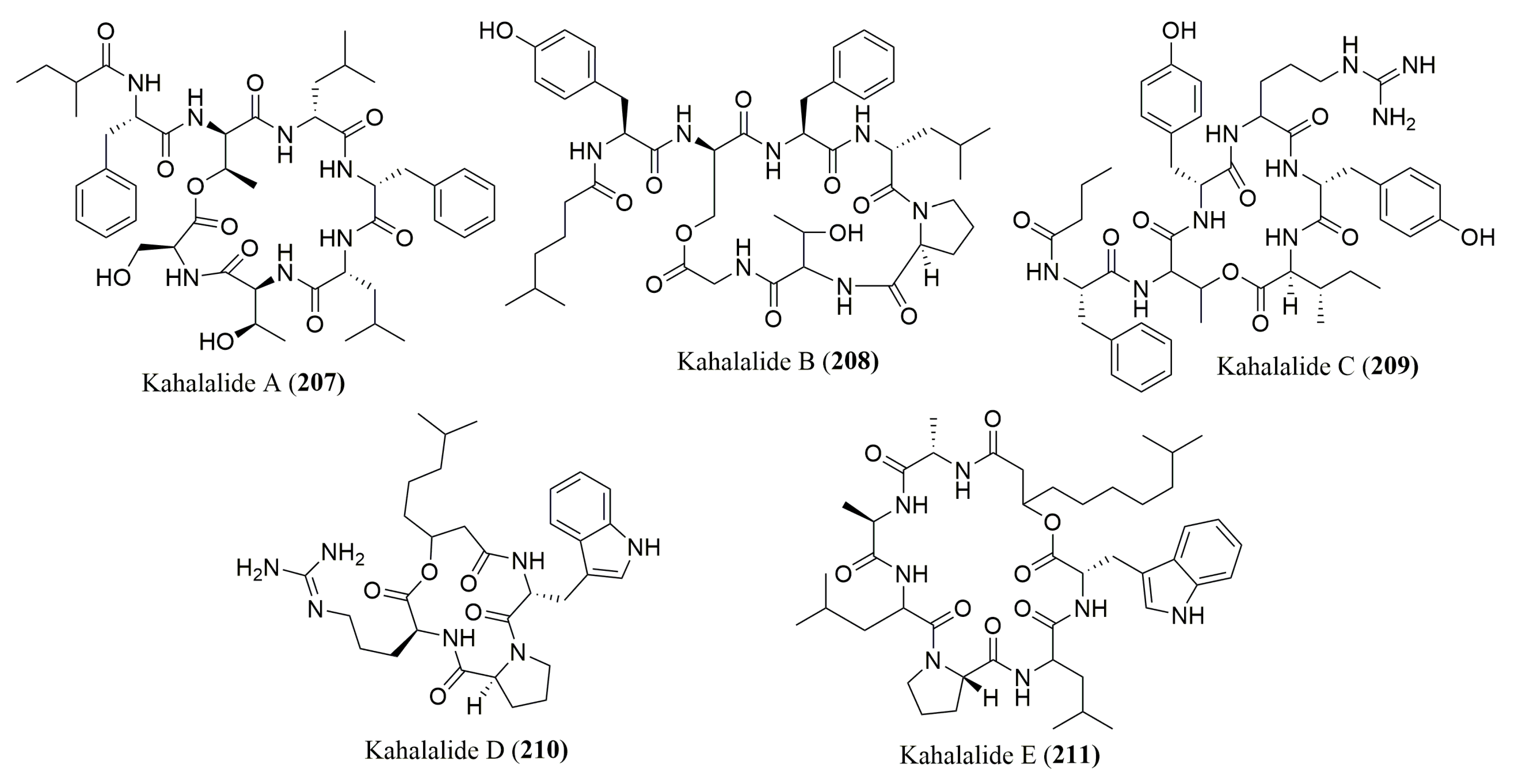
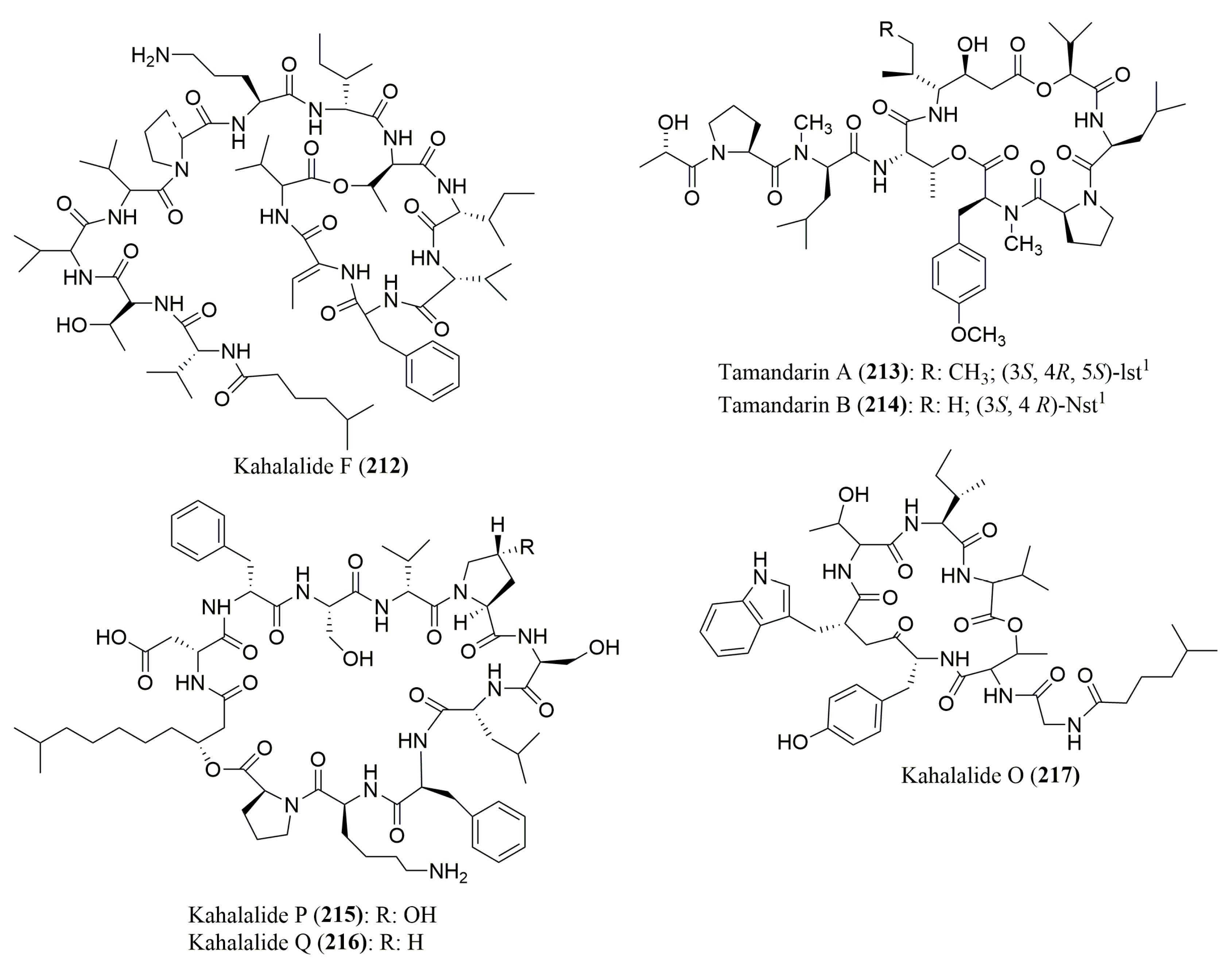
- Goetz, G.; Yoshida, W.Y.; Scheuer, P.J. The absolute stereochemistry of Kahalalide F. Tetrahedron 1999, 55, 7739–7746. [Google Scholar] [CrossRef]
- Vervoort, H.; Fenical, W.; Epifanio, R.A. Tamandarins A and B: New cytotoxic depsipeptides from a Brazilian ascidian of the family Didemnidae. J. Org. Chem. 2000, 65, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Dmitrenok, A.; Iwashita, T.; Nakajima, T.; Sakamoto, B.; Namikoshi, M.; Nagai, H. New cyclic depsipeptides from the green alga Bryopsis species; application of a carboxypeptidase hydrolysis reaction to the structure determination. Tetrahedron 2006, 62, 1301–1308. [Google Scholar] [CrossRef]
- Horgen, F.D.; delos Santos, D.B.; Goetz, G.; Sakamoto, B.; Kan, Y.; Nagai, H.; Scheuer, P.J. A new depsipeptide from the sacoglossan mollusk Elysia ornata and the green alga Bryopsis species. J. Nat. Prod. 2000, 63, 152–154. [Google Scholar] [CrossRef] [PubMed]
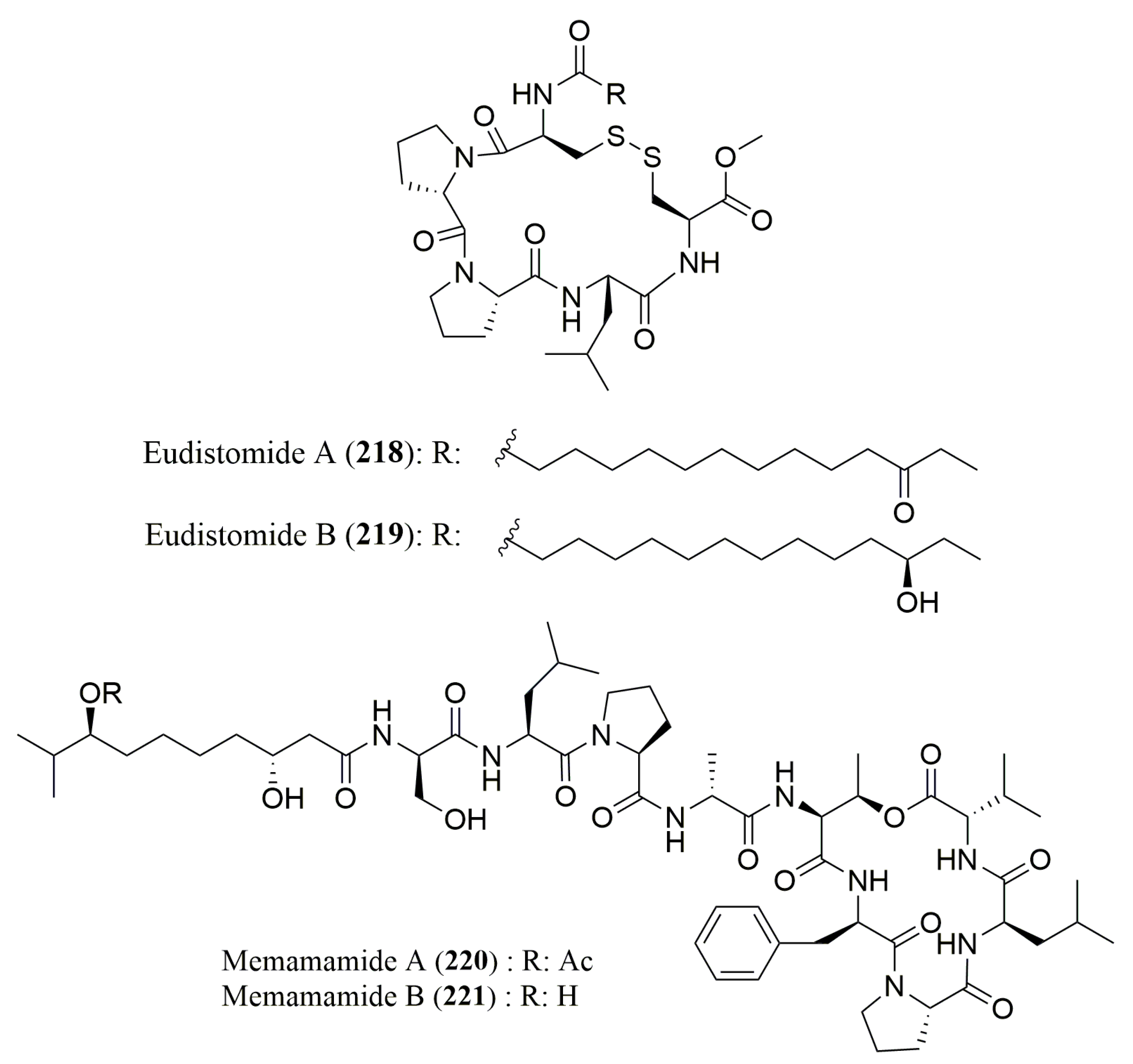
- Whitson, E.L.; Ratnayake, A.S.; Bugni, T.S.; Harper, M.K.; Ireland, C.M. Isolation, structure elucidation, and synthesis of eudistomides A and B, lipopeptides from a Fijian ascidian Eudistoma sp. J. Org. Chem. 2009, 74, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Ohno, O.; Sumimoto, S.; Matsubara, T.; Shimada, S.; Sato, T.; Suenaga, K. Mebamamides A and B, Cyclic Lipopeptides Isolated from the Green Alga Derbesia marina. J. Nat. Prod. 2015, 78, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Berthod, A.; Liu, Y.; Bagwill, C.; Armstrong, D.W. Facile liquid chromatographic enantioresolution of native amino acids and peptides using a teicoplanin chiral stationary phase. J. Chromatogr. A 1996, 731, 123–137. [Google Scholar] [CrossRef]
- Péter, A.; Török, G.; Armstrong, D. High-performance liquid chromatographic separation of enantiomers of unusual amino acids on a teicoplanin stationary phase. J. Chromatogr. A 1998, 793, 283–296. [Google Scholar] [CrossRef]

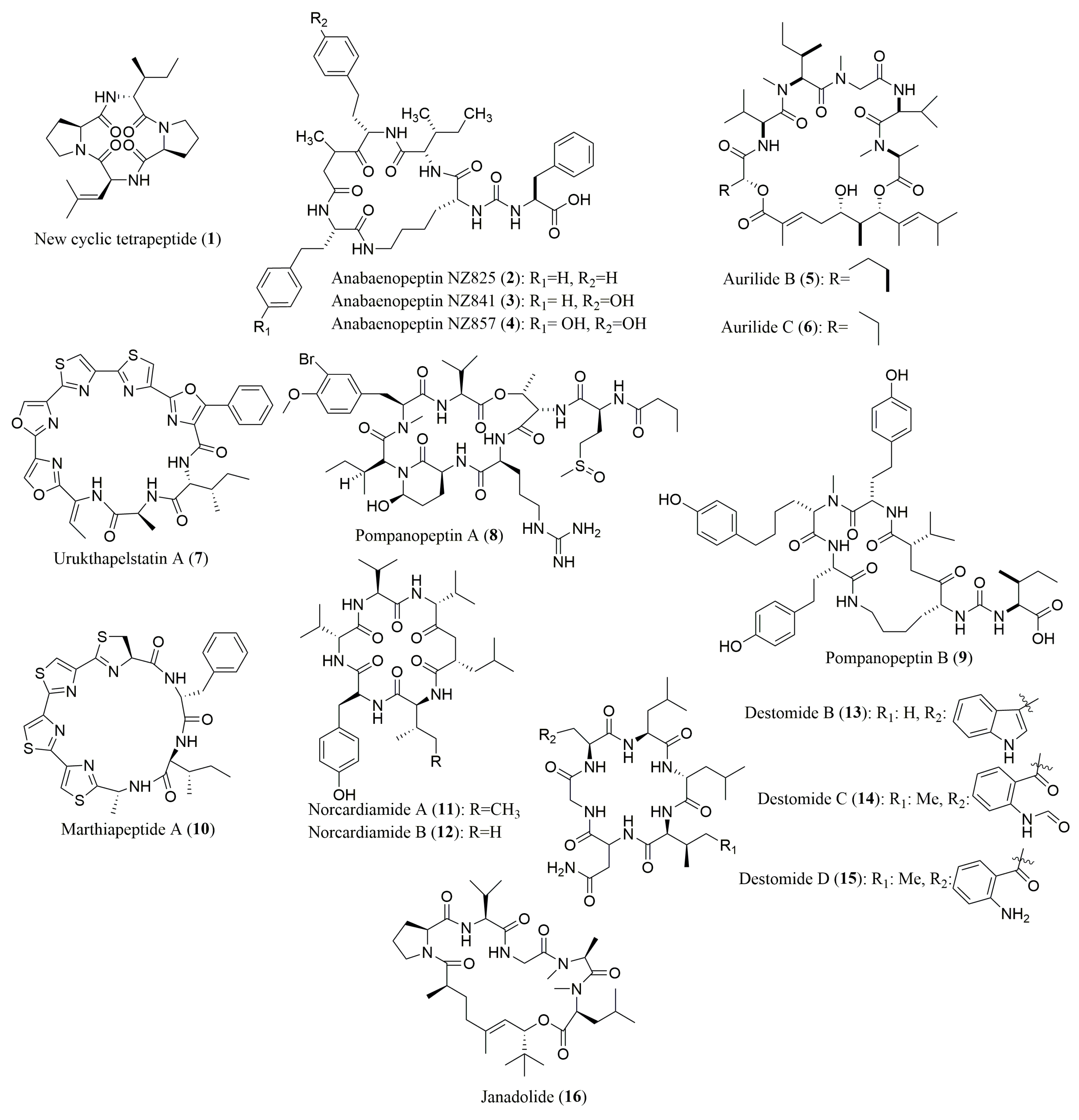






| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Refs. |
|---|---|---|---|---|---|
| Tetrapeptide (1) | Bacterium Nocardiopsis sp. | l-Ile, l-Leu, l-Pro | Marfey’s method (FDAA) combined with HPLC C18 (YMC-ODS-A) (4.5 × 250 mm) Flow rate: 0.8 mL/min; UV detection at 340 nm MP: ACN(aq) (0–50% (v/v)) with 0.1% TFA | Cytotoxicity toward the leukemia cell-line K-562 | [60] |
| Anabaenopeptins NZ825, NZ841, NZ857 (2–4) | Cyanobacterium Anabaena sp. | l-Ile, d-Lys, l-Phe; 2: l-Hph; 3: l-Hph, l-Hty; 4: l-Hty | Marfey’s method (FDAA) combined with HPLC Merck Chromolith performance RP-18e, (4.6 × 100 mm) MP: 50 mM TEAP buffer (pH 3)/ACN (9:1 to 1:1 v/v) | No inhibition of serine proteases | [61] |
| Aurilides B (5) and C (6) | Cyanobacterium Lyngbya majuscula | l-Val, N-Me-l-Ile, l-Ile | Ligand Exchange Type CSP; Phenomenex Chirex 3126 (D) (4.6 × 250 mm); Flow rate: 1.0 mL/min; UV detection at 254 nm; MP: 2 mM CuSO4 in ACN/H2O (5/95 v/v) or 2 mM CuSO4 in ACN/H2O (15/85 v/v) | Cytotoxicity against NCl-H460 and neuro-2a mouse neuroblastoma cell lines 5: also active against leukemia, renal, and prostate cancer cell lines | [62] |
| N-Me-l-Ala 6: N-Me-l-allo-Ile, d-Hiva | Marfey’s method (FDAA) combined with HPLC Microsob-MV C18 (4.6 × 250 mm) Flow rate: 1.0 mL/min; UV detection at 254 nm MP: 50 mM TEAP buffer pH 3/ACN (9:1 to 1:1 v/v) | ||||
| Urukthapelstatin A (7) | Marine Derived Mechercharimyces asporophorigenens YM11-542 | l-Ala | Marfey’s method (FDAA) combined with HPLC ODS-80Ts column (4.6 × 150 mm) Flow rate:1.0 mL/min; UV detection at 340 nm MP: MeOH, 0.1% TFA containing ACN or H2O | Growth inhibition of human lung cancer A549 cells, cytotoxicity against a human cancer cell line panel | [63,70] |
| d-allo-Ile | Ligand Exchange Type CSPSumichiral OA-5000 column (4.6 × 150 mm) Flow rate: 1.0 mL/min; UV detection at 254 nm MP: 5% IPA containing 2 mM CuSO4 | ||||
| Pompanopeptins A (8) and B (9) | Cyanobacterium Lyngbya confervoides | 8: l-Val, l-Thr, l-Met (O), S-Ahp, l-Ile, l-Arg 9: l-Ile | Ligand Exchange Type CSP; Phenomenex Chirex 3126 N,S-dioctyl-(d)-penicillamine, 5 µm (4.6 × 250 mm) Flow rate: 1.0 mL/min; UV detection at 254 nm MP: 2 mM CuSO4 or 2 mM CuSO4/ACN (95:5 v/v) | 8: Trypsin inhibitory activity | [64] |
| 8: N,O-diMe-Br-l-Tyr | Marfey’s method (FDLA) combined with HPLC-MS Phenomenex Synergi 4u Hydro RP 80A (2 × 340 nm) Flow rate: 0.15 mL/min; UV detection at 254 nm MP: ACN/HCOOH (10–90:0.1 v/v) in gradient | ||||
| 9: d-Lys, l-Val, d-Glu | Marfey’s method (FDLA) combined with HPLC-MS Alltech Altima HP C18 HL 54 (250 × 4.6 mm) Flow rate: 1.0 mL/min; PDA detection from 200–500 nm MP: ACN/aq TFA (30–70:0.1 v/v) in gradient | ||||
| Marthiapeptide A (10) | Deep sea-derived Marinactinospora thermotolerans SCSIO 00652 | l-Ile | Ligand Exchange Type CSP; MCIGELCR10W (4.6 × 150 mm); Flow rate: 0.5 mL/min; UV detection at 254 nm; MP: 2 mM CuSO4 solution | Antibacterial and cytotoxic activities | [65] |
| d-Phe, l-Ile | Marfey’s method (FDAA) combined with HPLC Zorbax SB-C8 column, 5 μm (2.1 × 30 mm) | ||||
| Nocardiamides A (11) and B (12) | Marine-derived Actinomycete Nocardiopsis sp. CNX037 | l-Tyr, d-Leu, d- and l-Val | Marfey’s method (FDAA or FDLA) combined with HPLC; Conditions not described | Antimicrobial activity and no cytotoxicity against HCT-116 cell line | [66] |
| 11: l-Ile | Ligand Exchange Type CSP; MCIGELCRS10W, (4.6 × 250 mm); Flow rate: 0.5 mL/min; UV detection at 254 nm; MP: 2 mM CuSO4/H2O | ||||
| Destomides B–D (13–15) | Deep sea-derived Streptomyces scopuliridis SCSIO ZJ46 | l-Asn, d-Leu 13: l-Trp, l-Val, l-Leu; 14: l-Gly, l-Ile, 15: l-Gly, l-Ile, l-Leu | Marfey’s method (FDAA) combined with HPLC Phenomenex ODS column, 5 µm (4.6 × 150 mm) Flow rate: 1.0 mL/min; UV detection at 340 nm MP: ACN:H2O:TFA (15:85:0.1 to 90:10:0.1) | 13: Antimicrobial activity against staphylococcus aureus ATCC 29213, Streptococcus pneumoniae NCTC 7466 and MRSE shhs-E1 13–15: no cytotoxicity | [67] |
