In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transfection, and Drug Treatments
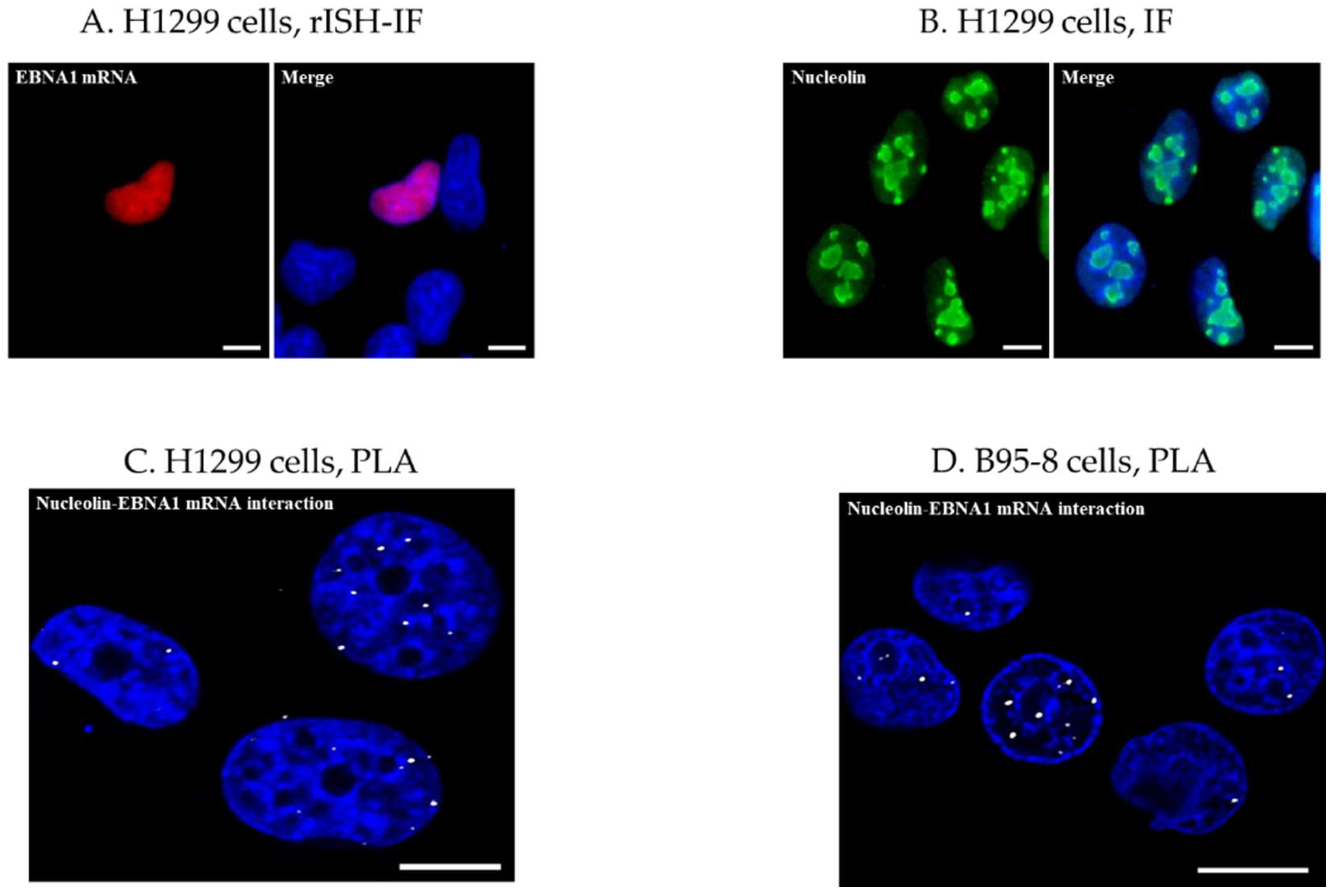
2.2. RNA In Situ Hybridization-Immunofluorescence (rISH-IF) and Immunofluorescence (IF)
2.3. Sequence of EBNA1 cDNA
2.4. Proximity Ligation Assay (PLA)
2.5. Microscopy and Image Analysis
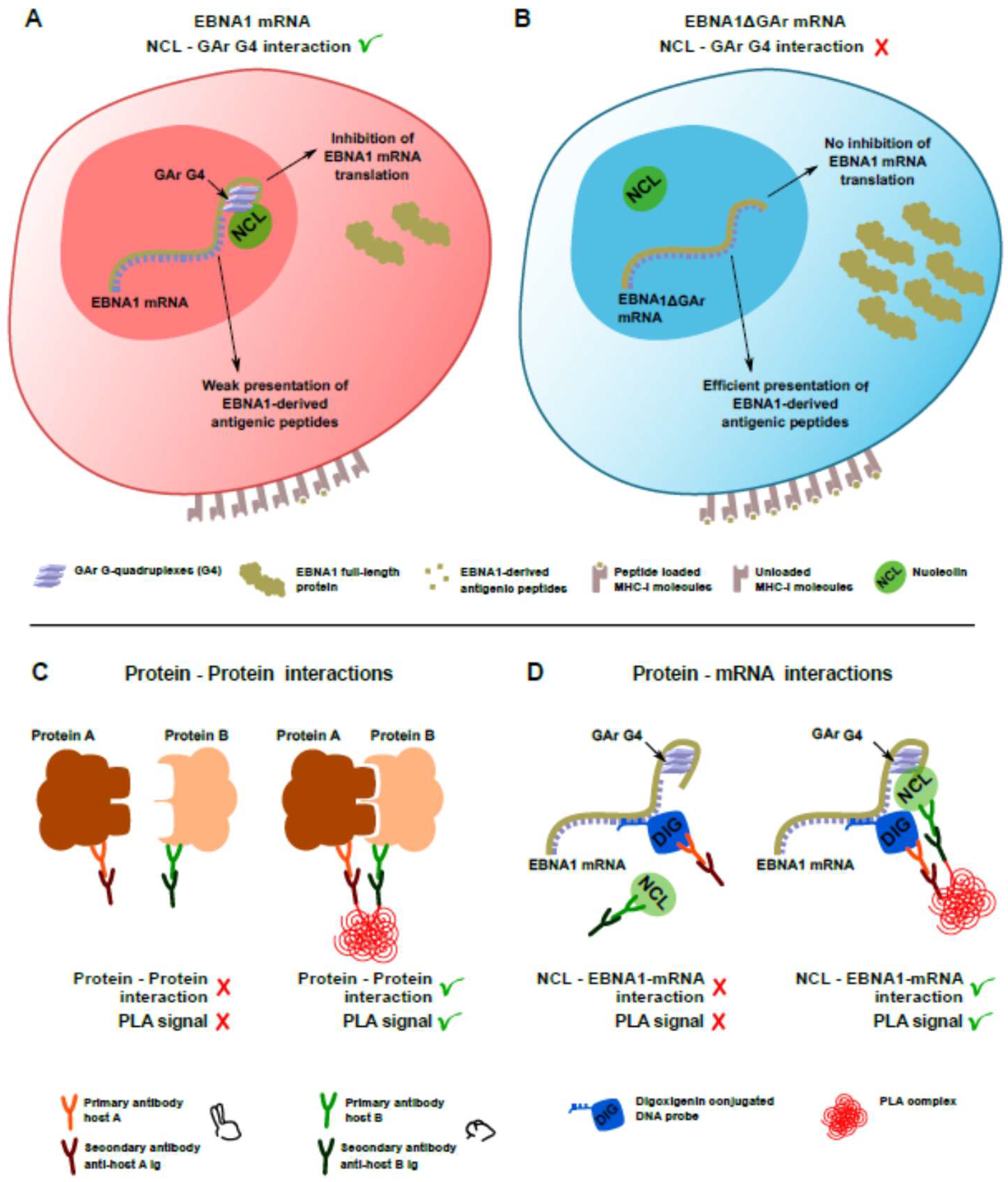
3. Results and Discussion
3.1. Detection of EBNA1 mRNA and NCL by rISH-IF and IF
3.2. Analysis of NCL-EBNA1 mRNA Interactions by Proximity Ligation
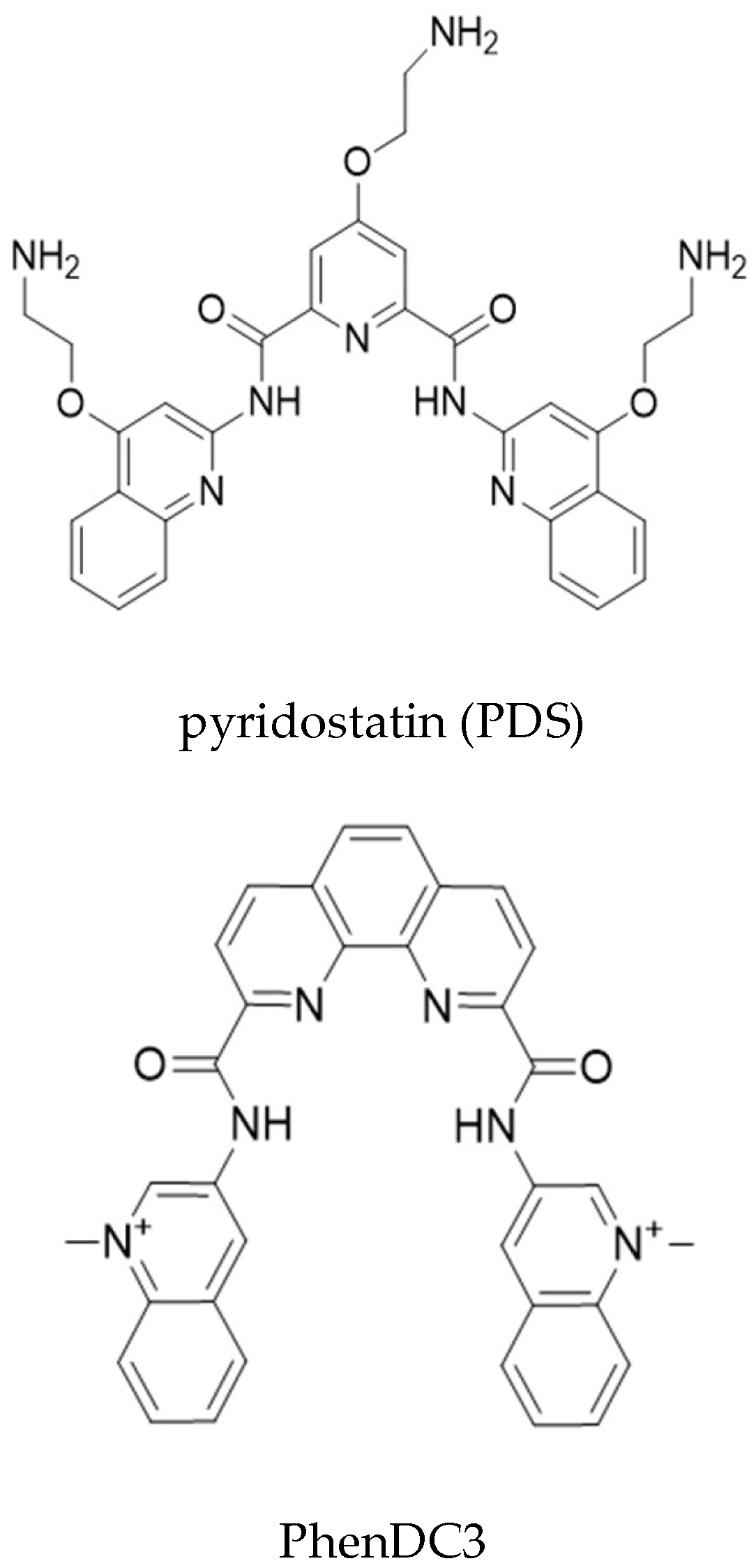
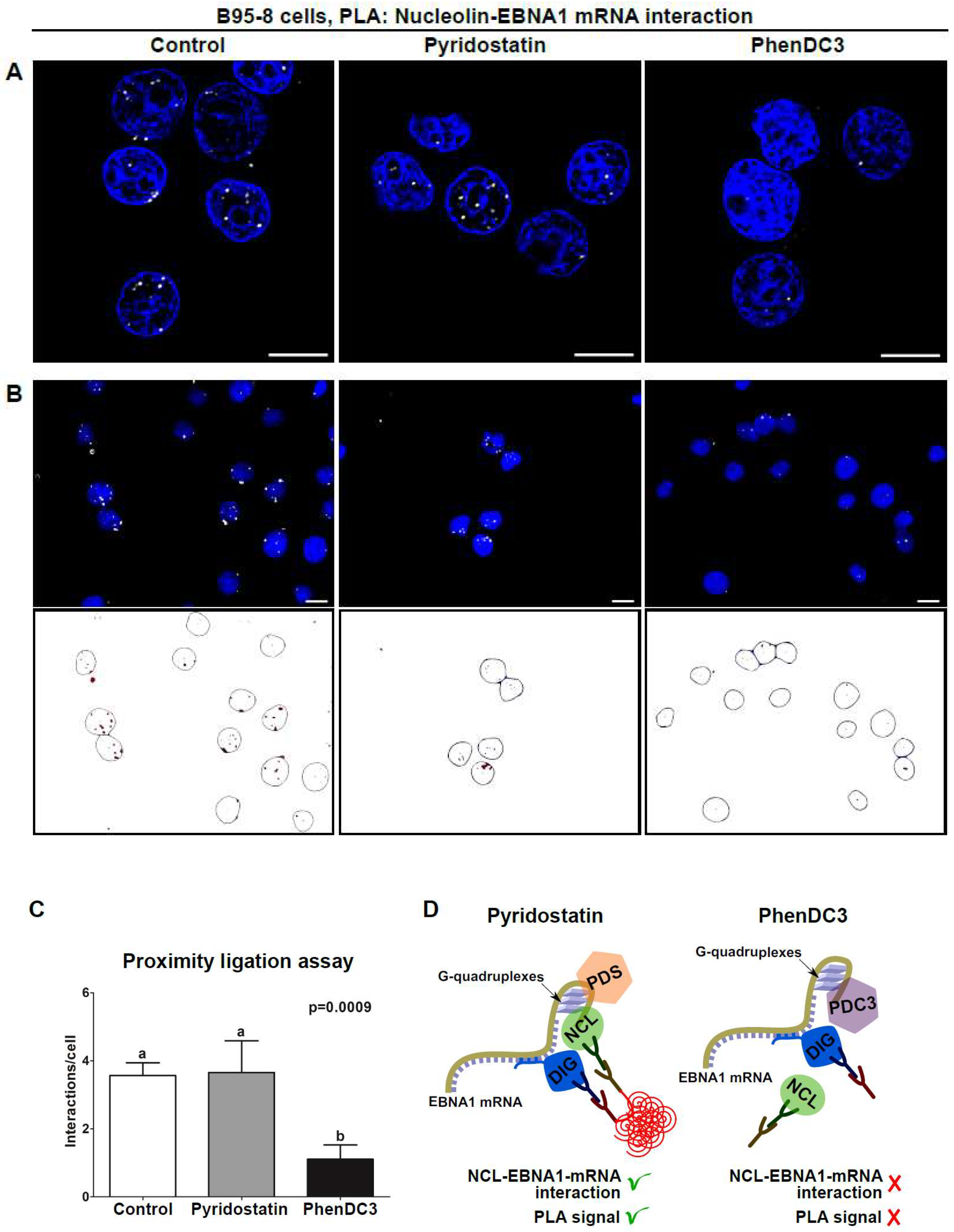
3.3. Use of Quantitative PLA to Evaluate the Effect of RNA-Binding Drugs on NCL-EBNA1 mRNA Interactions
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Beckmann, B.M.; Castello, A.; Medenbach, J. The expanding universe of ribonucleoproteins: Of novel RNA-binding proteins and unconventional interactions. Pflugers Arch. 2016, 468, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.L.; Ishizuka, T.; Sakamoto, T.; Fujimoto, K.; Uechi, T.; Kenmochi, N.; Xu, Y. Characterization of human telomere RNA G-quadruplex structures in vitro and in living cells using 19F NMR spectroscopy. Nucleic Acids Res. 2017, 45, 5501–5511. