From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme mARC
Abstract
1. Biological Relevance of Molybdenum Cofactor
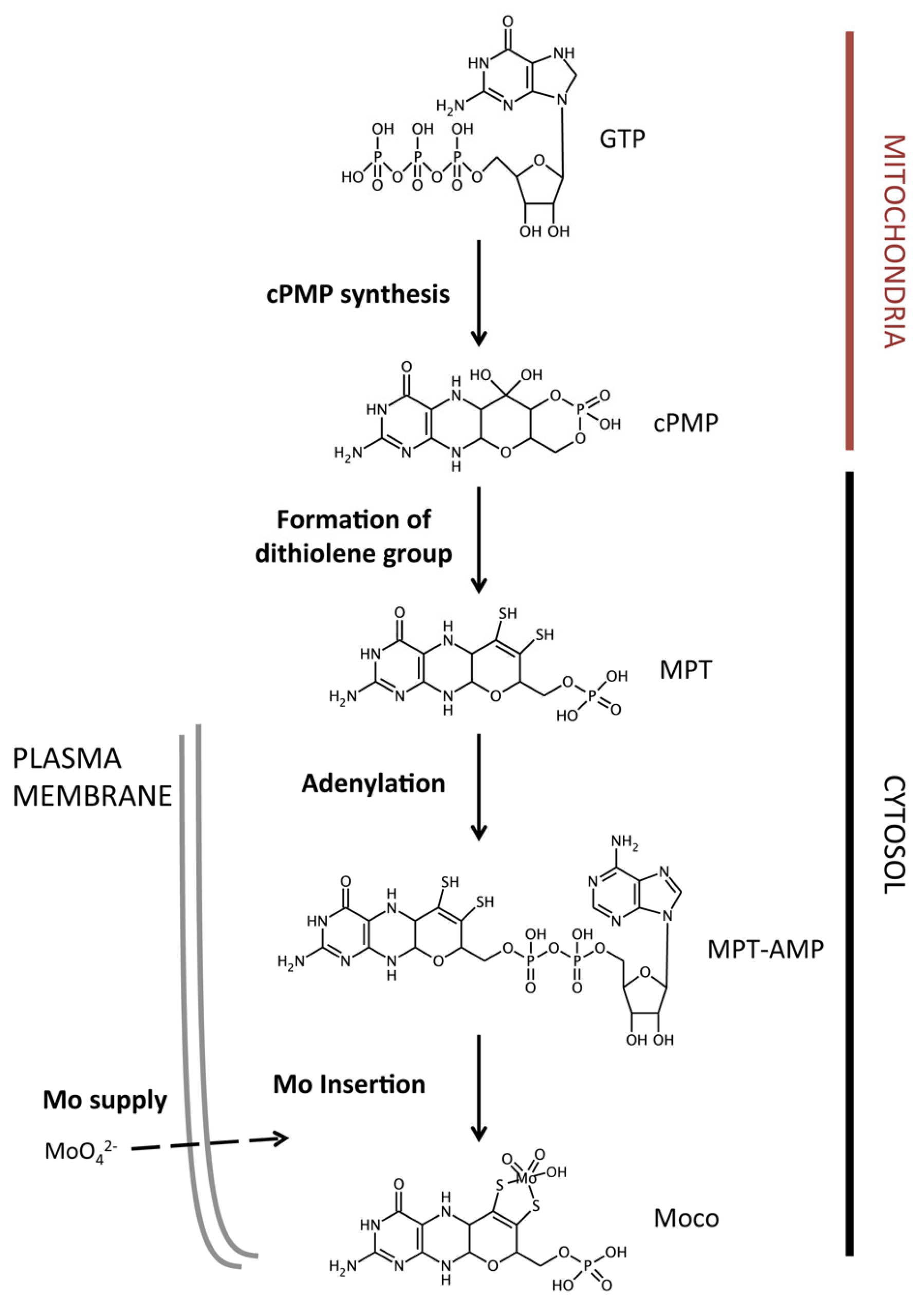
2. The Molybdenum Cofactor Biosynthetic Pathway
2.1. Molybdate Transport
2.2. Molybdopterin Synthesis
2.3. Molybdenum Insertion into Molybdopterin
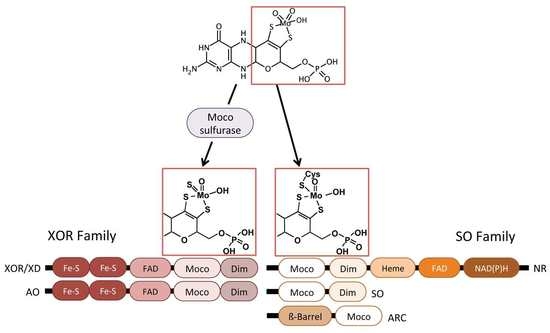
2.4. Molybdenum Cofactor Insertion
3. Eukaryotic Molybdenum Cofactor Enzymes
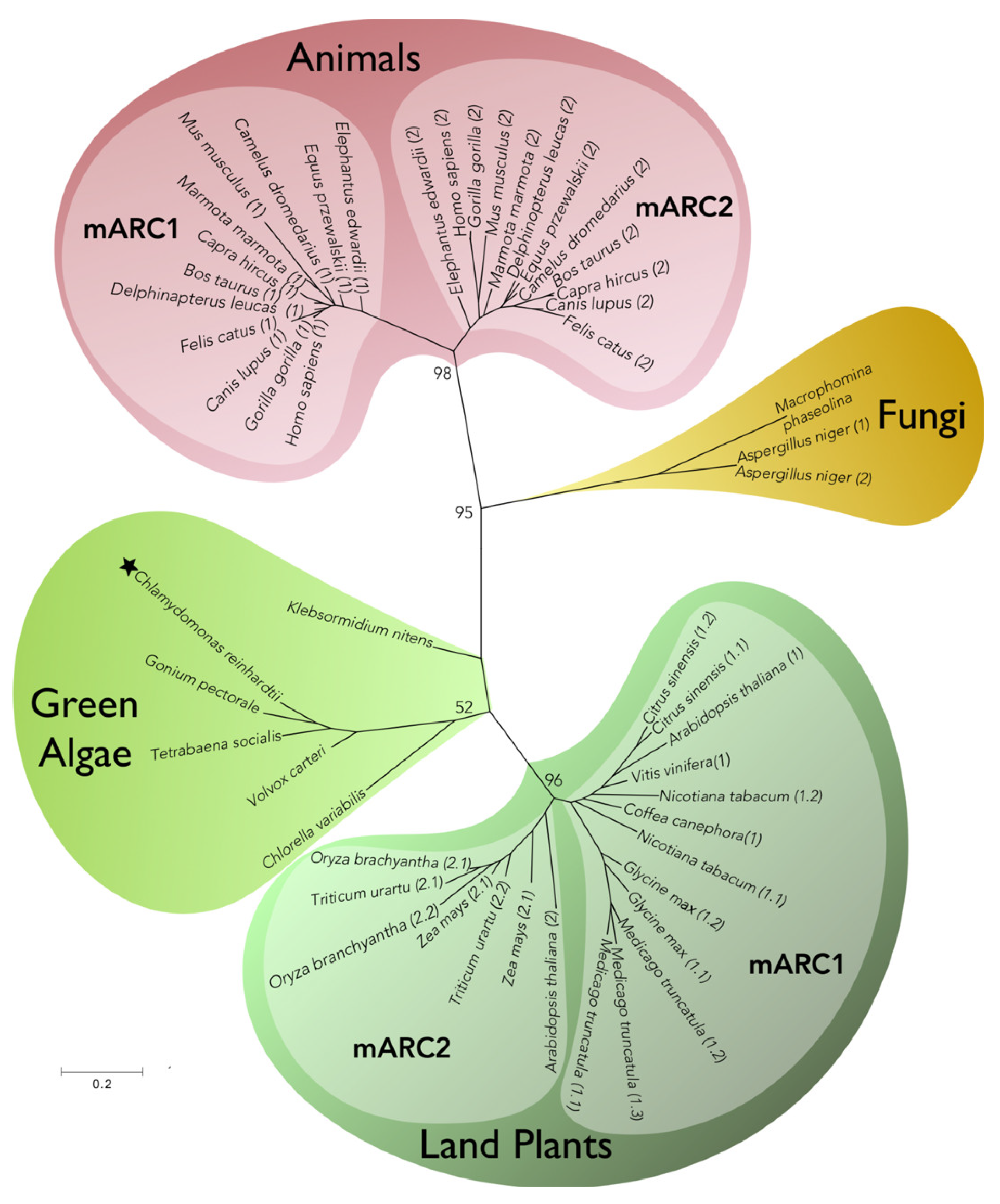
4. mARC More Than Just “One Enzyme”