| 15: l-Kyn | Ligand Exchange Type CSP; MCIGELCRS10W column, 3 µm (4.6 × 50 mm); Flow rate: 0.5 mL/min; UV detection at 254 nm; MP: 2 mM CuSO4 aqueous solution | ||||
| Janadolide (16) | Cyanobacterium Okeania sp. | N-Me-l-Leu, l-Pro, l-Val | Ligand Exchange Type CSP; Diacel CHIRALPAK (MA+) (4.6 × 50 mm); Flow rate: 1.0 mL/min; UV detection at 254 nm; MP: 2.0 mM CuSO4 | Antitrypanosomal activity | [68] |
| N-Me-l-Ala | Marfey’s method (FDAA) combined with HPLC Cosmosil Cholester (4.6 × 50 mm); Flow rate: 1.0 mL/min; UV detection at 340 nm MP: 0.02 M NaOAc(aq)/MeOH (45/55 v/v) | ||||
| Wewakazole B (17) | Cyanobacterium Moorea producens | l-Ala, l-Phe, l-Pro | Macrocyclic Antibiotic type CSP Chirobiotic TAG (2.1 × 250 mm); Flow rate: 0.3 mL/min; UV detection at 340 nm; MP: 0.1% aq. HCOOH and 1% (v/v) NH4OAc in MeOH | Cytotoxicity against MCF7 and human 460 lung cancer cell lines | [69] |
| l-Ile | Ligand Exchange type CSP; Sumichiral OA-5000 (4.6 × 150 mm); Flow rate: 1.0 mL/min; UV Detection at 254 nm; MP: MeOH/2.0 mM CuSO4 in H2O (5/95 v/v) |
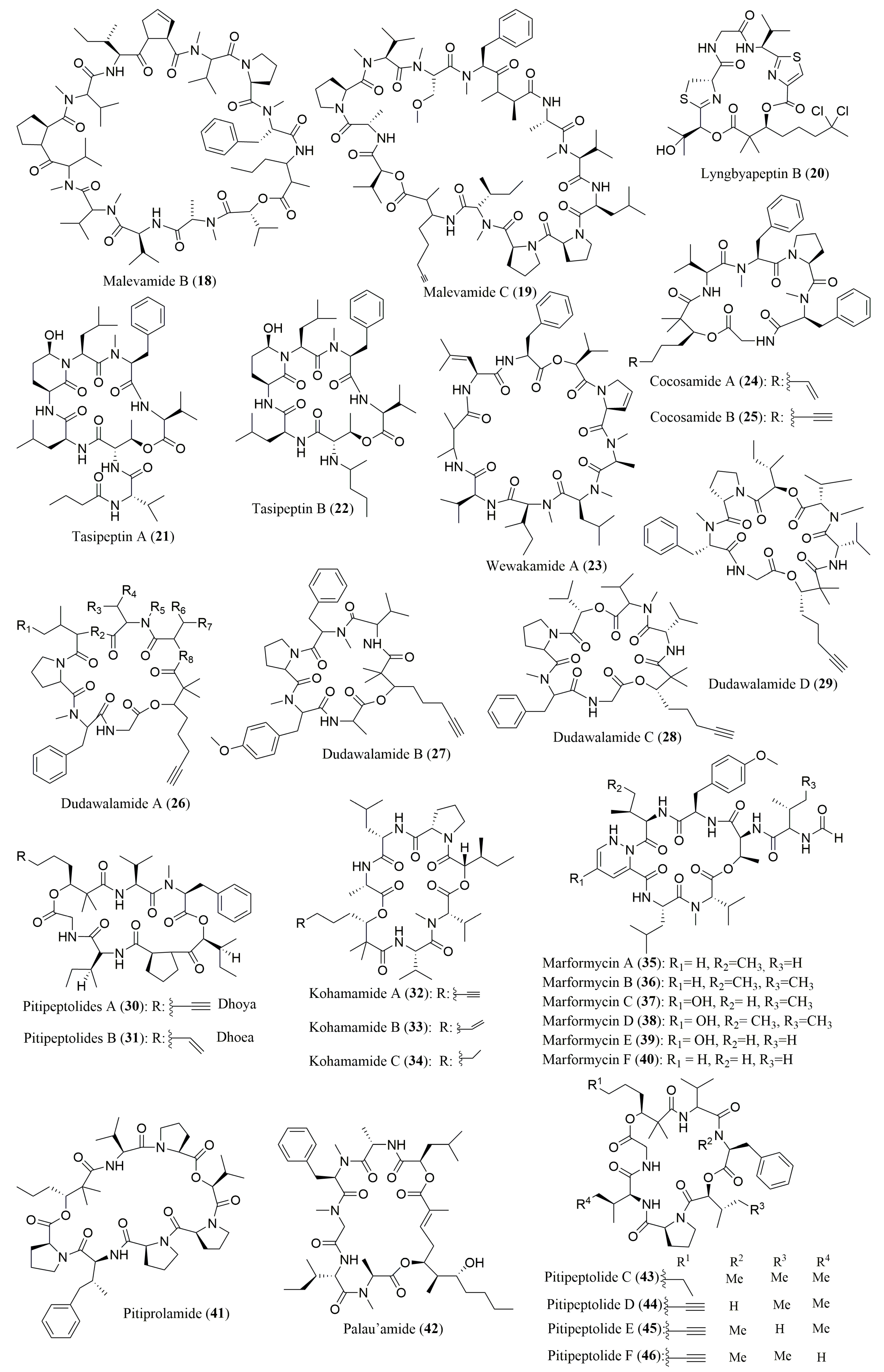
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Refs. |
|---|---|---|---|---|---|
| Malevamides B (18) and C (19) | Cyanobacterium Symploca laete-viridis | l-Pro, N-Me-l-Val, N-Me-l-Phe 18: l-Ile, N-Me-l-Ala, N-Me-d-Val, l-Val, (R)-Hiva; 19: l-Ala, N-diMe-l-Ser, l-Leu, N-Me-d-Ala, N-Me-l-Ile, (S)-Hiva | Ligand Exchange Type CSP; Chirex (D) Penicillamine, Phenomenex 00G-3126E0 (4.6 × 250 mm) MP: 1.7 mM CuSO4 in ACN/H2O (14:86 v/v), 1.9 mM CuSO4 in ACN/H2O (5:95 v/v) or 2.0 mM CuSO4 in H2O Flow rate: 1.0 and 0.8 mL/min; UV detection at 245 nm | Inactive against P-388, A-549 and HT-29 cancer cells | [71] |
| Lyngbyapeptin B (20) | Cyanobacterium Lyngbya majuscula | N-Me-l-Ile, N-Me-l-Leu, N,O-diMe-l-Tyr | Ligand Exchange Type CSP; Chirex (D) Penicillamine, Phenomenex 00G-3126E0 (4.6 × 250 mm) MP: 2 mM CuSO4 Flow: 0.8 mL/min; UV detection at 254 nm | Cytotoxicity against KB and LoVo cells | [72] |
| Tasipeptins A (21) and B (22) | Cyanobacterium Symploca sp. | l-Thr, l-Val, l-Leu, l-Glu, N-Me-l-Phe | Ligand Exchange Type CSP; Phenomenex Chirex Phase 3126 (D) (4.6 × 250 mm) MP: 2 mM CuSO4; 2 mM CuSO4/ACN (95:5 or 85:15 v/v) UV detection at 254 nm | Cytotoxicity toward KB cells | [73] |
| Wewakamide A (23) | Cyanobacteria Lyngbya semiplena and Lyngbya majuscula | l-M-Ala, l-Pro, l-Val, l-Me-Leu, l-Phe, l-Me-ILe, l-Hiv | Ligand Exchange Type CSP; Phenomenex Chirex 3126 (D) (4.6 × 250 mm); MP: 2 mM CuSO4 in H2O or 2 mM CuSO4 in ACN/H2O (15:85 or 5:95 v/v) Flow rate: 0.7, 0.8, 1.0 mL/min; UV detection at 254 nm | Brine shrimp toxicity | [74] |
| Cocosamide A (24) and B (25) | Cyanobacterium Lyngbya majuscula | l-Pro, l-Val, N-Me-l-Phe | Ligand Exchange Type CSP; Phenomenex Chirex (D), Penicillamine, 5 µm (4.6 × 250 mm) MP: 2.0 mM CuSO4/ACN (85:15 or 90:10 v/v) Flow rate: 1.0 mL/min; UV detection at 254 nm | Cytotoxicity against MCF-7 (breast cancer) and HT-29 (colon cancer) cells | [75] |
| Dudawalamides A–D (26–29) | Cyanobacterium Moorea producens | l-Dhoya, l-Hiva, l-Val 29: d-allo-Hiva | Ligand Exchange Type CSP; Chirex Phase 3126 (D) 5 µm (4.6 × 250 mm); MP: 2 mM CuSO4-ACN (95:5 or 85:15 v/v or 87.5:12.5 v/v/v), ACN-H2O-HCOOH (30:70:0.1 or 70:30:0.1 v/v/v) Flow rate: 0.8 mL/min; UV detection at 340 nm | Antiparasitic activity | [76] |
| Pitipeptolides A (30) and B (31) | Cyanobacterium Lyngbya majuscula | l-Gly, l-Pro, l-Val, l-Ile, N-Me-l-Phe, (2S,3S)-Hmp | Ligand Exchange Type CSP; Chiralpak MA (+) (4.6 × 50 mm); MP: 2 mM CuSO4: ACN (90:10 or 85:15 v/v) Flow rate: 1.0 mL/min; UV detection at 254 nm | Cytotoxic, antimycobacterial and elastase inhibitory activities | [77] |
| Kohamamides A–C (32–34) | Cyanobacterium Okeania sp. | l-Pro, l-Ala, l-Val, N-Me-l-Val, l-Leu; 32: l-Ile | Ligand Exchange Type CSP; Chiralpak MA (+) (4.6 × 250 mm); MP: 2 mM CuSO4, ACN: 2 mM CuSO4 (15:85 v/v); Flow rate: 1.0 mL/min; UV detection at 254 nm | No growth inhibition against HeLa and HL60 cells | [78] |
| Marformycins A–F (35–40) | Deep sea-derived Streptomyces drozdowiczii | 35: d-allo-Ile, l-Val; 36: d-allo-Ile, l-allo-Ile; 37: d-Val, l-allo-Ile; 38: d-allo-Ile, l-allo-Ile, l-Leu; 39 and 40: l-Thr, l-Val, d-Val, l-Leu | Ligand Exchange Type CSP; MCIGELCRS10W (4.6 × 50 mm); MP: 2 mM CuSO4 in H2O Flow rate: 0.5 mL/min; UV detection at 254 nm | Anti-infective activity against Micrococcus luteus | [79] |
| Pitiprolamide (41) | Cyanobacterium Lyngbya majuscula | l-Pro, l-Val | Macrocyclic Antibiotic Type CSP; Chirobiotic TAG (4.6 × 250 mm); MP: MeOH/10 mM NH4OAc (40:60 v/v) (pH 5.6) Flow rate: 0.5 mL/min | Cytotoxicity against CT116 and MCF7 cancer cell lines and antibacterial activity | [80] |
| Palau’amide (42) | Cyanobacterium Lyngbya sp. | l-Ala, l-Ile, N-Me-l-Ala, N-Me-d-Phe and d-hydroxyisocaproic acid | Ligand Exchange Type CSP; Phenomenex Chirex Phase 3126 (D) (4.6 × 250 mm) MP: 1 mM CuSO4; 2 mM CuSO4/ACN (95:5 or 85:15 v/v) Flow rate: 0.8 mL/min; UV detection at 254 nm | Cytotoxicity against KB cell line | [81] |
| Pitipeptolides C–F (43–46) | Cyanobacterium Lyngbya majuscula | l-Pro, l-Val, l-Ile, l-Phe, N-Me-l-Phe | Macrocyclic Antibiotic Type CSP; Chirobiotic TAG (4.6 × 250 mm); MP: MeOH/10 mM NH4OAc (40:60 v/v) (pH 5.6); Flow rate: 0.5 mL/min Detection by EIMS in positive ion mode (MRM scan) | 46: Active against Mycobacterium tuberculosis | [82] |
| Ulongapeptin (47) | Cyanobacterium Lyngbya sp. | l-lactic acid, l-Val, N-Me-l-Val, N-Me-d-Val, N-Me-d-Phe | Ligand Exchange Type CSP; Phenomenex Chirex Phase 3126 (D), 4.6 × 250 mm MP: 2 mM CuSO4; 2 mM CuSO4/ACN (95:5 or 85:15 v/v) Flow rate: 1.00 mL/min; UV detection at 254 nm | Cytotoxicity against KB cells | [83] |
