Discovery of a Natural Syk Inhibitor from Chinese Medicine through a Docking-Based Virtual Screening and Biological Assay Study
Abstract
1. Introduction
2. Results and Discussion
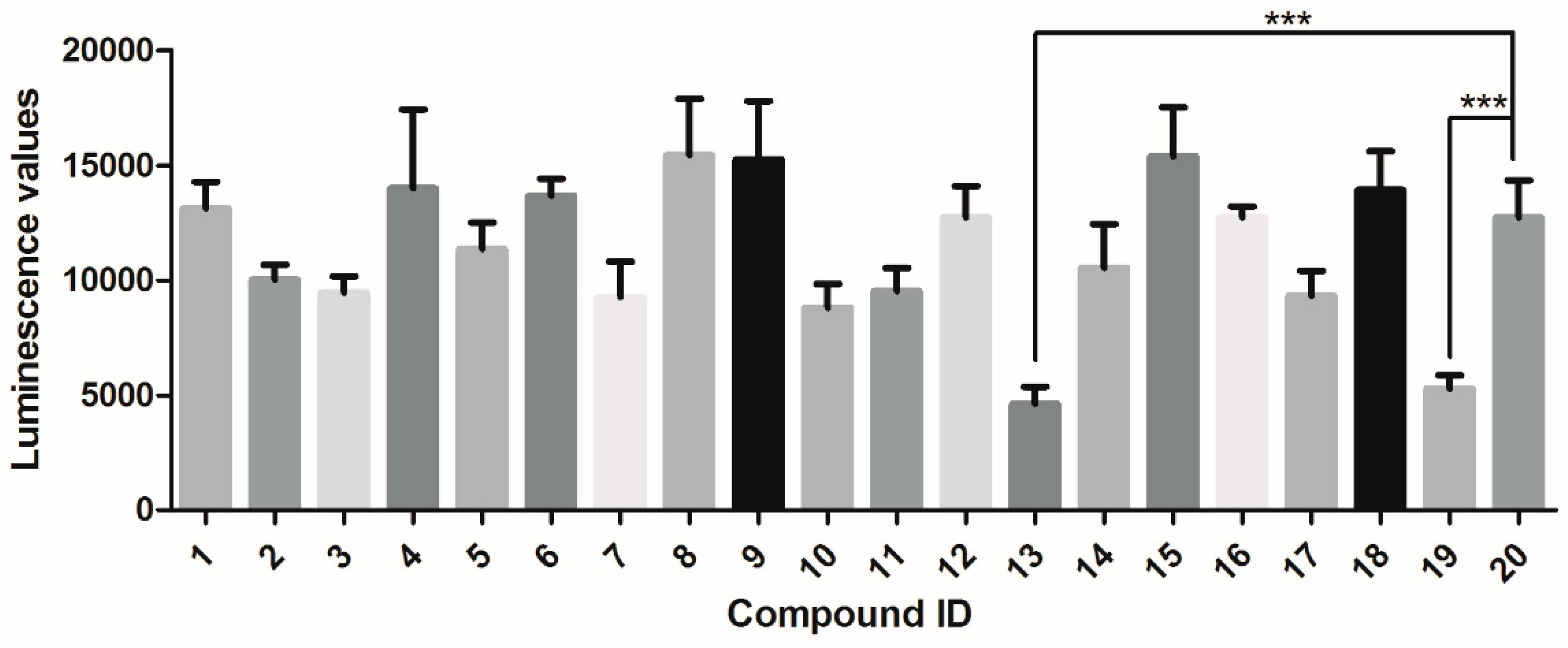
2.1. Docking-Based Virtual Screening Targeting Syk
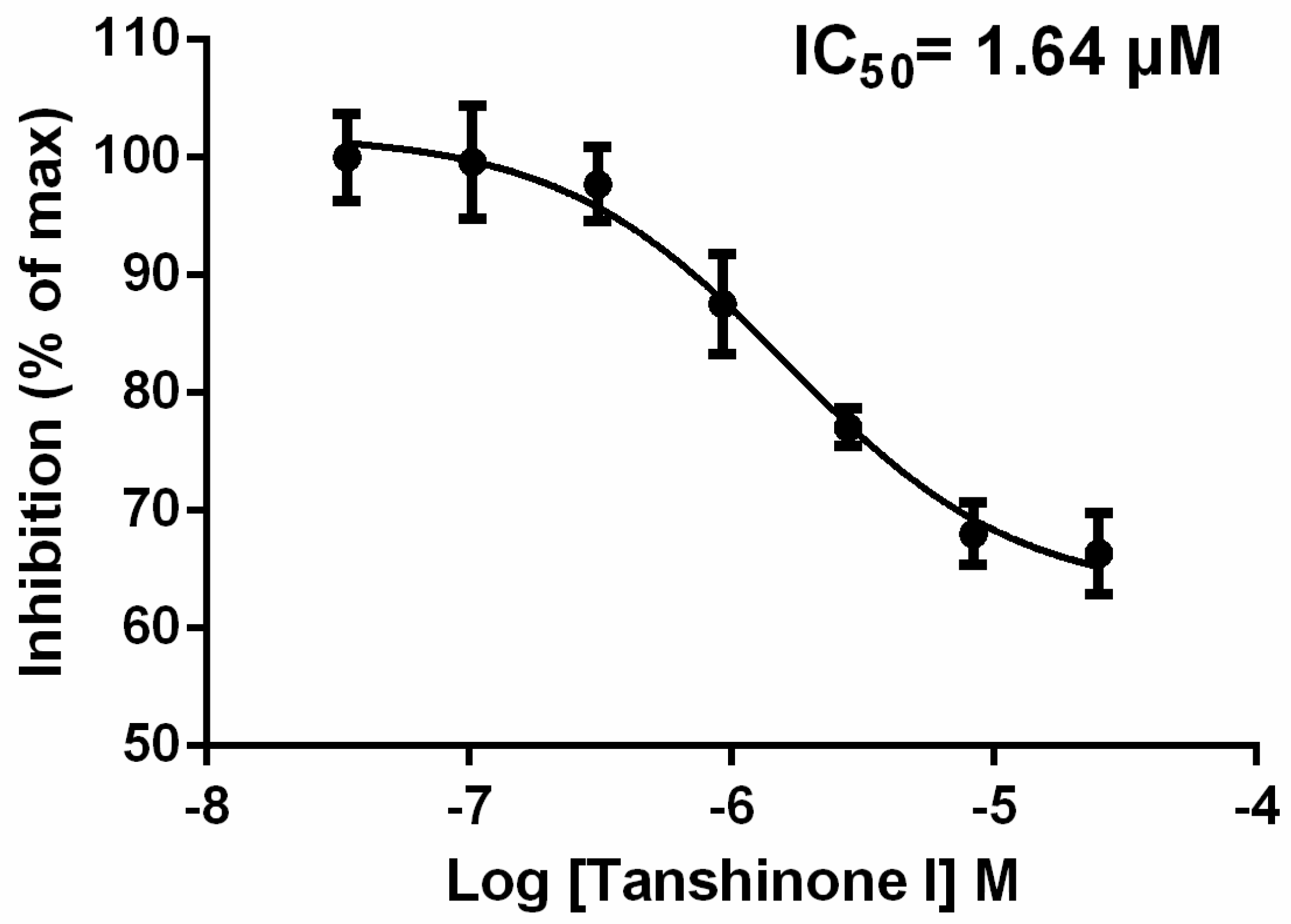
2.2. Tanshinone I Dose-Dependently Inhibited Syk Activity
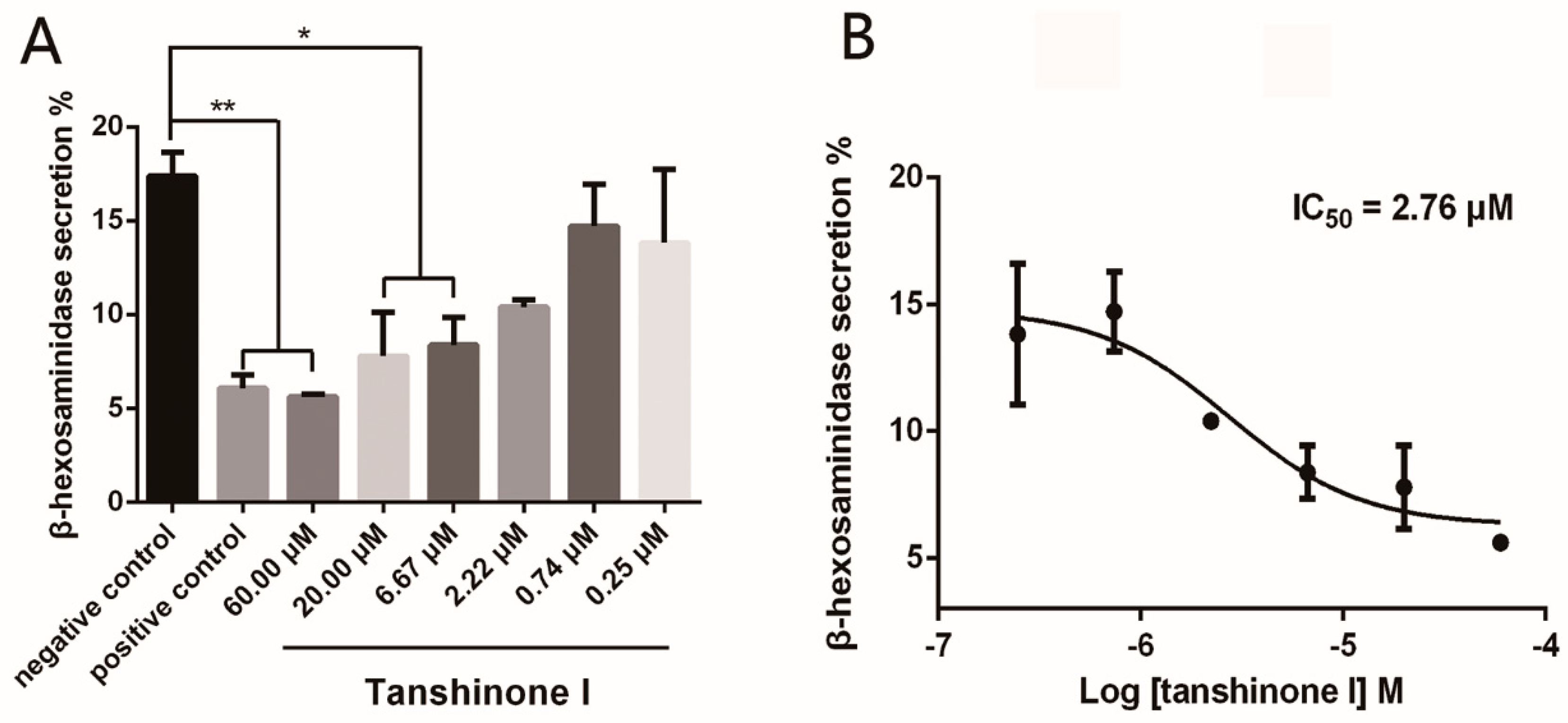
2.3. Tanshinone I Dose-Dependently Inhibited Mast Cell Degranulation