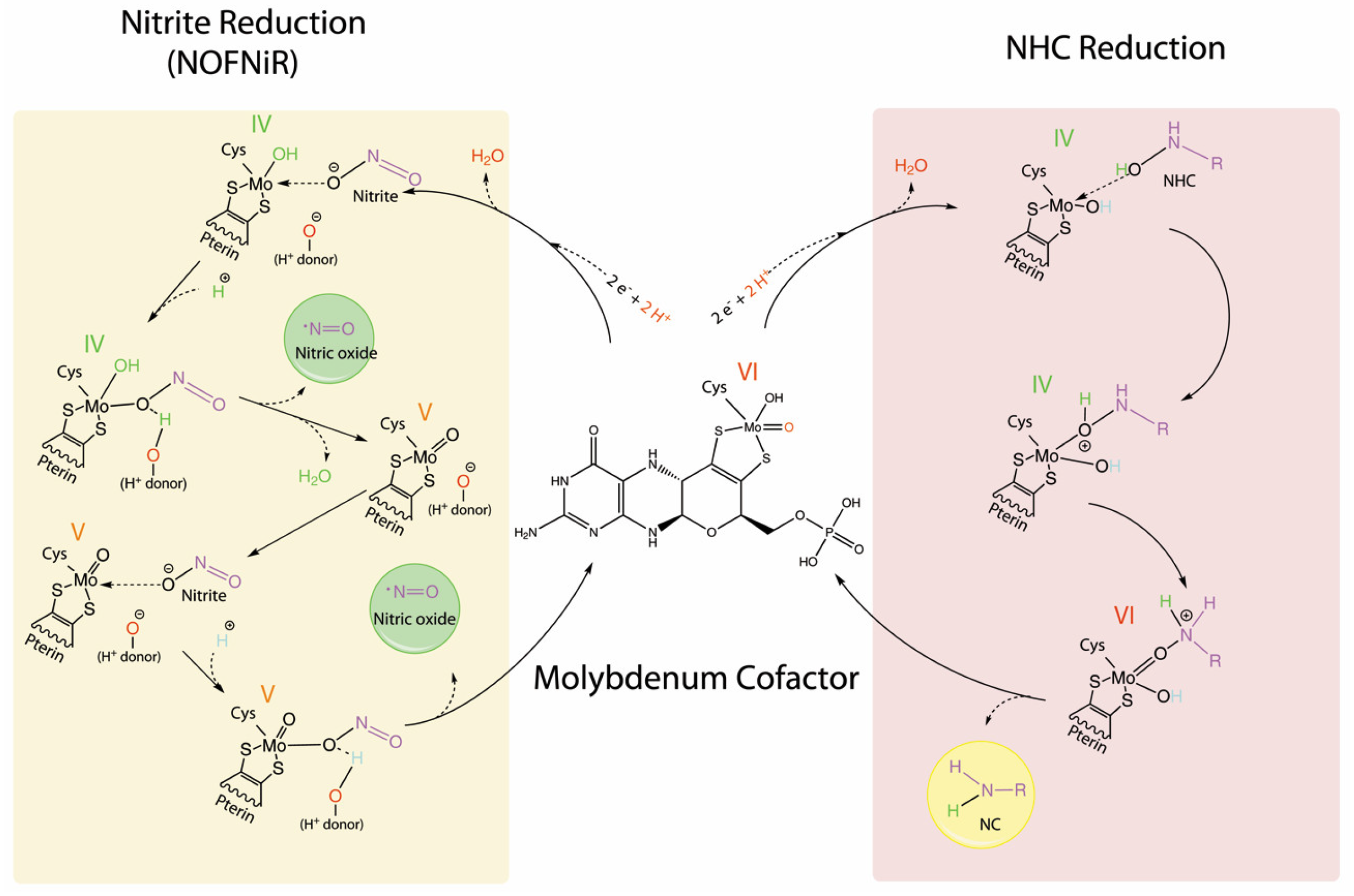
5. The NHC Reduction Capacity of mARC
6. The Nitrite Reduction Capacity of mARC
The mARC Reduction Mechanism of NHC Versus Nitrite
7. Are There Other mARC Substrates or Partners Still Uncovered?
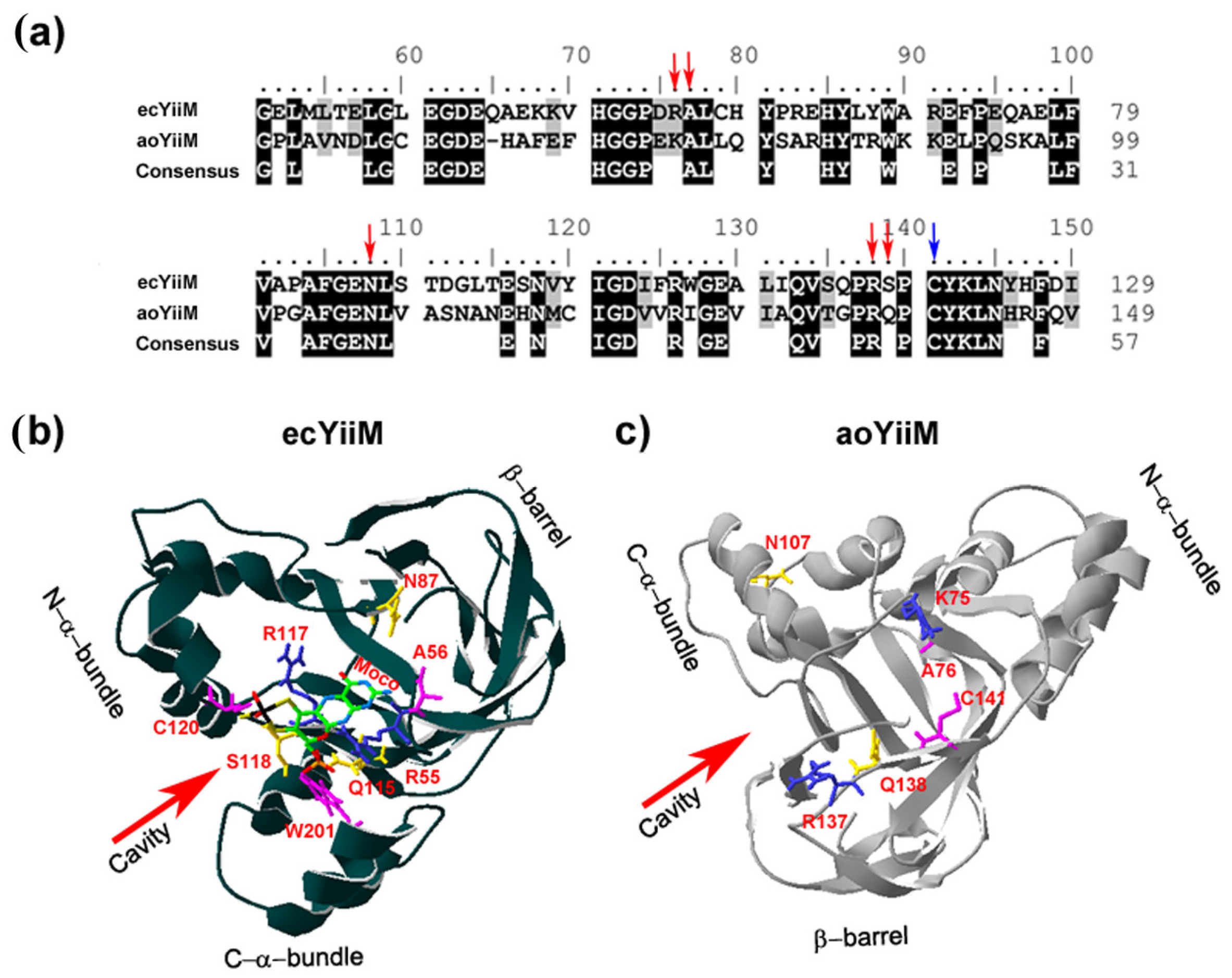
8. The Structural Modeling of a Eukaryotic MOSC Protein
9. Notes Added in Proof
Funding
Acknowledgments
Conflicts of Interest
References
- Arnon, D.I.; Stout, P.R. Molybdenum as an Essential Element for Higher Plants. Plant Physiol. 1939, 14, 599–602. [Google Scholar] [CrossRef] [PubMed]
- Leimkuhler, S.; Wuebbens, M.M.; Rajagopalan, K.V. The History of the Discovery of the Molybdenum Cofactor and Novel Aspects of its Biosynthesis in Bacteria. Coord. Chem. Rev. 2011, 255, 1129–1144. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Xu, Y.; Zhang, Y. Comparative genomics of molybdenum utilization in prokaryotes and eukaryotes. BMC Genom. 2018, 19, 691. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Jimenez, M.; Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Molybdenum metabolism in plants. Metallomics 2013, 5, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, B.N.; Gridley, K.L.; Ngaire Brady, J.; Phillips, T.; Tyerman, S.D. The role of molybdenum in agricultural plant production. Ann. Bot. 2005, 96, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A. Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr. Opin. Plant Biol. 2015, 25, 115–122. [Google Scholar] [CrossRef]
- Mendel, R.R.; Hansch, R. Molybdoenzymes and molybdenum cofactor in plants. J. Exp. Bot. 2002, 53, 1689–1698. [Google Scholar] [CrossRef]
- Verslues, P.E.; Zhu, J.K. Before and beyond ABA: Upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem. Soc. Trans. 2005, 33 Pt 2, 375–379. [Google Scholar] [CrossRef]
- Atwal, P.S.; Scaglia, F. Molybdenum cofactor deficiency. Mol. Genet. Metab. 2016, 117, 1–4. [Google Scholar] [CrossRef]
- Alvarez, H.M.; Xue, Y.; Robinson, C.D.; Canalizo-Hernandez, M.A.; Marvin, R.G.; Kelly, R.A.; Mondragon, A.; Penner-Hahn, J.E.; O’Halloran, T.V. Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation. Science 2010, 327, 331–334. [Google Scholar] [CrossRef]
- Llamas, A.; Tejada-Jimenez, M.; Fernandez, E.; Galvan, A. Molybdenum metabolism in the alga Chlamydomonas stands at the crossroad of those in Arabidopsis and humans. Metallomics 2011, 3, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R.; Leimkuhler, S. The biosynthesis of the molybdenum cofactors. J. Biol. Inorg. Chem. 2015, 20, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Bittner, F. Molybdenum metabolism in plants and crosstalk to iron. Front. Plant Sci. 2014, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Jimenez, M.; Llamas, A.; Sanz-Luque, E.; Galvan, A.; Fernandez, E. A high-affinity molybdate transporter in eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 20126–20130. [Google Scholar] [CrossRef] [PubMed]
- Tomatsu, H.; Takano, J.; Takahashi, H.; Watanabe-Takahashi, A.; Shibagaki, N.; Fujiwara, T. An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc. Natl. Acad. Sci. USA 2007, 104, 18807–18812. [Google Scholar] [CrossRef]
- Gasber, A.; Klaumann, S.; Trentmann, O.; Trampczynska, A.; Clemens, S.; Schneider, S.; Sauer, N.; Feifer, I.; Bittner, F.; Mendel, R.R.; et al. Identification of an Arabidopsis solute carrier critical for intracellular transport and inter-organ allocation of molybdate. Plant Biol. 2011, 13, 710–718. [Google Scholar] [CrossRef]
- Baxter, I.; Muthukumar, B.; Park, H.C.; Buchner, P.; Lahner, B.; Danku, J.; Zhao, K.; Lee, J.; Hawkesford, M.J.; Guerinot, M.L.; et al. Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet. 2008, 4, e1000004. [Google Scholar] [CrossRef]
- Duan, G.; Hakoyama, T.; Kamiya, T.; Miwa, H.; Lombardo, F.; Sato, S.; Tabata, S.; Chen, Z.; Watanabe, T.; Shinano, T.; et al. LjMOT1, a high-affinity molybdate transporter from Lotus japonicus, is essential for molybdate uptake, but not for the delivery to nodules. Plant J. 2017, 90, 1108–1119. [Google Scholar] [CrossRef]