| l-Val, N-Me-l-Val, N-Me-d-Val | Marfey’s method (FDLA) combined with HPLC YMC-Pack AQ-ODS (10 × 250 mm); MP: 50% ACN in 0.01 N TFA Flow rate: 2.5 mL/min; UV detection at 254 nm | ||||
| 2-hydroxy-3-methylvaleric acid N-Me-l-Ala | Ligand Exchange Type CSP; CHIRALPAK MA (+) (4.6 × 50 mm); MP: 1 mM CuSO4; 2 mM CuSO4/ACN (95:5 or 85:15 v/v) Flow rate: 0.7 mL/min; UV detection at 254 nm | ||||
| Largamides A–H (48–55) | Cyanobacterium Oscillatoria sp. | 48: l-Val, l-Thr, l-Ala, l-Leu, d-Gln, d-Tyr; 49: l-Val, l-Thr, l-Ala, l-Ahppa, d-Gln, d-Tyr; 50: l-Val, l-Thr, l-Ala, l-Ahpha, d-Gln, d-Tyr; 51: l-Val, l-Thr, l-Ala, l-Leu, l-Ahp, N-MeBr-l-Tyr, l-Ahppa; 52: l-Val, l-Thr, l-Ala, l-Leu, l-Ahp, N-MeCl-l-Tyr; 53: l-Val, l-Thr, l-Ala, l-Tyr, l-Ahp, N-MeCl-l-Tyr; 54: l-Val, l-Thr, l-Ala, l-hTyr, l-Ahp, N-MeCl-l-Tyr; 55: l-Val, l-Thr, l-Ala, l-Amppa, l-Gln, N-Me-l-Asn | Marfey’s method (FDLA) combined with HPLC Phenomenex Jupiter Proteo C12 column, 4 µm (4.6 × 150 mm); MP: ACN containing 0.01 M TFA Flow 0.5 mL/min; UV detection at 254 nm | 51–54: Chymotrypsin inhibition | [84] |
| d-Glyceric acid | Ligand Exchange Type CSP; Phenomenex Chirex 3126 (D) (4.6 × 150 mm); MP: 2 mM CuSO4:ACN (90/10 v/v); Flow 0.5 mL/min; UV detection at 254 nm | ||||
| Trungapeptins A–C (56–58) | Cyanobacterium Lyngbya majuscula | l-Val, l-N-MeVal, l-alloLeu, l-Pro | Marfey’s method (FDLA) combined with HPLC. Alltech Econosil C18; MP A:40% ACN with 0.04%TFA. MP B: 37.5% ACN with 0.05%TFA. Flow rate: 1.0 mL/min; UV detection at 254 nm | Brine shrimp toxicity and ichthyotoxicity | [85] |
| Phenyllactic acid (S) | Ligand Exchange Type CSP; CHIRALPAK MA (+) (4.6 × 50 mm); MP: 2 mM CuSO4/ACN (85:15) Flow rate: 0.5 mL/min; UV detection at 254 nm | ||||
| Carriebowmide (59) | Cyanbacterium Lyngbya polychroa | l-Ala, N-Me-l-Leu, N-Me-d-Phe, l-Phe, l-Met | Ligand Exchange Type CSP; Phenomenex, Chirex (D) Penicillamine, 5μm (4.6 × 250 mm) MP: 2.0 mM CuSO4-ACN (95:5, 90:10, or 85:15 v/v) Flow rate: 0.8 or 1.0 mL/min; UV detection at 254 nm | Lipophilic extract reduced feeding on agar food pellets | [86] |
| R-Hmba | Ligand Exchange Type CSP; Chiralpak MA (+) (4.6 × 250 mm); MP: 2.0 mM CuSO4-ACN (90:10 v/v) Flow rate:1.0 mL/min; UV detection at 254 nm | ||||
| l-Aba | Ligand Exchange Type CSP; Phenomenex, Chirex (D) Penicillamine, 5 μm (4.6 × 250 mm); MP: 2.0 mM CuSO4 Flow rate:1.0 mL/min; UV detection at 254 nm | ||||
| (2R,3R)-Amha | Marfey’s method (FDAA) combined with HPLC Atlantis, C18, (3.0 × 250 mm); MP: 50 mM NH4COOCH3(aq)-ACN (70:30 v/v) Flow rate: 1.0 mL/min; UV detection at 254 nm | ||||
| Symplocamide A (60) | Cyanobacterium Symploca sp. | l-Val, l-Thr, l-Ile, l-Cit, l-Gln, l-Btyr, l-But | Marfey’s method (FDAA) combined with HPLC Phenomenex Jupiter C18 column (4.6 × 250 mm) MP: ACN:H2O:HOAc (15:85:0.02 to 1:1:0.02 v/v/v) Flow rate: 0.5 mL/min; UV detection at 340 nm | Cytotoxicity and antimicrobial activities Chymotrypsin inhibitor | [87] |
| Kempopeptins A (61) and B (62) | Cyanobacterium Lyngbya sp. | 61: N-O-diMe-Br-l-Tyr | Marfey’s method (FDLA) combined with HPLC Conditions not described | 61: Elastase and chymotrypsin inhibition 62: Trypsin inhibition | [88] |
| 61: N-Me-l-Tyr, l-Val, l-Thr-2, l-Pro, l-Phe, l-Ahp, l-Leu 62: l-Lys, l-Thr, l-Val, l-Ile | Ligand Exchange Type CSP; Phenomenex Chirex Phase 3126 N,S-dioctyl-(d)-penicillamine column, 5 μm (4.6 × 250 mm); MP: 2 mM CuSO4 in H2O:ACN (95:5 v/v) or 2 mM CuSO4 Flow rate: 1.0 mL/min; UV detection at 254 nm | ||||
| Tiglicamides A–C (63–65) | Cyanobacterium Lyngbya confervoides | l-Ala, l-Thr, l-Val, d-Glu, d-Tyr; 63: l-Htyr; 65: l-Met (O) | Ligand Exchange Type CSP; Phenomenex, Chirex 3126, 5 μm (4.6 × 250 mm); Mobile Phase: 2 mM CuSO4 Flow rate: 1.0 mL/min; UV detection at 254 nm | Porcine pancreatic elastase inhibition | [89] |
| 65: l-Phe | Marfey’s method (FDLA) combined with HPLC Alltech Alltima HP C18, 5μm (4.6 × 250 mm) MP: 50–100% MeOH in 0.1% (v/v) aqueous TFA Flow rate: 0.8 mL/min; PDA detection at 200–500 nm | ||||
| Hantupeptin B (66) | Cyanobacterium Lyngbya majuscula | l-Pro, l-Val, N-Me-l-Val, N-Me-l-Ile | Marfey’s method (FDAA) combined with HPLC Phenomenex, Luna, 5 µm, (2.0 × 150 mm); MP: ACN in 0.1% (v/v) aqueous HCOOH; Flow rate: 0.2 mL/min | Cytotoxicity against MOLT-4 (leukemic) and MCF-7 (breast cancer) cell lines | [90] |
| l-3-phenyllactic acid (S) | Ligand Exchange Type CSP; Chiralpak MA (+) (4.6 × 500 mm) MP: 2 mM CuSO4/ACN (85:15 v/v) Flow rate: 0.7 mL/min; UV detection at 218 nm | ||||
| Palmyramide A (67) | Cyanobacterium (Lyngbya majuscula) and a red alga Centroceras sp. complex | l-Val, N-Me-l-Val, l-Pro | Marfey’s method (FDAA) combined with HPLC/MS on a Merck LiChrospher 100 RP-18 (4.0 × 125 mm) MP: ACN:H2O:HCOOH (30:70:0.1 to 70:30:0.1 v/v/v) or 2.0 mM CuSO4 in H2O Flow rate: 0.7 mL/min; UV detection at 254 nm | Sodium channel blocking activity in neuro-2a cells and cytotoxic activity in H-460 (human lung carcinoma) cells | [91] |
| l-Lac, l-Pla | Ligand Exchange Type CSP; Phenomenex Chirex 3126 (4.6 × 250 mm); Conditions not described | ||||
| Veraguamides A–G (68–74) | Cyanobacterium Symploca cf. hydnoides | 68–71, 73 and 74: l-Val, N-Me-l-Val, l-Pro; 70: (2S,3R) Br-Hmoya; 71: N-Me-l-Ile; 72: l-Ile, N-Me-l-Val, N-Me-l-Ile, l-Pro | Macrocyclic Antibiotic Type CSP; Chirobiotic TAG (4.6 × 250 mm); MP: MeOH/10 mM NH4OAc (40:60 v/v) (pH 5.6); Flow rate: 0.5 mL/min | Cytotoxic activity against HT29 (colorectal adenocarcinoma) and HeLa (cervical carcinoma) cell lines | [92] |
| 74: 2S:3R dpv 2R:3R Dml | Marfey’s method (FDAA) combined with HPLC-MS Phenomenex Synergi Hydro-RP (4.6 × 150 mm) MP: MeOH:H2O:HCOOH (40–100% MeOH: 0.1% HCOOH); Flow rate: 0.5 mL/min | ||||