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.L.; Xu, Y. Investigation of higher-order RNA G-quadruplex structures in vitro and in living cells by (19)F NMR spectroscopy. Nat. Protoc. 2018, 13, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.; Shaw, P.G.; Kim, M.S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P. Neurodegenerative diseases: G-quadruplex poses quadruple threat. Nature 2014, 507, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. RNA G-quadruplexes cause eIF4a-dependent oncogene translation in cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Granzhan, A.; Hamon, F.; Verga, D.; Teulade-Fichou, M.P. Visualizing the quadruplex: From fluorescent ligands to light-up probes. Top Curr. Chem. 2013, 330, 111–177. [Google Scholar] [PubMed]
- Mendoza, O.; Bourdoncle, A.; Boule, J.B.; Brosh, R.M., Jr.; Mergny, J.L. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Perreault, J.P.; Topisirovic, I.; Richard, S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation (Austin) 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Martins, R.P.; Angrand, G.; Quillevere, A.; Daskalogianni, C.; Voisset, C.; Teulade-Fichou, M.P.; Fahraeus, R.; Blondel, M. A yeast model for the mechanism of the Epstein-Barr virus immune evasion identifies a new therapeutic target to interfere with the virus stealthiness. Microb. Cell. 2017, 4, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Martins, R.P.; Billant, O.; Contesse, M.A.; Findakly, S.; Pochard, P.; Daskalogianni, C.; Beauvineau, C.; Guetta, C.; Jamin, C.; et al. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Delemos, E.; Mergny, J.L.; Teulade-Fichou, M.P.; Monchaud, D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Manoury, B.; Fahraeus, R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science 2003, 301, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded Nuclear Antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Soderberg, O.; Gullberg, M.; Jarvius, M.; Ridderstrale, K.; Leuchowius, K.J.; Jarvius, J.; Wester, K.; Hydbring, P.; Bahram, F.; Larsson, L.G.; et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 2006, 3, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.; Shankman, L.S.; Nguyen, A.T.; Owens, G.K. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods 2013, 10, 171–177. [Google Scholar] [CrossRef] [PubMed]
- de Klerk, E.; t Hoen, P.A. Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet. 2015, 31, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.F.; Parker, R. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014, 54, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Daskalogianni, C.; Pyndiah, S.; Apcher, S.; Mazars, A.; Manoury, B.; Ammari, N.; Nylander, K.; Voisset, C.; Blondel, M.; Fahraeus, R. Epstein-Barr virus-encoded EBNA1 and ZEBRA: Targets for therapeutic strategies against EBV-carrying cancers. J. Pathol. 2015, 235, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.B.; Manet, E.; Gruffat, H.; Busson, P.; Blondel, M.; Fahraeus, R. EBNA1: Oncogenic activity, immune evasion and biochemical functions provide targets for novel therapeutic strategies against Epstein-Barr virus-associated cancers. Cancers (Basel) 2018, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem 2014, 6, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T. Strategies for the design of drugs targeting RNA and RNA-protein complexes. Angew. Chem. Int. Ed. Engl. 2000, 39, 1890–1904. [Google Scholar] [CrossRef]
- Yangyuoru, P.M.; Di Antonio, M.; Ghimire, C.; Biffi, G.; Balasubramanian, S.; Mao, H. Dual binding of an antibody and a small molecule increases the stability of terra G-quadruplex. Angew. Chem. Int. Ed. Engl. 2015, 54, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Gabelica, V.; Maeda, R.; Fujimoto, T.; Yaku, H.; Murashima, T.; Sugimoto, N.; Miyoshi, D. Multiple and cooperative binding of fluorescence light-up probe thioflavin T with human telomere DNA G-quadruplex. Biochemistry 2013, 52, 5620–5628. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compound PhenDC3 are available from the authors. PDS is commercially available (Sigma-Aldrich, now Merck). |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prado Martins, R.; Findakly, S.; Daskalogianni, C.; Teulade-Fichou, M.-P.; Blondel, M.; Fåhraeus, R. In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules. Molecules 2018, 23, 3124. https://doi.org/10.3390/molecules23123124
Prado Martins R, Findakly S, Daskalogianni C, Teulade-Fichou M-P, Blondel M, Fåhraeus R. In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules. Molecules. 2018; 23(12):3124. https://doi.org/10.3390/molecules23123124
Chicago/Turabian StylePrado Martins, Rodrigo, Sarah Findakly, Chrysoula Daskalogianni, Marie-Paule Teulade-Fichou, Marc Blondel, and Robin Fåhraeus. 2018. "In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules" Molecules 23, no. 12: 3124. https://doi.org/10.3390/molecules23123124
APA StylePrado Martins, R., Findakly, S., Daskalogianni, C., Teulade-Fichou, M.-P., Blondel, M., & Fåhraeus, R. (2018). In Cellulo Protein-mRNA Interaction Assay to Determine the Action of G-Quadruplex-Binding Molecules. Molecules, 23(12), 3124. https://doi.org/10.3390/molecules23123124