- Tejada-Jimenez, M.; Gil-Diez, P.; Leon-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; Gonzalez-Guerrero, M. Medicago truncatula Molybdate Transporter type 1 (MtMOT1.3) is a plasma membrane molybdenum transporter required for nitrogenase activity in root nodules under molybdenum deficiency. New Phytol. 2017, 216, 1223–1235. [Google Scholar] [CrossRef]
- Gonzalez-Guerrero, M.; Escudero, V.; Saez, A.; Tejada-Jimenez, M. Transition Metal Transport in Plants and Associated Endosymbionts: Arbuscular Mycorrhizal Fungi and Rhizobia. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Gil-Diez, P.; Tejada-Jimenez, M.; Leon-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; Gonzalez-Guerrero, M. MtMOT1.2 is responsible for molybdate supply to Medicago truncatula nodules. Plant Cell Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Jimenez, M.; Galvan, A.; Fernandez, E. Algae and humans share a molybdate transporter. Proc. Natl. Acad. Sci. USA 2011, 108, 6420–6425. [Google Scholar] [CrossRef] [PubMed]
- Teschner, J.; Lachmann, N.; Schulze, J.; Geisler, M.; Selbach, K.; Santamaria-Araujo, J.; Balk, J.; Mendel, R.R.; Bittner, F. A novel role for Arabidopsis mitochondrial ABC transporter ATM3 in molybdenum cofactor biosynthesis. Plant Cell 2010, 22, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Santamaria-Araujo, J.A.; Fischer, B.; Otte, T.; Nimtz, M.; Mendel, R.R.; Wray, V.; Schwarz, G. The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J. Biol. Chem. 2004, 279, 15994–15999. [Google Scholar] [CrossRef] [PubMed]
- Santamaria-Araujo, J.A.; Wray, V.; Schwarz, G. Structure and stability of the molybdenum cofactor intermediate cyclic pyranopterin monophosphate. J. Biol. Inorg. Chem. 2012, 17, 113–122. [Google Scholar] [CrossRef]
- Hoff, T.; Schnorr, K.M.; Meyer, C.; Caboche, M. Isolation of two Arabidopsis cDNAs involved in early steps of molybdenum cofactor biosynthesis by functional complementation of Escherichia coli mutants. J. Biol. Chem. 1995, 270, 6100–6107. [Google Scholar] [CrossRef]
- Reiss, J.; Cohen, N.; Dorche, C.; Mandel, H.; Mendel, R.R.; Stallmeyer, B.; Zabot, M.T.; Dierks, T. Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat. Genet. 1998, 20, 51–53. [Google Scholar] [CrossRef]
- Kruse, I.; Maclean, A.E.; Hill, L.; Balk, J. Genetic dissection of cyclic pyranopterin monophosphate biosynthesis in plant mitochondria. Biochem. J. 2018, 475, 495–509. [Google Scholar] [CrossRef]
- Kaufholdt, D.; Gehl, C.; Geisler, M.; Jeske, O.; Voedisch, S.; Ratke, C.; Bollhoner, B.; Mendel, R.R.; Hansch, R. Visualization and quantification of protein interactions in the biosynthetic pathway of molybdenum cofactor in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 2005–2016. [Google Scholar] [CrossRef]
- Stallmeyer, B.; Drugeon, G.; Reiss, J.; Haenni, A.L.; Mendel, R.R. Human molybdopterin synthase gene: Identification of a bicistronic transcript with overlapping reading frames. Am. J. Hum. Genet. 1999, 64, 698–705. [Google Scholar] [CrossRef]
- Matthies, A.; Rajagopalan, K.V.; Mendel, R.R.; Leimkuhler, S. Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc. Natl. Acad. Sci. USA 2004, 101, 5946–5951. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Tejada-Jimenez, M.; Gonzalez-Ballester, D.; Higuera, J.J.; Schwarz, G.; Galvan, A.; Fernandez, E. Chlamydomonas reinhardtii CNX1E reconstitutes molybdenum cofactor biosynthesis in Escherichia coli mutants. Eukaryot. Cell 2007, 6, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Mendel, R.R.; Schwarz, G. Synthesis of adenylated molybdopterin: An essential step for molybdenum insertion. J. Biol. Chem. 2004, 279, 55241–55246. [Google Scholar] [CrossRef]
- Llamas, A.; Otte, T.; Multhaup, G.; Mendel, R.R.; Schwarz, G. The Mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. A novel route toward metal cofactor assembly. J. Biol. Chem. 2006, 281, 18343–18350. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Schulze, J.; Bittner, F.; Eilers, T.; Kuper, J.; Bollmann, G.; Nerlich, A.; Brinkmann, H.; Mendel, R.R. The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 2000, 12, 2455–2472. [Google Scholar] [CrossRef] [PubMed]