| Porpoisamides A (75) and B (76) | Cyanobacterium Lyngbya sp. | 75 and 76: l-Ala, l-Pro, N-Me-d-Phe, (2S,3S)-Hmpa | Ligand Exchange Type CSP; Phenomenex Chirex 3126 (4.6 × 250 mm); MP: 5% or 15% ACN in 2 mM CuSO4 in H2O; Flow rate:1.0 mL/min | Cytotoxicity against HCT 116 (colorectal carcinoma) and U2OS (osteosarcoma) cells | [93] |
| 75: (2S,3R)-Amoa 76: (2R,3R)-Amoa | Ligand Exchange Type CSP; Chiralpak MA (+) (4.6 × 50 mm); MP: 15% ACN in 2 mM CuSO4 in H2O Flow rate: 1.0 mL/min | ||||
| (2R,3R) 3-NH2-2-Me-octanoic acid | Marfey’s method (FDAA) combined with HPLC YMC-Pack AQ-ODS (10 × 250 mm) MP: ACN:H2O: N-TFA (57:43:0.1 v/v/v) Flow rate: 2.5 mL/min; UV detection at 340 nm | ||||
| 76: (2S)-Hiva | Ligand Exchange Type CSP; CHIRALPAK MA (+) (4.6 × 50 mm); MP: ACN/2 mM CuSO4 (10:90 v/v) Flow: 1.0 mL/min; UV detection at 254 nm | ||||
| Companeramides A (77) and B (78) | Cyanobacterial assemblage collected from Coiba National Park, Panama | 77: l-Ala, N-Me-l-Ala, l-Pro, l-Ile, N-Me-l-Leu, and N-Me-l-Val; 78: l-Pro, N-Me-l-Val, l-Val, l-Ile, d- and N-Me-l-Ala | Marfey’s method (FDAA) combined with HPLC C18 column (3.9 × 150 mm) MP: 40 mM NH4OAc (pH 5.2):ACN (9:1 to 1:1 v/v) Flow rate: 1.0 mL/min; UV detection at 340 nm | Antiplasmodial activity against Plasmodium falciparum | [94] |
| S-Hiva | Ligand Exchange Type CSP; Phenomenex Chirex 3126 (D) (4.6 × 250 mm); MP: CuSO4/ACN Flow: 1.0 mL/min; UV detection at 254 nm | ||||
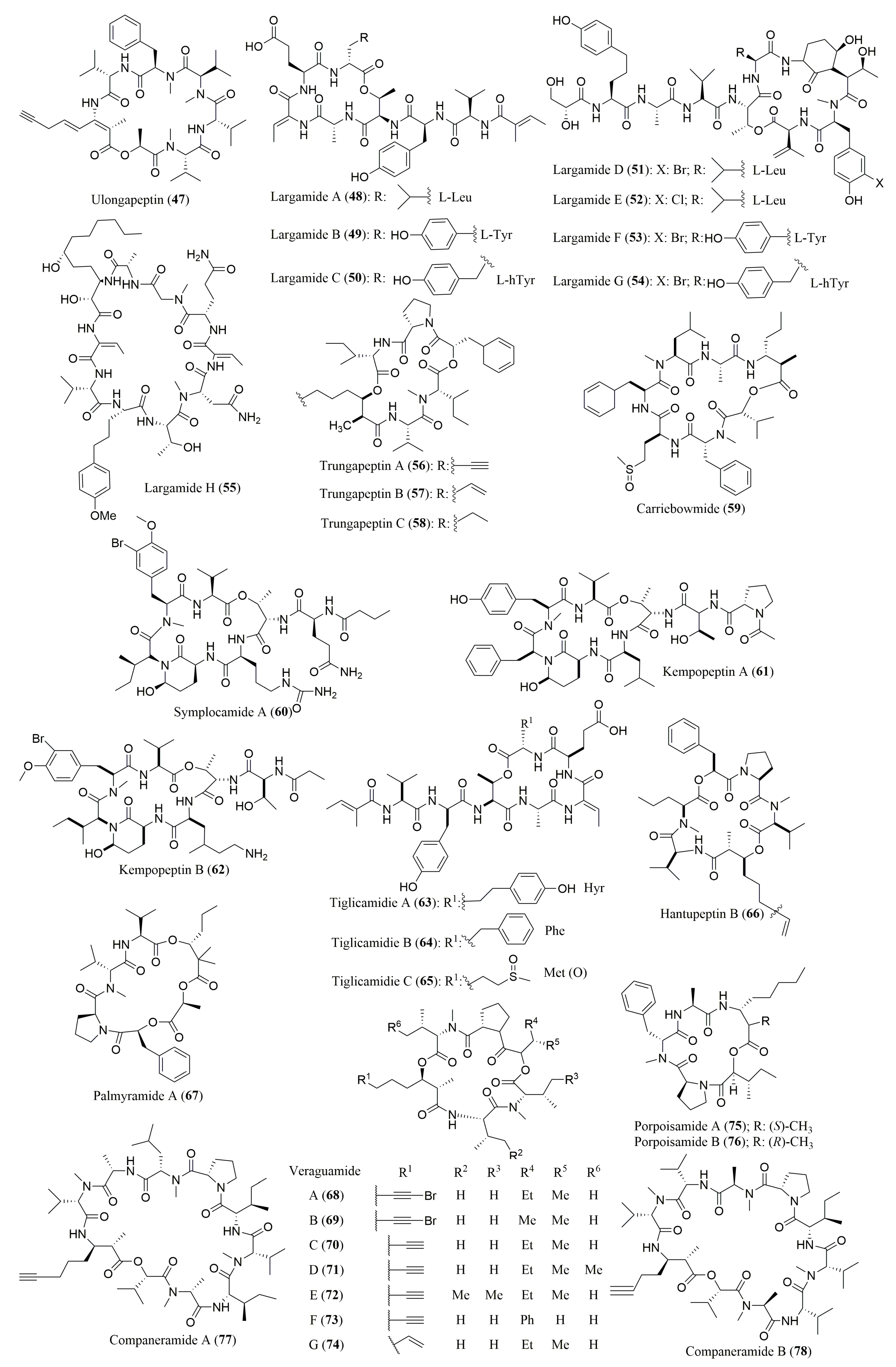
| Piperazimycins A–C (79–81) | Fermentation broth of a Streptomyces sp. | (S)-AMNA, (S,S)-OHPip1, (R,R)-γOHPip2, 79: (S)-αMeSer | Marfey’s method (FDAA) combined with HPLC C18; MP: ACN in H2O (10–100%) Flow rate: 1.0 mL/min; UV detection: 210, 254, 340 nm | 79: Active against diverse cancer cell lines | [95] |
| Grassypeptolides D (82) and E (83) | Red sea cyanobacterium Leptolyngbya sp. | d-allo-Thr, N-Me-d-Leu, l-Thr, N-Me-l-Leu | Marfey’s method (FDAA) combined with HPLC Gemini C18 110 A, 5 µm (4.6 × 250 mm) | Cytotoxicity against HeLa and mouse neuro-2a blastoma cells | [96] |
| l-PLa, N-Me-l-Val, l-Pro, N-Me-l-Phe, (2S)-MeCysA, d-Aba, l-Cya, (2R,3R)-Maba | Marfey’s method (FDAA) combined with HPLC Kinetex XB-C18, 110 A, 2.6 µm (4.6 × 100 mm) MP: ACN:H2O:HCOOH (30:70:0.1 to 70:30:0.1 v/v/v) or ACN:H2O:TFA (30:70:0.1 to 70:30:0.1 v/v/v); Flow rate: 0.2 mL/min; UV detection at 340 nm and ESIMS | ||||
| Fijimycins A–C (84–86) | Fermentation broth of Streptomyces sp. strain CNS-575 | 84: d-PhSar, l-Ala, l-DiMe-Leu, Sar, d-Hyp, d-Leu, l-Thr; 85: l-N-MeLeu, l-Ala, l-DiMeLeu, Sar, d-Hyp, d-Leu, l-Thr; 86: l-PhSar, l-Ser, l-DiMeLeu, Sar, d-Hyp, d-Leu, l-Thr | Marfey’s method (FDAA) combined with HPLC C18 column, Luna (4.6 × 100 mm) MP: ACN:H2O:TFA (10:90:1 to 50:50:1 v/v/v) Flow rate: 0.7 mL/min; UV detection at 340 nm | Antibacterial activity against three MRSA strains of Staphylococcus aureus | [97] |
| Itralamides A (87) and B (88), and Carriebowmide sulfone (89) | Cyanobacterium Lyngbya majuscula | 87: l-Ala, d-Ala, N-Me-l-Ala, N-Me-d-Phe, N-Me-l-Thr, N-Me-l-Val | Marfey’s method (FDLA) combined with HPLC Eclipse XDB-18, Agilent (4.6 × 150 mm) MP: ACN:H2O:HCOOH (20:80:0.1 to 80:20:0.1 v/v/v) Flow rate: 0.8 mL/min; Detection by ESI-MS | 88: Cytotoxicity against HEK293 (human embryonic kidney) cell line | [98] |
| 88: N-Me-l-Ala, N-Me-d-Phe, N-Me-l-Thr, d-Val | Marfey’s method (FDLA) combined with HPLC Luna C18, Phenomenex, 5 µm (4.6 × 250 mm) MP: ACN:H2O:HCOOH (20:80:0.1 to 90:10:0.1 v/v/v) Flow rate: 0.8 mL/min | ||||
| 89: (2S,3R)-AMHA | Marfey’s method (FDLA) combined with HPLC-PDA dC18, 5 µm (3.0 × 250 mm); MP: ACN:H2O:HCOOH (0:100:0.1 to 50:50:0.1 v/v/v); Flow rate: 0.3 mL/min | ||||
| Viequeamide A (90) | Marine button cyanobacterium Rivularia sp. | l-Val, l-Thr, N-Me-l-Val, l-Pro | Marfey’s method (FDLA) combined with HPLC Conditions not described | Highly toxic to H460 (human lung cancer) cells | [99] |
| Ngercheumicin F–I (91–94) | Photobacterium related to P. halotolerans | l-Ser, l-allo-Thr, d-Ser, d-Thr, l-Leu, d-Leu | Marfey’s method (FDLA) combined with HPLC Dionex RSLC Ultimate 300 with a diode array detector Kinetex C18 column, 2.6 µm at 60 °C (2.1 × 150 mm) ACN:H2O:TFA (0:100:0.1 to 50:50:0.1 v/v/v) Flow rate: 0.8 mL/min | 91–93: rnaIII inhibiting activities | [100] |
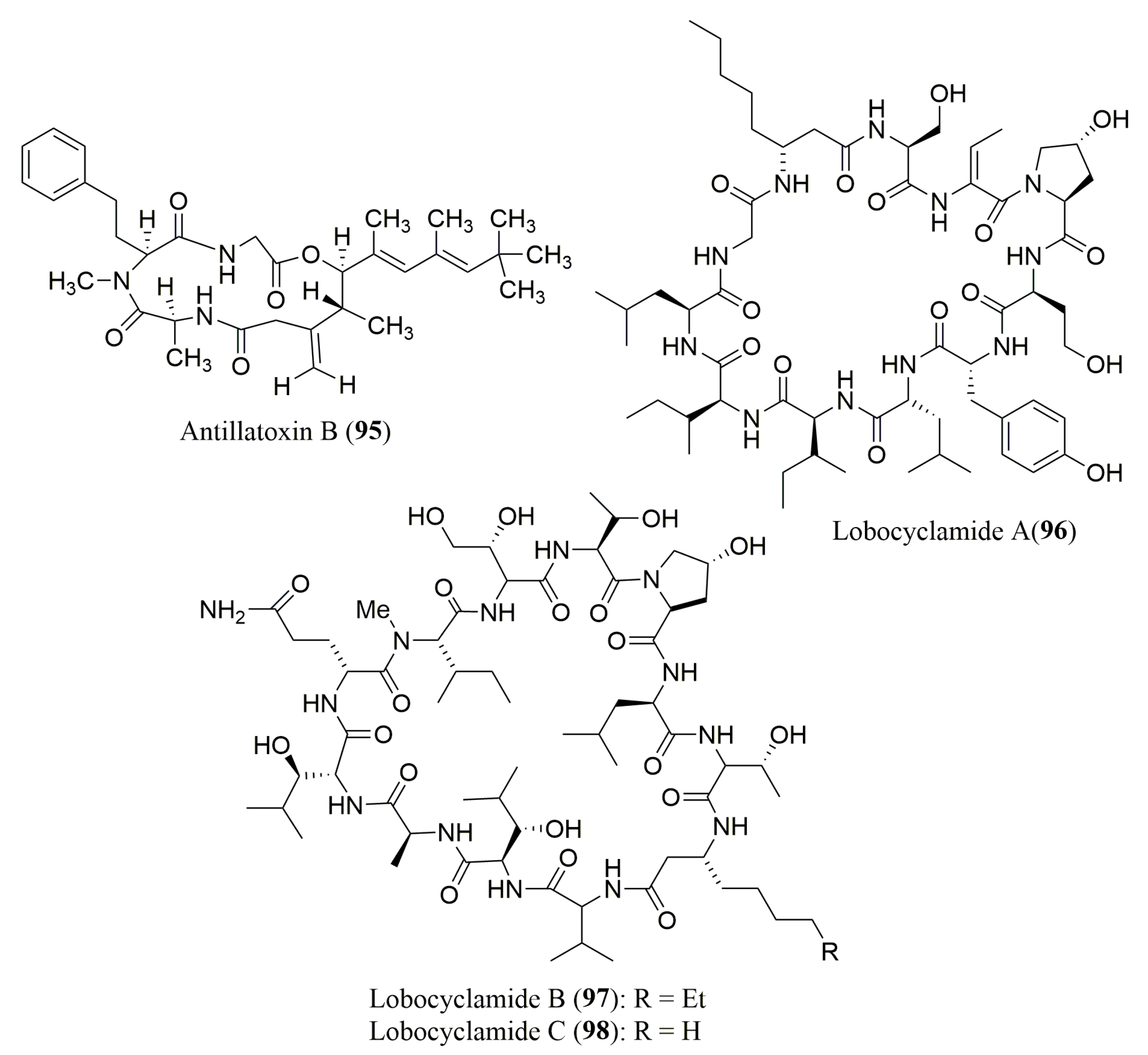
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
| Antillatoxin B (95) | Cyanobacterium Lyngbya majuscula | N-Me-l-Hph | Marfey’s method (FDAA) combined with HPLC Waters Nova-Pak C18 (3.9 × 150 mm), MP: 10 to 50% ACN in H2O with 0.05% TFA, UV detection at 340 nm | Sodium channel-activating and ichthyotoxic activities | [101] |
| Lobocyclami-des A–C (96–98) | Cyanobacterium Lyngbya confervoides | 96: S-Ile, S-allo-Ile, S-Leu, R-β-Aoa, S-Ser, R-Tyr, S-Hse, R-Hpr | Ligand Exchange Type CSP Chirex 3126 (d)-penicillamine column; MP: 2 mM aq CuSO4/ACN (1:99, 95:5 or 86:14 v/v); Flow rate: 1.15–1.20 mL/min, UV detection at 254 nm | Antifungal activity against a panel of Candida sp. | [102] |
| 97: S-Ala, S-Thr, N-Me-S-Ile, R-Aoa, R-Ada, 2R,3R-4-OH-Hth, 2R,3S-3-OH-Leu, trans-3-OH-Pro | Marfey’s method (FDAA) combined with HPLCC18 column (4.8 × 250 mm); MP: ACN: 0.1% aq. TFA buffer (pH 3) (1:9 to 1:1 v/v) Flow rate: 1.0 mL/min; UV detection at 340 nm |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
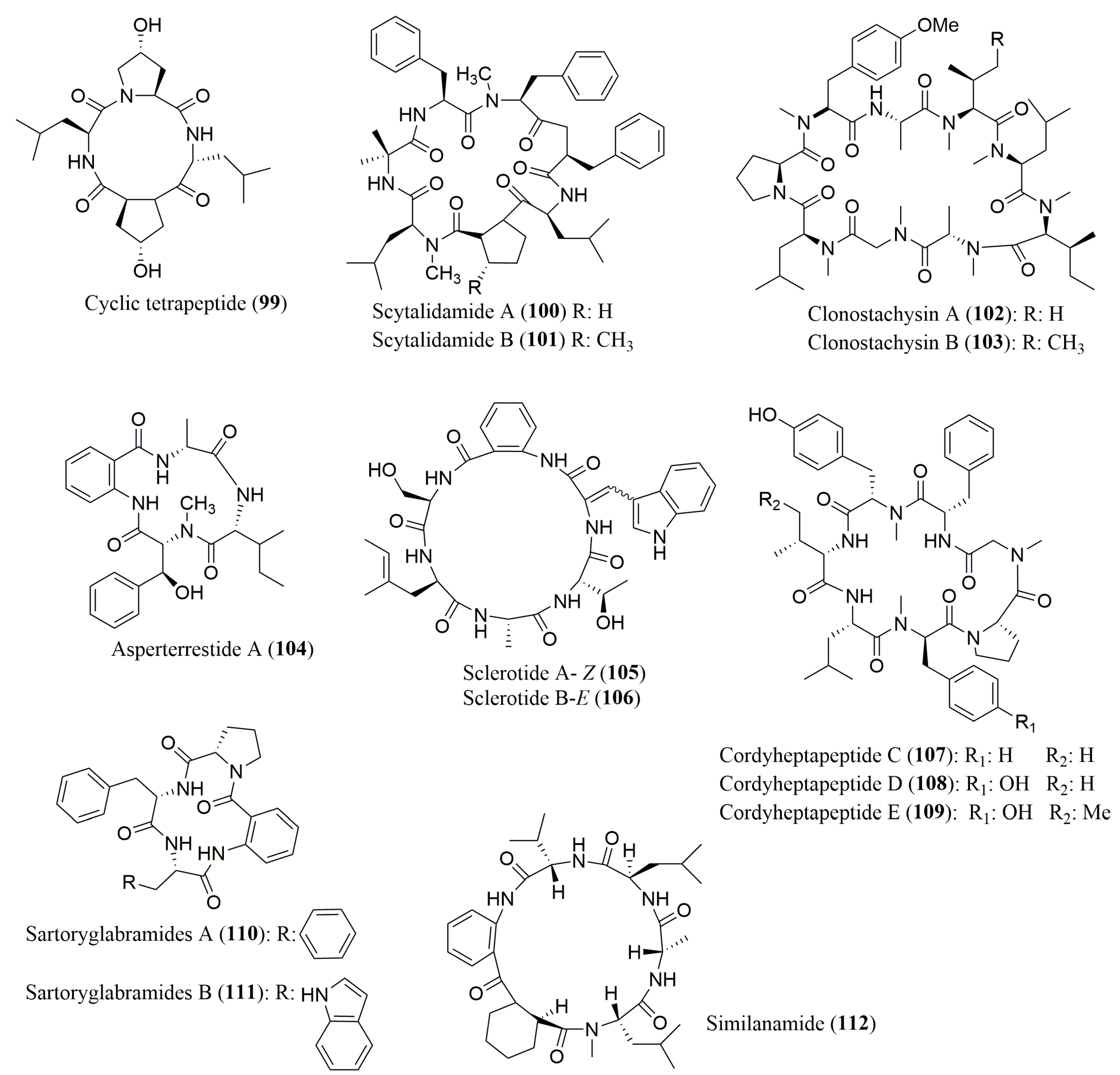
| Cyclo-(l-leucyl-trans-4-hydroxyl-l-prolyl-d-leucyl-trans-4-hydroxy-l-proline) (99) | Marine mangrove-derived fungi Phomopsis sp. K38 and Alternaria sp. E33 | 4-OH-l-Pro, d-Leu, l-Leu | Marfey’s method (FDAA) combined with LC/MS Alltima C18 column, 5 μm; (4.6 × 250 mm) MP: MeOH:H2O:HCOOH (60:40:0.05 to 10:90:0.05 v/v/v); Flow rate: 0.6 mL/min | Inhibition against four crop-threatening fungi | [105] |