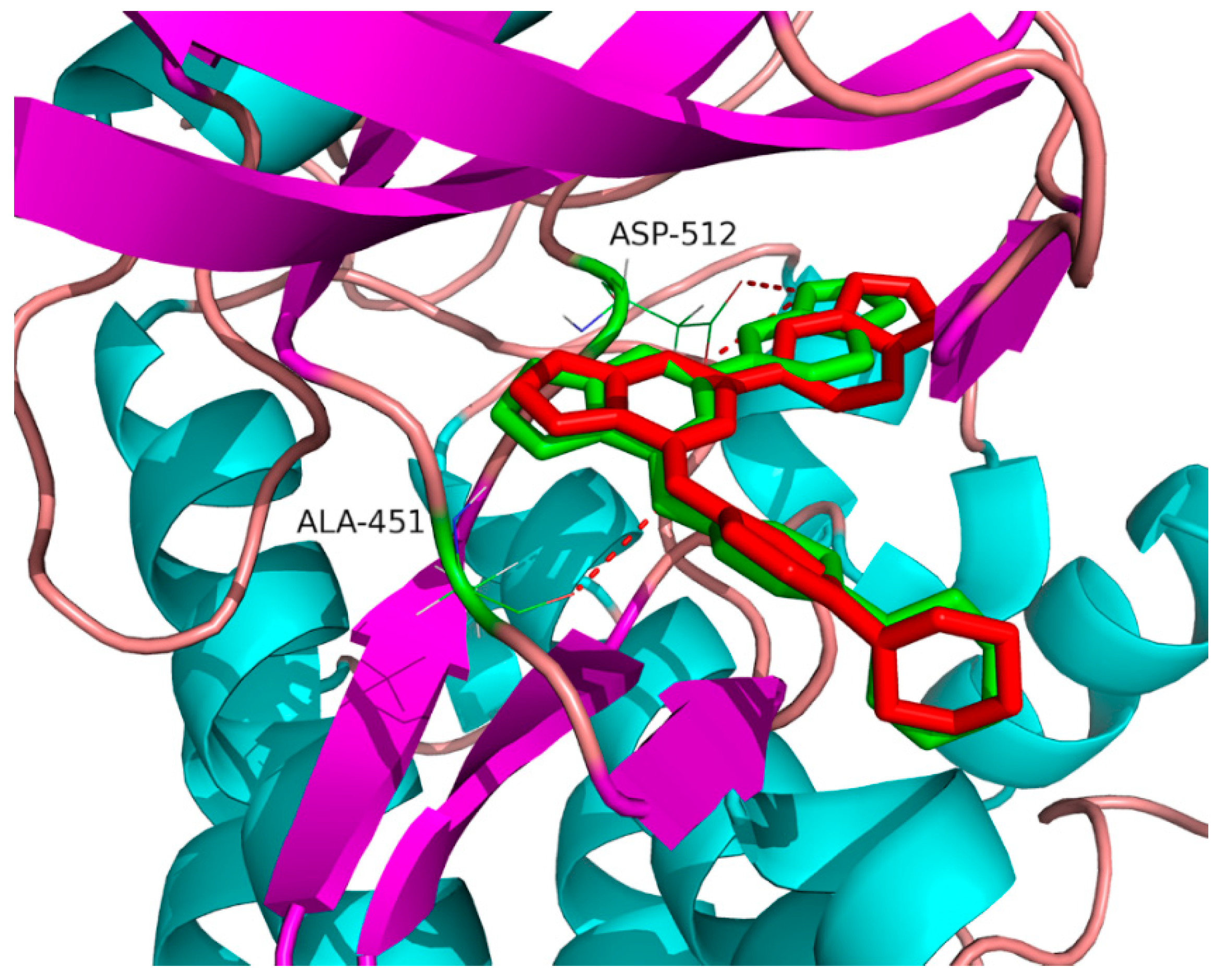
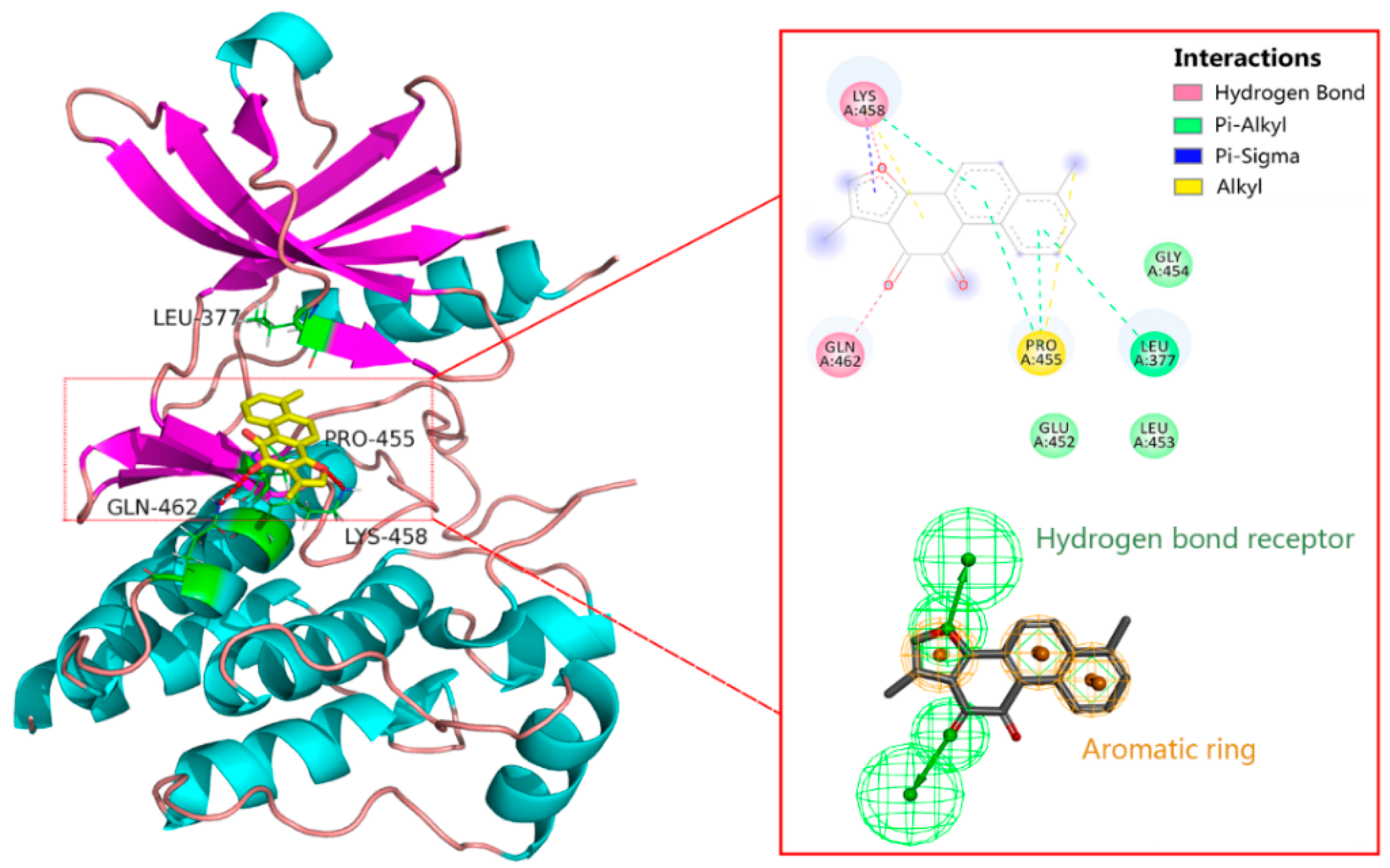
2.4. Binding Site of Tanshinone I in Syk Model
3. Materials and Methods
3.1. Molecular Docking
3.2. In Vitro Kinase Inhibition Assays
3.3. Cell Culture
3.4. β-Hexosaminidase Release Assay
3.5. Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barker, M.D.; Liddle, J.; Atkinson, F.L.; Wilson, D.M.; Dickson, M.C.; Ramirez-Molina, C.; Lewis, H.; Davis, R.P.; Somers, D.O.; Neu, M.; et