- Kaufholdt, D.; Baillie, C.K.; Bikker, R.; Burkart, V.; Dudek, C.A.; von Pein, L.; Rothkegel, M.; Mendel, R.R.; Hansch, R. The molybdenum cofactor biosynthesis complex interacts with actin filaments via molybdenum insertase Cnx1 as anchor protein in Arabidopsis thaliana. Plant Sci. 2016, 244, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Bittner, F.; Oreb, M.; Mendel, R.R. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 40381–40384. [Google Scholar] [CrossRef]
- Ichida, K.; Matsumura, T.; Sakuma, R.; Hosoya, T.; Nishino, T. Mutation of human molybdenum cofactor sulfurase gene is responsible for classical xanthinuria type II. Biochem. Biophys. Res. Commun. 2001, 282, 1194–1200. [Google Scholar] [CrossRef]
- Wollers, S.; Heidenreich, T.; Zarepour, M.; Zachmann, D.; Kraft, C.; Zhao, Y.; Mendel, R.R.; Bittner, F. Binding of sulfurated molybdenum cofactor to the C-terminal domain of ABA3 from Arabidopsis thaliana provides insight into the mechanism of molybdenum cofactor sulfuration. J. Biol. Chem. 2008, 283, 9642–9650. [Google Scholar] [CrossRef]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Calo, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef]
- Ataya, F.S.; Witte, C.P.; Galvan, A.; Igeno, M.I.; Fernandez, E. Mcp1 encodes the molybdenum cofactor carrier protein in Chlamydomonas reinhardtii and participates in protection, binding, and storage functions of the cofactor. J. Biol. Chem. 2003, 278, 10885–10890. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Llamas, A.; Tejada-Jimenez, M.; Schrader, N.; Kuper, J.; Ataya, F.S.; Galvan, A.; Mendel, R.R.; Fernandez, E.; Schwarz, G. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 2006, 281, 30186–30194. [Google Scholar] [CrossRef] [PubMed]
- Kruse, T.; Gehl, C.; Geisler, M.; Lehrke, M.; Ringel, P.; Hallier, S.; Hansch, R.; Mendel, R.R. Identification and biochemical characterization of molybdenum cofactor-binding proteins from Arabidopsis thaliana. J. Biol. Chem. 2010, 285, 6623–6635. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.P.; Igeno, M.I.; Mendel, R.; Schwarz, G.; Fernandez, E. The Chlamydomonas reinhardtii MoCo carrier protein is multimeric and stabilizes molybdopterin cofactor in a molybdate charged form. FEBS Lett. 1998, 431, 205–209. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, A.; Galvan, A.; Fernandez, E. Nitrate Reductase Regulates Plant Nitric Oxide Homeostasis. Trends Plant Sci. 2017, 22, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Hille, R. The Mononuclear Molybdenum Enzymes. Chem. Rev. 1996, 96, 2757–2816. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Luque, E.; Ocana-Calahorro, F.; de Montaigu, A.; Chamizo-Ampudia, A.; Llamas, A.; Galvan, A.; Fernandez, E. THB1, a truncated hemoglobin, modulates nitric oxide levels and nitrate reductase activity. Plant J. 2015, 81, 467–479. [Google Scholar] [CrossRef]
- Feng, C.; Tollin, G.; Enemark, J.H. Sulfite oxidizing enzymes. Biochim. Biophys. Acta 2007, 1774, 527–539. [Google Scholar] [CrossRef]
- Kisker, C.; Schindelin, H.; Rees, D.C. Molybdenum-cofactor-containing enzymes: Structure and mechanism. Annu. Rev. Biochem. 1997, 66, 233–267. [Google Scholar] [CrossRef]
- Cohen, H.J.; Fridovich, I.; Rajagopalan, K.V. Hepatic sulfite oxidase. A functional role for molybdenum. J. Biol. Chem. 1971, 246, 374–382. [Google Scholar] [PubMed]
- Randewig, D.; Hamisch, D.; Herschbach, C.; Eiblmeier, M.; Gehl, C.; Jurgeleit, J.; Skerra, J.; Mendel, R.R.; Rennenberg, H.; Hansch, R. Sulfite oxidase controls sulfur metabolism under SO2 exposure in Arabidopsis thaliana. Plant Cell Environ. 2012, 35, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Luniak, N.; Witt, C.; Wustefeld, Y.; Wachter, A.; Mendel, R.R.; Hansch, R. Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol. 2004, 45, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Eilers, T.; Schwarz, G.; Brinkmann, H.; Witt, C.; Richter, T.; Nieder, J.; Koch, B.; Hille, R.; Hansch, R.; Mendel, R.R. Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J. Biol. Chem. 2001, 276, 46989–46994. [Google Scholar] [CrossRef] [PubMed]