| Scytalidamides A (100) and B (101) | Marine Fungus of the genus Scytalidium | l-Phe, N-Me-l-Phe, l-Leu, N-Me-l-Leu, l-Pro, 3-Me-l-Pro | Marfey’s method (FDLA) combined with HPLC Agilent Hypersil ODS column, 5 μm (4.6 × 100 mm); MP: ACN 25 to 65%; Flow rate: 0.7 mL/min | Cytotoxicity against HCT-116 and NCI 60 cell lines | [106] |
| Clonostachysins A (102) and B (103) | Marine sponge-derived fungus Clonostachys rogersoniana strain HJK9 | N-Me-l-Ile, N-Me-l-Leu, l-Pro, l-Gly, N-Me-l-Tyr, N-Me-l-Ala 102: N-Me-l-Val; 103: N-Me-l-Ile | Marfey’s method (FDLA) combined with LC-ESI MS/MS; Conditions not described | Inhibitory effect on dinoflagellate Prorocentrum micans | [107] |
| Asperterrestide A (104) | Marine-derived fungus Aspergillus terreus SCSGAF0162 | d-Ala | Marfey’s method (FDAA) combined with HPLC Alltima C18 column, 5 μm (4.6 × 250 mm); MP: ACN:H2O:TFA (15:85:0.1 to 90:10:0.1 v/v/v); Flow rate: 0.5 mL/min; UV detection at 254 nm | Cytotoxicity against U937 and MOLT4 human carcinoma cell lines and inhibitory effects on influenza virus | [108] |
| Ligand Exchange Type CSP; MCI GELCRS 10 W (4.6 × 50 mm); MP: 2 mM CuSO4:H2O solution Flow rate: 1.0 mL/min; UV detection at 254 nm | |||||
| Sclerotides A (105) and B (106) | Marine-derived fungus, Aspergillus sclerotiorum PT06-1 | l-Thr, l-Ala, d-Phe, d-Ser | Crown Ether CSP; Crownpak CR (+); MP: aq HClO4 pH 2.0; Flow rate: 0.4 mL/min; UV detection at 200 nm | 105 and 106: Antifungal activity 106: Cytotoxicity and antibacterial activity | [109] |
| Cordyheptapeptides C–E (107–109) | Marine-derived fungus Acremonium persicinum SCSIO 115 | N-Me-l-Tyr, l-Phe, l-Pro, l-Leu 107–109: N-Me-d-Phe, l-Val 109: N-Me-l-Gly, N-Me-d-Tyr, l-allo-Ile | Crown Ether Chiral CSP; Crownpak CR (+) MP: 2.0 mM CuSO4:ACN (95:5 v/v) Flow rate: 1.0 mL/min; UV detection at 254 nm | 107 and 109: Cytotoxicity against SF-268, MCF-7, and NCI-460 tumor cell lines | [110] |
| Similanamide (110) | Marine sponge-associated fungus Aspergillus similanensis KUFA 0013 | l-Ala, d-Leu, l-Val, N-Me-l-Leu, d-pipecolic acid | Macrocyclic Antibiotic Type CSP; Chirobiotic T, 5 μm (4.6 × 150 mm); MP: MeOH:H2O:CH3COOH (70:30:0.02 v/v/v); Flow rate: 1.0 mL/min; UV detection at 210 nm | Cytotoxicity against MCF-7, NCI-H460 and A373 tumor cell lines | [111] |
| Sartoryglabramide A (111) and B (112) | Marine sponge-associated fungus Neosartorya glabra KUFA 0702 | l-Phe, l-Pro 112: l-Trp | Macrocyclic Antibiotic Type CSP; Chirobiotic T, 5 μm (4.6 × 150 mm); MP: MeOH:H2O (80:20 v/v) Flow rate: 1.0 mL/min; UV detection at 210 nm | Neither antibacterial nor antifungal activity | [112] |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
| Exumolides A (113) and B (114) | Fungus of the genus Scytalidium sp. | l-Pro, l-Phe, N-Me-l-Leu | Marfey’s method (FDAA) combined with HPLC Hewlett Packard 1090 Diode Array, 5 µm (10 × 250 mm); MP: 10–50% aq ACN (0.1% TFA) Flow rate: 1.0 mL/min; UV detection at 340 nm | Antimicroalgal activity against unicellular chlorophyte Dunaliella sp | [113] |
| Guangomide A (115) | Sponge-derived fungus | N-Me-d-Phe | Marfey’s method (FDAA) combined with HPLC Alltech Altima C18 column, 5 µm (10 × 250 mm) MP: ACN:H2O (4:1 to 1:1 v/v); Flow rate: 1.0 mL/min; UV detection at 340 nm | Antibacterial activity against Staphylococcus epidermidis and Enterococcus durans | [114] |
| Destruxin E chlorohydrin (116) and pseudodestruxin C (117) | Marine-derived fungus Beauveria felina | N-Me-l-Val 116: N-Me-l-Ala, l-Ile 117: l-Phe | Marfey’s method (FDAA) combined with HPLC C18 column, 5 µm (4.6 × 250 mm); MP: 10–20% ACN in 0.1 M NH4OAc (pH = 5) Flow rate: 1.0 mL/min; UV detection at 340 nm | Cytotoxicity in NCI’s 60 cell line panel | [115] |
| Zygosporamide (118) | Marine-derived fungus Zygosporium masonii | l-Phe, l-Leu, d-Leu | Marfey’s method (FDAA) combined with HPLC C18, Agilent column, 5 µm (4.6 × 250 mm) MP: 10–50% ACN (0.1% TFA) Flow rate: 1.0 mL/min; UV detection at 340 nm | Cytotoxicity in RXF 393 and SF-268 cancer cell lines | [116] |
| Petriellin A (119) | Coprophilous fungus Petriella sordida | N-Me-l-Ile, N-Me-l-Thr d-Phenyllactate | Marfey’s method (FDAA) combined with HPLC C18 column (4.6 × 250 mm); Conditions not described; UV detection at 260 nm | Antifungal activity | [117] |
| Alternaramide (120) | Marine derived fungus Alternaria sp. SF-5016 | l-Pro, d-Phe | Marfey’s method (FDAA) combined with HPLC Capcell Pak C18 column; MP: 30–60% ACN in H2O (0.1% HCOOH); Flow rate: 1.0 mL/min | Antibacterial activity against Bacillus subtilis and Staphylococcus aureus | [118] |
| Petrosifungins A (121) and B (122) | Penicillum brevicompac-tum | l-Val, l-Pro, l-Thr, l-pipecolinic acids | Marfey’s method (FDAA) combined with HPLC C18 column, Waters, 5 µm (2.1 × 150 mm); MP: H2O or ACN (0.05% TFA); Flow rate: 1.0 mL/min | Not described | [119] |
| Oryzamides A–E (123–127) | Sponge-Derived fungus Nigrospora oryzae PF18 | l-Ala, d-Leu, l-Val 123: l-Leu; 124: l-Tyr 125 and 126: l-Met; 127: l-Phe | Marfey’s method (FDLA) combined with UHPLC Acquity UHPLC BEH column, 1.7 µm (2.1 × 250 mm); MP: 10–100% ACN in H2O with 0.1% HCOOH; Flow rate: 0.5 mL/min; UV detection at 360 nm | No cytotoxicity, antibacterial, antiparasitic, and NF-kB activities | [122] |
| Spicellamide A (128) and B (129) | Marine-derived fungus Spicellum roseum | N-Me-d-Phe, N-Me-l-Ala, l-Ala | Marfey’s method (FDAA) combined with HPLC C18 column; Macherey-Nagel Nucleodur 100, 5 µm (2.0 × 125 mm); MP: MeOH:H2O (10:90 v/v to 100% MeOH) or 100% MeOH with NH4Ac, 2 mmol | 129: Cytotoxicity | [123] |
| l-2-hydroxyisocaproic acid | Ligand Exchange Type CSP; Phenomenex Chirex 3126 N,S-dioctyl-(d)-penicillamine (4.6 × 50 mm) MP: 2 mM CuSO4 in ACN:H2O (15:85 v/v) Flow rate: 1.0 mL/min; UV detection at 254 nm | ||||
| Depsipeptides 1962A (130) and 1962B (131) | Endophytic fungus Kandelia candel | l-Tyr, l-Val, d-Leu, (S)-O-Leu | Crown Ether CSP; Crownpak CR (+) column (0.4 × 150 mm), MP: 2 mM CuSO4 aq. solutions Flow rate: 0.5 mL/min; UV detection at 200 nm | 131: Activity against MCF-7 tumor cell line | [124] |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
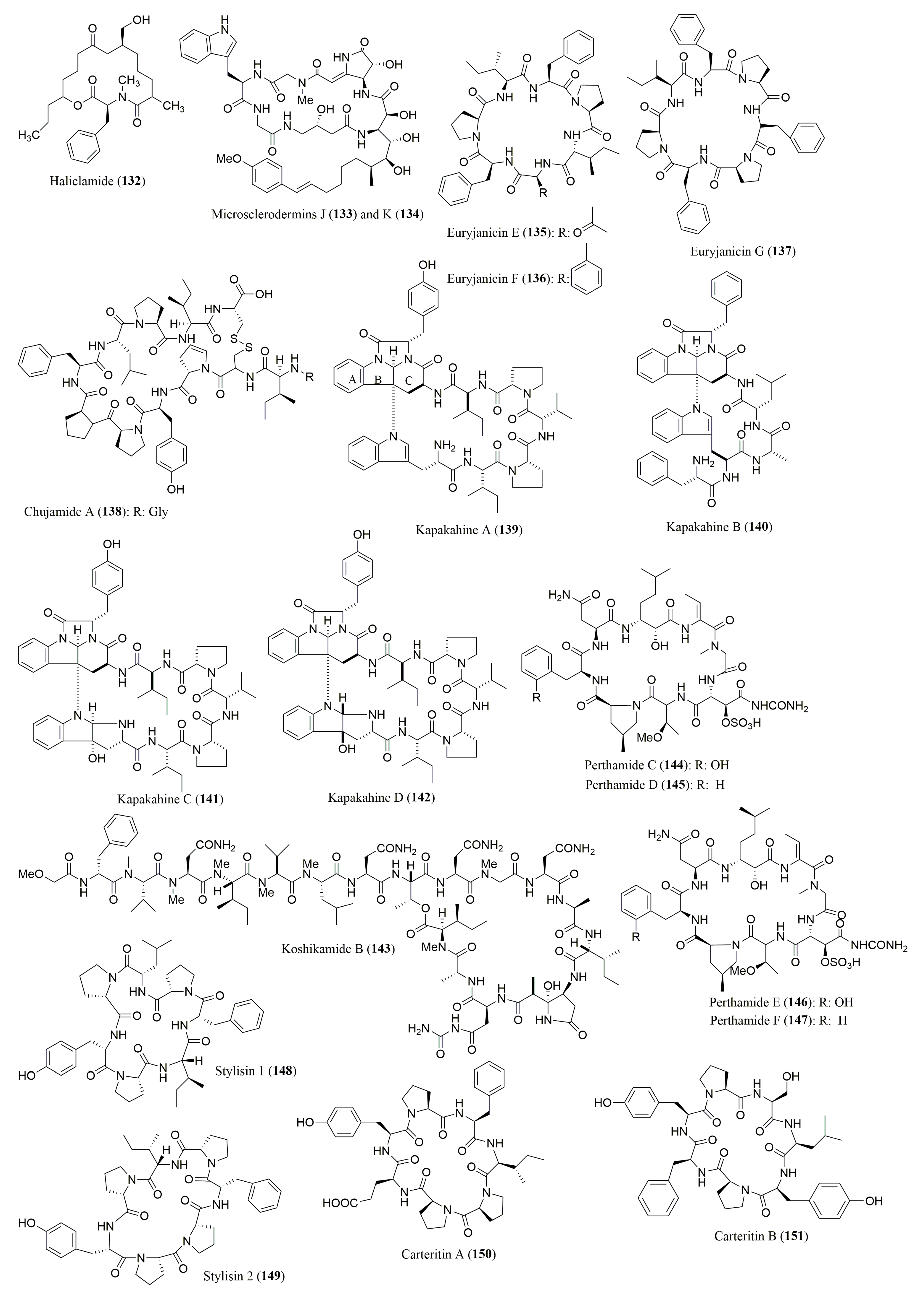
| Haliclamide (132) | Vanuatu marine sponge Haliclona sp. | N-Me-l-Phe | Marfey’s method (FDAA) combined with HPLC Vydac C18; MP: ACN in H2O with 0.1% TFA (9:1 to 1:1 v/v); UV detection at 340 nm | Cytotoxicity against NSCLC-N6 carcinoma cell line | [129] |
| Microsclerodermins J (133) and K (134) | Deep water sponge Microscleroderma herdmani | l-Ile, l-Thr 133: l-Phe, l-Gly 134: l-Val, l-Ala | Marfey’s method (FDAA) combined with HPLC C18 column, 5 µm (4.6 × 150 mm) Flow rate: 1.0 mL/min; UV detection at 340 nm | Activity against opportunistic pathogenenic fungi | [130] |
| Euryjanicins E–G (135–137) | The Caribbean Sponge Prosuberites laughlini | l-Pro, l-Ile, l-Phe 135: l-Asp | Marfey’s method (FDAA) combined with HPLC C18 column, 5 µm (4.6 × 150 mm) Flow rate: 1.0 mL/min; UV detection at 340 nm | No significant activity cytotoxicity against the National Cancer Institute 60 tumor cell line panel | [131] |
| Chujamide A (138) | Marine sponge Suberites waedoensis | l-Pro, l-Tyr, l-Cys, l-Leu, l-Phe l-Ile (S) | Marfey’s method (FDAA) combined with HPLC ESI-LC/MS YMC ODS-A column, 5 µm (4.6 × 250 mm) MP: H2O:ACN (80:20 to 30:70 v/v) Flow rate: 0.7 mL/min; UV detection at 360 nm | Weak cytotoxicity against A549 and K562 cell lines | [132] |
| Kapakahines A–D (139–142) | Marine Sponge Cribrochalina olemda | l-Val, l-Ile, l-Leu, l-Trp, l-Phe, l-Ala, l-Pro, l-Try | Marfey’s method (FDAA) combined with HPLC Cosmosil C18-MS column, 5 µm (4.6 × 250 mm) MP 37.5% ACN in 0.05% TFA or 20% or 38% ACN in 50 mM NH4OAc | 139–141: Cytotoxicity against P388 cell line 139: Inhibition against protein phosphatase | [133] |
| Koshikamide B (143) | Marine sponge Theonella sp. | d-Phe, l-Thr, N-Me-l-Val, N-Me-l-Asn, N-Me-l-Leu | Marfey’s method (FDAA) combined with HPLC ODS HPLC (10 × 250 mm); MP: ACN:H2O:TFA (25:75:0.05 to 55:45:0.05 v/v/v); Flow rate: 1.0 mL/min; UV detection at 340 nm | Cytotoxicity against P388 and HCT-116 tumor cell lines | [134] |
| Perthamides C (144) and D (145) | Solomon Lithistid sponge Theonella swinhoei | l-Asp, l-ThrOMe, (2R,3S)-βOHAsp, l-Phe | Marfey’s method (FDAA) combined with HPLC/MS Proteo C18 column (1.8 × 25 mm) MP: 10–50% aq ACN with 5% HCOOH and 0.05% TFA Flow rate: 0.15 mL/min | Anti-inflammatory activity | [135] |
| Perthamides E (146) and F (147) | Polar extracts of the sponge Theonella swinhoei | 146: l-ThrOMe 147: l-Phe | Marfey’s method (FDAA) combined with HPLC Proteo C18 column, (1.8 × 25 mm); MP: 10–50% aq ACN with 5% HCOOH and 0.05% TFA Flow rate: 0.15 mL/min | 147: IL-8 release inhibition | [136] |
| Stylisins 1 (148) and 2 (149) | Jamaican sponge Stylissa caribica | l-Pro, l-Tyr, l-Ile 148: l-Leu, l-Phe | Marfey’s method (FDAA) combined with HPLC HPLC water Nova Pack column (3.9 × 150 mm) MP: TEAP buffer (pH 3.0 ± 0.02):ACN (90 to 60% TEAP) UV detection at 340 nm | No antimicrobial, antimalarial, anticancer, anti-HIV-1, anti-Mtb and anti-inflammatory activities | [137] |
| Carteritins A (150) and B (151) | Marine sponge Stylissa carteri | 150: l-Pro, l-Phe, l-Ile, l-Pro (trans), l-Pro (cis), l-Glu, l-Tyr; 151: l-Pro (trans), l-Leu, l-Tyr, l-Pro(cis) | Marfey’s method (FDAA) combined with HPLC Cosmosil C18 MS (4.6 × 250 mm); MP: H2O:TFA (100:0.1) to ACN:H2O:TFA (50:50:0.1 v/v/v); Flow rate: 1.0 mL/min; UV detection at 340 nm | 150: Cytotoxicity against HeLa, HCT116, and RAW264 cells | [139] |
| Stylissatins B–D (152–154) | Marine sponge Stylissa massa | l-Pro, l-Phe, l-Leu 152: l-His 153–154: l-Asp, l-Val | Marfey’s method (FDAA) combined with HPLC Thermo BDS Hypersil C18 column, 5 μm (4.6 × 150 mm); MP: 30–70% MeOH:H2O (H3PO4) Flow rate: 1.0 mL/min; UV detection at 340 nm | 152: Inhibitory effects against a panel of human tumor cell lines including HCT-116, HepG2, BGC-823, NCI-H1650, A2780, and MCF7 | [138] |
| Callyaerin G (155) | Indonesian sponge Callyspongia aerizusa | l-Pro, l-Leu, l-Phe, l-FGly | Marfey’s method (FDAA) combined with HPLC/MS; Conditions not described | Cytotoxicity against L5178Y, Hela, and PC12 | [140] |
| Reniochalistatins A–E (156–160) | Marine sponge Reniochalina stalagmitis | l-Pro, l-Phe, l-Val, l-Leu, l-Ile, l-Tyr | Ligand Exchange Type CSP; MCI GELCRS 10 W (4.6 × 50 mm); MP: 2 mM CuSO4:H2O solution Flow rate: 1.0 mL/min; UV detection at 254 nm | 160: Cytotoxicity against RPMI-8226, MGC-803, HL-60, HepG2, and HeLa | [141] |
| 156: l-Asn 160: l-Trp | Marfey’s method (FDAA) combined with HPLC YMC-Park Pro C18, 5 µm (4.6 × 250 mm) MP: 2 mM CuSO4:H2O solution Flow rate: 1.0 mL/min; UV detection at 254 nm | ||||
| Phakellistatins 15–18 (161–164) | South china sea sponge Phakellia fusca | l-Pro 161: l-Trp, l-Ile, l-Leu, l-Thr; 162: l-Phe, l-Asp, l-Ser, l-Arg, l-Ala, l-Val, l-Thr, l-Tyr; 163: l-Trp, l-Val, l-Leu, l-Ile; 164: l-Tyr, l-Ile, l-Phe | Ligand-exchange type CSP; Chirex 3126 (d)-penicillamine column (4.6 × 150 mm) MP: aq 2 mM CuSO4:MeOH (85:15 to 70:30 v/v) or aq 1 mM/0.5 mM CuSO4; Flow rate: 0.5 or 1.0 mL/min | 161: Cytotoxicity against P388 cancer cell line 162: Cytotoxicity against P388 and BEL-7402 cancer cell lines | [142] |
| Peptide | Source | aa Composition | Chromatographic conditions | Biological activities | Ref. |
|---|---|---|---|---|---|
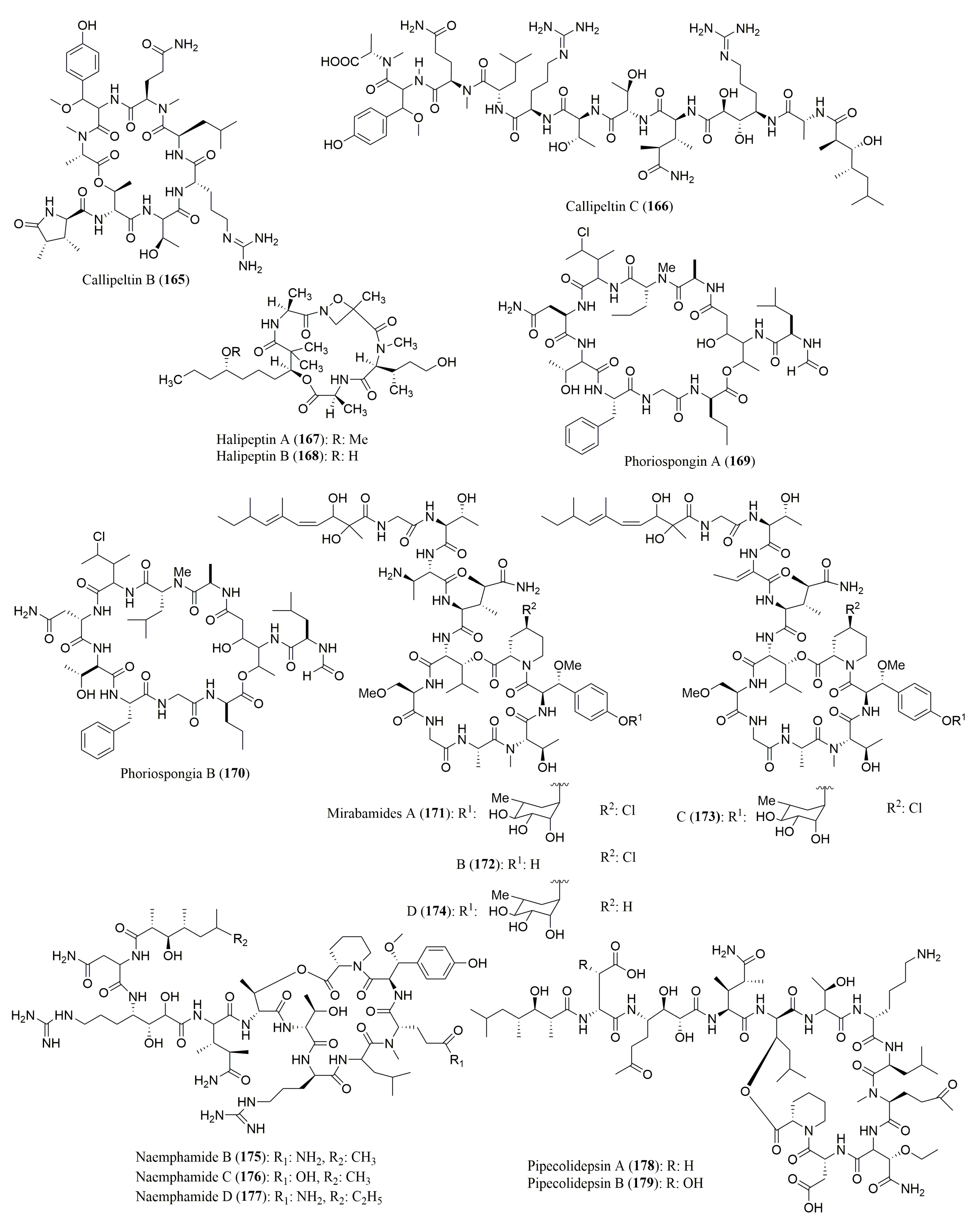
| Callipeltins B (165) and C (166) | Callipelta sp. | l-Ala, d-Arg, l-Thr, N-Me-l-Ala, l-Leu | Marfey’s method (FDAA) combined with HPLC; Column not described; MP: TEAP (50 nM, pH 3.0):ACN 90–50% TEAP Flow rate: 2.0 mL/min; UV detection at 340 nm | Cytotoxicity 166: Growth inhibitory activity against Candida albicans | [143] |