- Gerin, S.; Mathy, G.; Blomme, A.; Franck, F.; Sluse, F.E. Plasticity of the mitoproteome to nitrogen sources (nitrate and ammonium) in Chlamydomonas reinhardtii: The logic of Aox1 gene localization. Biochim. Biophys. Acta 2010, 1797, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Yesbergenova, Z.; Yang, G.; Oron, E.; Soffer, D.; Fluhr, R.; Sagi, M. The plant Mo-hydroxylases aldehyde oxidase and xanthine dehydrogenase have distinct reactive oxygen species signatures and are induced by drought and abscisic acid. Plant J. 2005, 42, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Zarepour, M.; Kaspari, K.; Stagge, S.; Rethmeier, R.; Mendel, R.R.; Bittner, F. Xanthine dehydrogenase AtXDH1 from Arabidopsis thaliana is a potent producer of superoxide anions via its NADH oxidase activity. Plant Mol. Biol. 2010, 72, 301–310. [Google Scholar] [CrossRef]
- Ma, X.; Wang, W.; Bittner, F.; Schmidt, N.; Berkey, R.; Zhang, L.; King, H.; Zhang, Y.; Feng, J.; Wen, Y.; et al. Dual and Opposing Roles of Xanthine Dehydrogenase in Defense-Associated Reactive Oxygen Species Metabolism in Arabidopsis. Plant Cell 2016, 28, 1108–1126. [Google Scholar] [CrossRef]
- Rodriguez-Trelles, F.; Tarrio, R.; Ayala, F.J. Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc. Natl. Acad. Sci. USA 2003, 100, 13413–13417. [Google Scholar] [CrossRef]
- Coelho, C.; Mahro, M.; Trincao, J.; Carvalho, A.T.; Ramos, M.J.; Terao, M.; Garattini, E.; Leimkuhler, S.; Romao, M.J. The first mammalian aldehyde oxidase crystal structure: Insights into substrate specificity. J. Biol. Chem. 2012, 287, 40690–40702. [Google Scholar] [CrossRef]
- Garattini, E.; Mendel, R.; Romao, M.J.; Wright, R.; Terao, M. Mammalian molybdo-flavoenzymes, an expanding family of proteins: Structure, genetics, regulation, function and pathophysiology. Biochem. J. 2003, 372 Pt 1, 15–32. [Google Scholar] [CrossRef]
- Seo, M.; Peeters, A.J.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Pavlov, Y.I.; Dunn, R.L.; Schaaper, R.M. Hypersensitivity of Escherichia coli Delta(uvrB-bio) mutants to 6-hydroxylaminopurine and other base analogs is due to a defect in molybdenum cofactor biosynthesis. J. Bacteriol. 2000, 182, 3361–3367. [Google Scholar] [CrossRef]
- Havemeyer, A.; Bittner, F.; Wollers, S.; Mendel, R.; Kunze, T.; Clement, B. Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 2006, 281, 34796–34802. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. The Chlamydomonas reinhardtii molybdenum cofactor enzyme crARC has a Zn-dependent activity and protein partners similar to those of its human homologue. Eukaryot. Cell 2011, 10, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.I.; Castruita, M.; Malasarn, D.; Urzica, E.; Erde, J.; Page, M.D.; Yamasaki, H.; Casero, D.; Pellegrini, M.; Merchant, S.S.; et al. The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol. Cell. Proteom. 2013, 12, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Wahl, B.; Reichmann, D.; Niks, D.; Krompholz, N.; Havemeyer, A.; Clement, B.; Messerschmidt, T.; Rothkegel, M.; Biester, H.; Hille, R.; et al. Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J. Biol. Chem. 2010, 285, 37847–37859. [Google Scholar] [CrossRef]
- Klein, J.M.; Busch, J.D.; Potting, C.; Baker, M.J.; Langer, T.; Schwarz, G. The mitochondrial amidoxime-reducing component (mARC1) is a novel signal-anchored protein of the outer mitochondrial membrane. J. Biol. Chem. 2012, 287, 42795–42803. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. Study of Different Variants of Mo Enzyme crARC and the Interaction with Its Partners crCytb5-R and crCytb5-1. Int. J. Mol. Sci. 2017, 18, 670. [Google Scholar] [CrossRef]
- Anantharaman, V.; Aravind, L. MOSC domains: Ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including Molybdenum cofactor sulfurases. FEMS Microbiol. Lett. 2002, 207, 55–61. [Google Scholar] [CrossRef]
- Giles, L.J.; Ruppelt, C.; Yang, J.; Mendel, R.R.; Bittner, F.; Kirk, M.L. Molybdenum site structure of MOSC family proteins. Inorg. Chem. 2014, 53, 9460–9462. [Google Scholar] [CrossRef] [PubMed]
- Kubitza, C.; Ginsel, C.; Bittner, F.; Havemeyer, A.; Clement, B.; Scheidig, A.J. T4 lysozyme-facilitated crystallization of the human molybdenum cofactor-dependent enzyme mARC. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74 Pt 6, 337–344. [Google Scholar] [CrossRef]
- Llamas, A.; Chamizo-Ampudia, A.; Tejada-Jimenez, M.; Galvan, A.; Fernandez, E. The molybdenum cofactor enzyme mARC: Moonlighting or promiscuous enzyme? Biofactors 2017, 43, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Moonlighting proteins. Trends Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef]
- Franco-Serrano, L.; Hernández, S.; Calvo, A.; Severi, M.A.; Ferragut, G.; Pérez-Pons, J.; Piñol, J.; Pich, Ò.; Mozo-Villarias, Á.; Amela, I.; et al. MultitaskProtDB-II: An update of a database of multitasking/moonlighting proteins. Nucleic Acids Res. 2018, 46, D645–D648. [Google Scholar] [CrossRef] [PubMed]
- Gruenewald, S.; Wahl, B.; Bittner, F.; Hungeling, H.; Kanzow, S.; Kotthaus, J.; Schwering, U.; Mendel, R.R.; Clement, B. The fourth molybdenum containing enzyme mARC: Cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 2008, 51, 8173–8177. [Google Scholar] [CrossRef]
- D’Arrigo, A.; Manera, E.; Longhi, R.; Borgese, N. The specific subcellular localization of two isoforms of cytochrome b5 suggests novel targeting pathways. J. Biol. Chem. 1993, 268, 2802–2808. [Google Scholar] [PubMed]
- Islinger, M.; Lüers, G.H.; Li, K.W.; Loos, M.; Völkl, A. Rat liver peroxisomes after fibrate treatment. A survey using quantitative mass spectrometry. J. Biol. Chem. 2007, 282, 23055–23069. [Google Scholar] [CrossRef] [PubMed]
- Da Cruz, S.; Xenarios, I.; Langridge, J.; Vilbois, F.; Parone, P.A.; Martinou, J.C. Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 2003, 278, 41566–41571. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Leroy, P.; Pavlov, Y.I.; Schaaper, R.M. YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol. Microbiol. 2008, 68, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Wang, J.; Schaaper, R.M. Role for CysJ flavin reductase in molybdenum cofactor-dependent resistance of Escherichia coli to 6-N-hydroxylaminopurine. J. Bacteriol. 2010, 192, 2026–2033. [Google Scholar] [CrossRef] [PubMed]
- Krompholz, N.; Krischkowski, C.; Reichmann, D.; Garbe-Schönberg, D.; Mendel, R.R.; Bittner, F.; Clement, B.; Havemeyer, A. The mitochondrial Amidoxime Reducing Component (mARC) is involved in detoxification of N-hydroxylated base analogues. Chem. Res. Toxicol. 2012, 25, 2443–2450. [Google Scholar] [CrossRef] [PubMed]
- Plitzko, B.; Havemeyer, A.; Kunze, T.; Clement, B. The pivotal role of the mitochondrial amidoxime reducing component 2 in protecting human cells against apoptotic effects of the base analog N6-hydroxylaminopurine. J. Biol. Chem. 2015, 290, 10126–10135. [Google Scholar] [CrossRef] [PubMed]
- Kozmin, S.G.; Schaaper, R.M. Molybdenum cofactor-dependent resistance to N-hydroxylated base analogs in Escherichia coli is independent of MobA function. Mutat. Res. 2007, 619, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Ott, G.; Plitzko, B.; Krischkowski, C.; Reichmann, D.; Bittner, F.; Mendel, R.R.; Kunze, T.; Clement, B.; Havemeyer, A. Reduction of sulfamethoxazole hydroxylamine (SMX-HA) by the mitochondrial amidoxime reducing component (mARC). Chem. Res. Toxicol. 2014, 27, 1687–1695. [Google Scholar] [CrossRef] [PubMed]
- Mikula, M.; Rubel, T.; Karczmarski, J.; Goryca, K.; Dadlez, M.; Ostrowski, J. Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct. Integr. Genom. 2011, 11, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Havemeyer, A.; Lang, J.; Clement, B. The fourth mammalian molybdenum enzyme mARC: Current state of research. Drug Metab. Rev. 2011, 43, 524–539. [Google Scholar] [CrossRef]
- Abbenante, G.; Fairlie, D.P. Protease inhibitors in the clinic. Med. Chem. 2005, 1, 71–104. [Google Scholar] [CrossRef]
- Peterlin-Masic, L.; Cesar, J.; Zega, A. Metabolism-directed optimisation of antithrombotics: The prodrug principle. Curr. Pharm. Des. 2006, 12, 73–91. [Google Scholar] [CrossRef]