| Halipeptiins A (167) and B (168) | Haliclona species | l-Ala | Marfey’s method (FDAA) combined with HPLC Vydac C18 column; MP: H2O (0.1% TFA):ACN (0:1 to 1:1 v/v) UV detection at 340 nm | 168: Anti-inflammatory activity | [144] |
| Phoriospongin A (169) and B (170) | Phoriospongia sp. and Callyspongia bilamellata | d-Asp, d-allo-Thr, d-Ala, l-Phe, d-Leu, d-nor-Val, N-Me-d-nor-Val 170: N-Me-l-Leu | Marfey’s method (FDAA) combined with HPLC C18 column, 5 µm (4.6 × 250 mm) Flow rate: 1.0 mL/min; UV detection at 340 nm | Nematocidal activity against the parasite Haemonchus contortus | [145] |
| Mirabamides A–D (171–174) | Siliquarias-pongia mirabilis | N-Me-l-Thr, l-Thr, l-Ala, d-3-OMeAla, (2R,3R)-3-OH-Leu (3S,4R)-diMe-l-Glu, (2S,3R)-diaminobutanoic acid; 174: l-HPr | Marfey’s method (FDAA) combined with HPLC Phenomenex Jupiter Proteo C12 column, 4 µm (4.6 × 150 mm) MP: 25–70% ACN; Flow rate: 0.5 mL/min | 171: Anti-HIV activity 173 and 174: Antibacterial activity 171–173: Antifungal activity | [146] |
| Neamphamides B (175), C (176) and D (177) | Neamphius huxleyi | d-Arg, l-Asn, l-Hpr, l-Leu, d-allo-Thr 175 and 177: N-Me-l-Gln 176: N-Me-l-Glu | Marfey’s method (FDAA) combined with HPLC Phenomenex Luna Column C18, 3 µm (2.0 × 150 mm) MP: H2O:ACN:HCOOH (100:0:0.1 to 0:100:0.1 v/v/v) UV detection at 340 nm | Growth inhibition of human cell lines: A549, HeLa, LNCaP, PC3, and NFF | [147] |
| Pipecolidepsins A (178) and B (179) | Homophymia lamellosa | d-Asp, l-Leu, d-Lys, d-allo-Thr, (3S,4R) diMe-l-Glu, (2S,3S)-EtO-Asp, N-Me-l-Glu, l-Pip | Marfey’s method (FDAA) combined with HPLC Symmetry C18, 5 µm (4.6 × 150 mm); MP: 20–50% ACN (0.04% TFA) in H2O (0.04% TFA); Flow rate: 0.8 mL/min | Cytotoxicity against three human tumor cell lines (A-549, HT-29, and MDA-MB-231) | [148] |
| Stellatolide A (180) | Ecionemia acervus | N-Me-l-Ala, l-Leu, N-Me-l-Gln, N-Me-d-Ser, d-allo-Thr | Marfey’s method (FDAA) combined with HPLC Hewlett-Packard Hypersil BDS-C18, 4 µm (4.0 × 100 mm); MP: H2O (0.1% TFA):ACN (90:10 to 50:50 v/v); Flow rate: 1.0 mL/min | In in vitro antiproliferative activity | [149] |
| Cyclolithistide A (181) | Theonella swinhoei | nor-S-Val, S-Phe, S-Gln, N-Me-S-Leu, S-Ala, S-Allo-S-Thr | Marfey’s method (FDAA) combined with HPLC ODS (4.6 × 250 mm); MP: 100% H2O Flow rate: 2.0 mL/min; UV detection at 210 nm | Antifungal activity against Candida albicans (ATCC 24433) | [150] |
| Nagahamide A (182) | Theonella swinhoei | l-Val, l-Ser, 3S-AHBA | Marfey’s method (FDAA) combined with HPLC ODS column (4.6 × 250 mm); Conditions not described | Antibacterial activity | [151] |
| Theopapuamides B (183) and C (184), Celebesides A–C (185–187) | Siliquarias-pongia mirabilis | 185: l-βMeAsn | Marfey’s method (FDAA) combined with HPLC/MS Phenomenex Jupiter Proteo C12 column, 4 µm (4.6 × 150 mm) MP: 25–70% ACN with 0.01 M TFA; Flow rate: 0.5 mL/min | 185: Inhibits HIV-1 Entry 183–185: Cytotoxic to human colon tumor cell line (HCT-116) 183 and 185: Antifungal activity against Candida albicans | [152] |
| Ligand Exchange Type CSP Phenomenex column, Chirex Phase 3126 (D) (4.6 × 150 mm); MP: 1 mM CuSO4:ACN (95:5 v/v)Flow rate: 0.5 mL/min; UV detection at 254 nm | |||||
| Theopapuamide (188) | Lithistid sponge Theonella swinhoei | N-Me-l-Leu, d-Asp, l-Leu, N-Me-l-Glu | Ligand Exchange Type CSP Chirex Phase 3126 (D), 5 µm (4.6 × 250 mm); MP: IPA: 2 mM CuSO4 (5:95 v/v) Flow rate: 1.0 mL/min; UV detection at 254 nm | Cytotoxicity against CEM-TART and HCT-cell lines | [153] |
| d-allo-Thr | Marfey’s method (FDAA) combined with HPLCPhenomenex C18, 5 µm (4.6 × 250 mm); MP: 10–50% ACN in H2O (0.05% TFA); Flow rate: 1.0 mL/min; UV detection at 340 nm | ||||
| Mutremdamide A (189) and Koshikamides C–H (190–195) | Theonella swinhoei and Theonella cupola | 189: N-Me-l-Val; 190: N-Me-l-Val, N-Me-l-Asn, l-Asn, N-Me-l-Leu, l-Pro, N-Me-allo-l-Ile, d-Phe | Marfey’s method (FDAA) combined with HPLC LC-MS analysis using a C12 column, 4 µm (4.6 × 250 mm); MP: ACN with 0.01% TFA; Flow rate: 0.5 mL/min | 189–195: Anti-HIV-1 activity | [154] |
| 191 and 192: N-Me-allo-l-Ile, N-Me-l-Val; 192–194: N-Me-allo-l-Ile, l-Ala 1, d-Ala2, l-Asn | Marfey’s method (FDAA) combined with HPLC LC-MS, C18 column, 4 µm (4.6 × 250 mm); MP: 20 mM buffer (AF):ACN (3:1 to 3:7 v/v); Flow rate: 0.5 mL/min | ||||
| 195: N-Me-allo-l-Ile | Chiral HPLC (column not described); MP: 1 mM CuSO4:ACN (95:5 v/v); Flow rate: 0.5 mL/min; UV detection at 254 nm |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activity | Ref. |
|---|---|---|---|---|---|
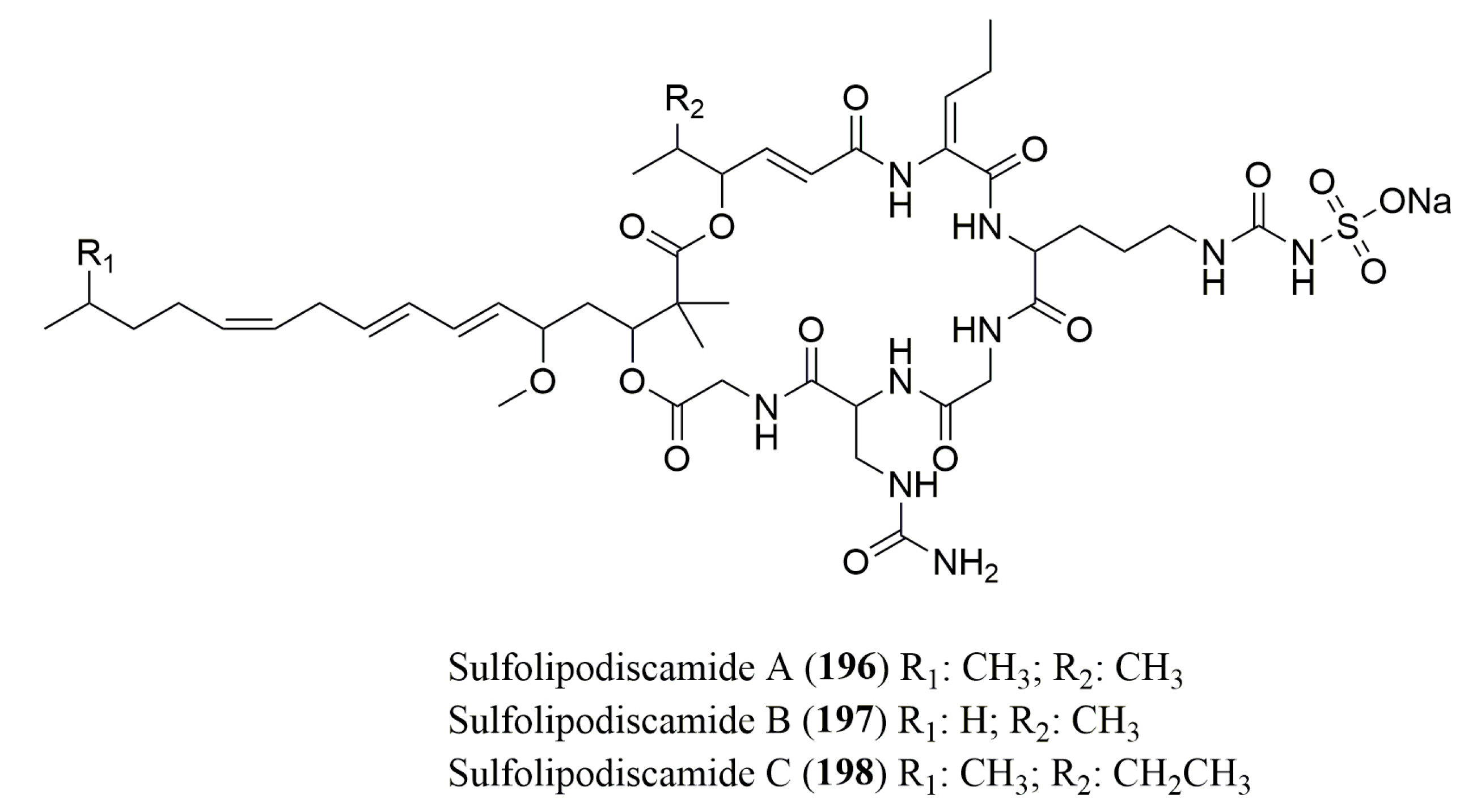
| Sulfolipo-discamides A–C (196–198) | Sponge Discoderma kiiensis | l-Uda, l-Gly | Marfey’s method (FDAA) combined with HPLC Cosmosil C18-MSII column (4.6 × 250 mm); MP: 100 mM NaClO4 in 60% ACN Flow rate: 0.8 mL/min | 196: Cytotoxicity against P388 cell line | [155] |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
| Didomolamides A (199) and B (200) | Ascidian Didemnum molle | l-Thr, l-Ala, l-Phe 200: l-Tzl | Marfey’s method (FDAA) combined with HPLC; MP: 50 mM (TEAP) buffer pH 3: ACN (9:1 to 1:1 v/v); Flow rate: 1.0 mL/min; UV detection at 340 nm | Cytotoxicity against A549, HT29 MEL28 tumor cell lines | [157] |
| Mollamides B (201) and C (202) | Tunicate Didemnum molle | l-Thr, l-Ile, l-Pro 201: l-Val, l-Phe 202: l-Ser, l-Leu | Marfey’s method (FDAA) combined with HPLC; MP: 50 mM TEAP, pH 3.0: ACN (90:10 to 60:40 v/v) or 40 mM NH4OAc, 70% ACN, and 30% MeOH (98:2 to 66:34 v/v) Flow rate: 1.0 or 0.8 mL/min; UV detection at 340 nm | 201: Activity against HIV, Plasmodium falciparum, Lieshmania donovan, and cytotoxicity against H460, MCF7, SF-268 cell lines | [158] |
| Antatollamides A (203) and B (204) | Ascidian Didemnum-molle | l-Ile, l-Phe, l-Val, l-Pro, d-Ala | Marfey’s method (FDLA) combined with HPLC/MSHypersil Gold C18 column, 1.9 µm (2.1 × 50 mm); MP: H2O 0.1%; HCOOH:ACN (85:15 to 55:45 v/v) Flow rate: 0.5 mL/min | 203: Weak cytotoxicity against a chronic lymphocytic leukemia cell line | [159] |