- Ott, G.; Havemeyer, A.; Clement, B. The mammalian molybdenum enzymes of mARC. J. Biol. Inorg. Chem. 2015, 20, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, D.; Elg, M. The pharmacodynamics and pharmacokinetics of the oral direct thrombin inhibitor ximelagatran and its active metabolite melagatran: A mini-review. Thromb. Res. 2003, 109 (Suppl. 1), S9–S15. [Google Scholar] [CrossRef]
- Neve, E.P.; Köfeler, H.; Hendriks, D.F.; Nordling, Å.; Gogvadze, V.; Mkrtchian, S.; Näslund, E.; Ingelman-Sundberg, M. Expression and Function of mARC: Roles in Lipogenesis and Metabolic Activation of Ximelagatran. PLoS ONE 2015, 10, e0138487. [Google Scholar] [CrossRef]
- Froriep, D.; Clement, B.; Bittner, F.; Mendel, R.R.; Reichmann, D.; Schmalix, W.; Havemeyer, A. Activation of the anti-cancer agent upamostat by the mARC enzyme system. Xenobiotica 2013, 43, 780–784. [Google Scholar] [CrossRef]
- Jakobs, H.H.; Froriep, D.; Havemeyer, A.; Mendel, R.R.; Bittner, F.; Clement, B. The mitochondrial amidoxime reducing component (mARC): Involvement in metabolic reduction of N-oxides, oximes and N-hydroxyamidinohydrazones. ChemMedChem 2014, 9, 2381–2387. [Google Scholar] [CrossRef] [PubMed]
- Ginsel, C.; Plitzko, B.; Froriep, D.; Stolfa, D.A.; Jung, M.; Kubitza, C.; Scheidig, A.J.; Havemeyer, A.; Clement, B. The Involvement of the Mitochondrial Amidoxime Reducing Component (mARC) in the Reductive Metabolism of Hydroxamic Acids. Drug Metab. Dispos. 2018, 46, 1396–1402. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Girreser, U.; Havemeyer, A.; Bittner, F.; Clement, B. Detoxification of Trimethylamine N-Oxide by the Mitochondrial Amidoxime Reducing Component mARC. Chem. Res. Toxicol. 2018, 31, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef]
- Feelisch, M.; Martin, J.F. The early role of nitric oxide in evolution. Trends Ecol. Evol. 1995, 10, 496–499. [Google Scholar] [CrossRef]
- Lamattina, L.; García-Mata, C.; Graziano, M.; Pagnussat, G. Nitric oxide: The versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 2003, 54, 109–136. [Google Scholar] [CrossRef]
- Kotthaus, J.; Wahl, B.; Havemeyer, A.; Schade, D.; Garbe-Schönberg, D.; Mendel, R.; Bittner, F.; Clement, B. Reduction of N(ω)-hydroxy-L-arginine by the mitochondrial amidoxime reducing component (mARC). Biochem. J. 2011, 433, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Cosby, K.; Partovi, K.S.; Crawford, J.H.; Patel, R.P.; Reiter, C.D.; Martyr, S.; Yang, B.K.; Waclawiw, M.A.; Zalos, G.; Xu, X.; et al. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat. Med. 2003, 9, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Tejero, J.; Gladwin, M.T. The globin superfamily: Functions in nitric oxide formation and decay. Biol. Chem. 2014, 395, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Tiso, M.; Tejero, J.; Basu, S.; Azarov, I.; Wang, X.; Simplaceanu, V.; Frizzell, S.; Jayaraman, T.; Geary, L.; Shapiro, C.; et al. Human neuroglobin functions as a redox-regulated nitrite reductase. J. Biol. Chem. 2011, 286, 18277–18289. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Azarova, N.A.; Font, M.D.; King, S.B.; Hogg, N.; Gladwin, M.T.; Shiva, S.; Kim-Shapiro, D.B. Nitrite reductase activity of cytochrome c. J. Biol. Chem. 2008, 283, 32590–32597. [Google Scholar] [CrossRef] [PubMed]
- Stöhr, C.; Strube, F.; Marx, G.; Ullrich, W.R.; Rockel, P. A plasma membrane-bound enzyme of tobacco roots catalyses the formation of nitric oxide from nitrite. Planta 2001, 212, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.; Schwarz, G. Nitrite-dependent nitric oxide synthesis by molybdenum enzymes. FEBS Lett. 2018. [Google Scholar] [CrossRef]
- Mur, L.A.; Mandon, J.; Persijn, S.; Cristescu, S.M.; Moshkov, I.E.; Novikova, G.V.; Hall, M.A.; Harren, F.J.; Hebelstrup, K.H.; Gupta, K.J. Nitric oxide in plants: An assessment of the current state of knowledge. AoB PLANTS 2013, 5, pls052. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.E.; Tejero, J.; Sun, B.; Gauthier, M.C.; Thomas, J.; Ragireddy, V.; Merchant, B.A.; Wang, J.; Azarov, I.; Basu, P.; et al. Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 2014, 289, 10345–10358. [Google Scholar] [CrossRef]
- Campbell, W.H. Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci. 2001, 58, 194–204. [Google Scholar] [CrossRef]
- Rockel, P.; Strube, F.; Rockel, A.; Wildt, J.; Kaiser, W.M. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 2002, 53, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Moura, J.J. Nitrite reduction by xanthine oxidase family enzymes: A new class of nitrite reductases. J. Biol. Inorg. Chem. 2011, 16, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Moura, J.J. Nitrite reduction by molybdoenzymes: A new class of nitric oxide-forming nitrite reductases. J. Biol. Inorg. Chem. 2015, 20, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Giles, L.J.; Ruppelt, C.; Mendel, R.R.; Bittner, F.; Kirk, M.L. Oxyl and hydroxyl radical transfer in mitochondrial amidoxime reducing component-catalyzed nitrite reduction. J. Am. Chem. Soc. 2015, 137, 5276–5279. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R.; Bittner, F. Cell biology of molybdenum. Biochim. Biophys. Acta 2006, 1763, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Neve, E.P.; Nordling, A.; Andersson, T.B.; Hellman, U.; Diczfalusy, U.; Johansson, I.; Ingelman-Sundberg, M. Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 2012, 287, 6307–6317. [Google Scholar] [CrossRef] [PubMed]
- Andersson, S.; Hofmann, Y.; Nordling, A.; Li, X.Q.; Nivelius, S.; Andersson, T.B.; Ingelman-Sundberg, M.; Johansson, I. Characterization and partial purification of the rat and human enzyme systems active in the reduction of N-hydroxymelagatran and benzamidoxime. Drug Metab. Dispos. 2005, 33, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.N.; Rossios, C.; Al-Kafaji, G.; Shah, A.; Page, R.A. Glucose regulation of CDK7, a putative thiol related gene, in experimental diabetic nephropathy. Biochem. Biophys. Res. Commun. 2007, 357, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, X.; Wang, P.; Liu, Y.; Zhang, Z.; Zhao, G.; Xu, H.; Zhu, J.; Qin, X.; Chen, S.; et al. Novel association strategy with copy number variation for identifying new risk Loci of human diseases. PLoS ONE 2010, 5, e12185. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Padilla, M.N.; Begara-Morales, J.C.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitro-fatty acids in plant signaling: New key mediators of nitric oxide metabolism. Redox Biol. 2017, 11, 554–561. [Google Scholar] [CrossRef]
- Martin-Montalvo, A.; Sun, Y.; Diaz-Ruiz, A.; Ali, A.; Gutierrez, V.; Palacios, H.H.; Curtis, J.; Siendones, E.; Ariza, J.; Abulwerdi, G.A.; et al. Cytochrome. NPJ Aging Mech. Dis. 2016, 2, 16006. [Google Scholar] [CrossRef] [PubMed]
- Namgung, B.; Kim, J.H.; Song, W.S.; Yoon, S.I. Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site. Sci. Rep. 2018, 8, 3304. [Google Scholar] [CrossRef] [PubMed]
- Kubitza, C.; Bittner, F.; Ginsel, C.; Havemeyer, A.; Clement, B.; Scheidig, A.J. Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes. Proc. Natl. Acad. Sci. USA 2018, 115, 11958–11963. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tejada-Jimenez, M.; Chamizo-Ampudia, A.; Calatrava, V.; Galvan, A.; Fernandez, E.; Llamas, A. From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme mARC. Molecules 2018, 23, 3287. https://doi.org/10.3390/molecules23123287
Tejada-Jimenez M, Chamizo-Ampudia A, Calatrava V, Galvan A, Fernandez E, Llamas A. From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme mARC. Molecules. 2018; 23(12):3287. https://doi.org/10.3390/molecules23123287
Chicago/Turabian StyleTejada-Jimenez, Manuel, Alejandro Chamizo-Ampudia, Victoria Calatrava, Aurora Galvan, Emilio Fernandez, and Angel Llamas. 2018. "From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme mARC" Molecules 23, no. 12: 3287. https://doi.org/10.3390/molecules23123287
APA StyleTejada-Jimenez, M., Chamizo-Ampudia, A., Calatrava, V., Galvan, A., Fernandez, E., & Llamas, A. (2018). From the Eukaryotic Molybdenum Cofactor Biosynthesis to the Moonlighting Enzyme mARC. Molecules, 23(12), 3287. https://doi.org/10.3390/molecules23123287