| Sanguinamide A (205) | Nudibranch Hexabranchs sanguineus | l-Pro, l-Ile, l-Ala, l-Phe | Marfey’s method (FDLA) combined with HPLC Agilent Zorbax SB-Aq C18 column, 5 µm (4.6 × 250 mm) MP: 80% (H2O: 0.1% HCOOH), 20% (ACN) | Antifungal activity | [160] |
| Gamakamide E (206) | Oysters Crassostrea giga | l-Met(O), N-Me-l-Phe, l-Leu, d-Lys, l-Phe | Marfey’s method (FDLA) combined with HPLC Conditions not described | No growth inhibition abilities | [161] |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
| Kahalalides A–F (207–212) | Mollusk Elysia rufescens | 207: d-Val-5; 208: l-Val-1, d-Val-2, d-allo-Thr-1; 209: l-Val-3, d-Val-4, l-Thr-2; 210: d-Val-2, d-allo-Thr-1 | Marfey’s method (FDLA) combined with HPLC COSMOSIL 5C18-AR MP: ACN:H2O:TFA (42:48:0.05 v/v/v) or ACN:H2O:50 mM NH4OAc (20:80:0.01 v/v/v) | 207: Antimalarial activity 211: Activity against RSV II virus | [162] |
| Table 10. Cont.Tamandarins A (213) and B (214) | Ascidian of the family Didemni-dae | 213: S-Lac, l-Pro, N-Me-d-Leu, l-Thr, (3S,4R,5S)-Ist 214: S-Lac, l-Pro, N-Me-d-Leu, l-Thr 3S,4R)-Nst | Marfey’s method (FDAA) combined with HPLC Hewlett-Packard ODS Hypersil 5 µm (4.6 × 200 mm); MP: 0.1% TFA in H2O or MeOH; Flow rate: 1.0 mL/min; UV detection at 340 nm | 213: Cytotoxicity against various human cancer cell lines | [163] |
| KahalalidesP (215) and Q (216) | Green alga Bryopsis sp. | l-Asp, l-Val, d-Leu, l-Ser, l-Hyp, l-Pro, l-Lys | Marfey’s method (FDAA) combined with HPLC COSMOSIL 5C18-AR-II (4.6 × 250 mm); MP: 0.1 M NH4OAc pH 3 or 90% aq ACN | No antimicrobial and no hemolytic activities | [164] |
| Kahalalide O (217) | Mollusk Elysia ornata and green alga Bryopsis sp. | l-Ile, l-Thr, d-allo-Thr, d-Tyr, l-Val | Ligand Exchange Type CSP Chirex (D) Penicillamine Column (4.6 × 250 mm); MP: 1.9 mM CuSO4 in ACN:H2O (5:95) or 2.0 mM CuSO4 in H2O; UV detection at 254 nm | No growth inhibition of P-388, A549, HT29 and MEL28 cancer cell lines | [165] |
| d-Trp | Marfey’s method (FDAA) combined with HPLC COSMOSIL 5C18-AR; MP: ACN:H2O:TFA (37.5:62.5:0.05 v/v/v); Flow rate: 1.0 mL/min UV detection at 254 nm |
| Peptide | Source | aa Composition | Chromatographic Conditions | Biological Activities | Ref. |
|---|---|---|---|---|---|
| Eudistomides A (218) and B (219) | Ascidian Eudistoma sp. | l-Pro, l-Ala, l-Leu 219: l-Cyp | Ligand Exchange Type CSP Phenomenex Chirex 3126 (D) (4.6 × 250 mm); MP: 2 mM CuSO4, 2 mM CuSO4:ACN (95:5 or 85:15 v/v); Flow rate: 1.0 mL/min UV detection at 254 nm | No activity reported | [166] |
| Mebamamides A (220) and B (221) | Green algae Derbesia marina | l-Leu, l-Pro, d-Ala, l-Thr, l-Val, d-Phe, d-Ser | Ligand Exchange Type CSP Diacel CHIRALPAK (MA+) (4.6 × 50 mm); MP: 2.0 mM CuSO4, Flow rate: 1.0 mL/min; UV detection at 254 nm | No growth inhibitory activity against HeLa and HL60 cell lines | [167] |
| Retention Time (min) | Retention Time (min) | ||
|---|---|---|---|
| d-Trp (A) | 5.20 | d-pipecolic acid (B) | 14.67 |
| l-Trp (A) | 4.51 | Acidic hydrolysate of 110 (B) | 6.59, 7.20, 8.09, 8.83, 9.67, 10.57, 14.69 |
| l-Val (B) | 6.60 | Acidic hydrolysate of 110 + dl-Val (co-injection) (B) | 6.61, 7.31, 8.30, 8.10, 8.84, 9.70, 10.50, 14.95 |
| d-Val (B) | 8.32 | Acidic hydrolysate of 110 + dl-Ala (co-injection) (B) | 6.59, 7.19, 8.04, 8.81, 9.37, 9.70, 10.50, 14.90 |
| l-Ala (B) | 7.16 | Acidic hydrolysate of 110+ dl-Leu (co-injection) (B) | 6.60, 6.76, 7.26, 8.04, 8.83, 9.67, 10.54, 15.02 |
| d-Ala (B) | 9.36 | Acidic hydrolysate of 110 + dl-pipecolic acid (co-injection) (B) | 6.58, 7.20, 8.09, 8.64, 8.84, 9.77, 10.64, 14.64 |
| l-Leu (B) | 6.78 | Acidic hydrolysate of 110 + N-Me-l-Leu (co-injection) (B) | 6.59, 7.20, 8.09, 8.83, 9.67, 10.57, 14.69 |
| d-Leu (B) | 9.67 | Acidic hydrolysate of 111 (A) | 1.91, 2.55, 2.86, 3.49, 3.89, 6.79 |
| N-Me-l-Leu (B) | 8.09 | Acidic hydrolysate of 111 + dl-Phe (co-injection) (A) | 1.87, 2.50, 2.89, 3.68, 5.01, 6.82 |
| l-Phe (A) | 3.81 | Acidic hydrolysate of 111 + dl-Pro (co-injection) (A) | 1.96, 2.60, 2.96, 3.52, 3,92, 6.70, 21.09 |
| d-Phe (A) | 5.00 | Acidic hydrolysate of 112 (A) | 1.93, 3.07, 3.80, 4.29, 4.60, 6.62 |
| l-Pro (A) | 6.72 | Acidic hydrolysate of 112 + dl-Phe (co-injection) (A) | 1.90, 3.10, 3.78, 4.39, 5.04, 6.70 |
| d-Pro (A) | 20.10 | Acidic hydrolysate of 112 + dl-Pro (co-injection) (A) | 2.04, 3.02, 3.72, 4.30, 4.60, 6.66, 19.40 |
| l-pipecolic acid (B) | 8.68 | Acidic hydrolysate of 112 + dl-Trp (co-injection) (A) | 1.93, 2.99, 3.70, 4.29, 4.60, 5.07, 6.33 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phyo, Y.Z.; Ribeiro, J.; Fernandes, C.; Kijjoa, A.; Pinto, M.M.M. Marine Natural Peptides: Determination of Absolute Configuration Using Liquid Chromatography Methods and Evaluation of Bioactivities. Molecules 2018, 23, 306. https://doi.org/10.3390/molecules23020306
Phyo YZ, Ribeiro J, Fernandes C, Kijjoa A, Pinto MMM. Marine Natural Peptides: Determination of Absolute Configuration Using Liquid Chromatography Methods and Evaluation of Bioactivities. Molecules. 2018; 23(2):306. https://doi.org/10.3390/molecules23020306
Chicago/Turabian StylePhyo, Ye’ Zaw, João Ribeiro, Carla Fernandes, Anake Kijjoa, and Madalena M. M. Pinto. 2018. "Marine Natural Peptides: Determination of Absolute Configuration Using Liquid Chromatography Methods and Evaluation of Bioactivities" Molecules 23, no. 2: 306. https://doi.org/10.3390/molecules23020306
APA StylePhyo, Y. Z., Ribeiro, J., Fernandes, C., Kijjoa, A., & Pinto, M. M. M. (2018). Marine Natural Peptides: Determination of Absolute Configuration Using Liquid Chromatography Methods and Evaluation of Bioactivities. Molecules, 23(2), 306. https://doi.org/10.3390/molecules23020306