al. Discovery of potent and selective Spleen Tyrosine Kinase inhibitors for the topical treatment of inflammatory skin disease. Bioorg. Med. Chem. Lett. 2018, 28, 3458–3462. [Google Scholar] [CrossRef] [PubMed]
- Grodzki, A.C.; Moon, K.D.; Berenstein, E.H.; Siraganian, R.P. FcepsilonRI-induced activation by low antigen concentrations results in nuclear signals in the absence of degranulation. Mol. Immunol. 2009, 46, 2539–2547. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Tsokos, G.C. Spleen tyrosine kinase: An Src family of non-receptor kinase has multiple functions and represents a valuable therapeutic target in the treatment of autoimmune and inflammatory diseases. Autoimmunity 2010, 43, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Buyanravjikh, S.; Han, S.; Lee, S.; Jeong, A.L.; Ka, H.I.; Park, J.Y.; Boldbaatar, A.; Lim, J.S.; Lee, M.S.; Yang, Y. Cryptotanshinone inhibits IgEmediated degranulation through inhibition of spleen tyrosine kinase and tyrosineprotein kinase phosphorylation in mast cells. Mol. Med. Rep. 2018, 18, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, J.W.; Sharman, J.; Sweetenham, J.; Johnston, P.B.; Vose, J.M.; LaCasce, A.; Schaefer-Cutillo, J.; De Vos, S.; Sinha, R.; Leonard, J.P.; et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood 2010, 115, 2578–2585. [Google Scholar] [CrossRef] [PubMed]
- Tabeling, C.; Herbert, J.; Hocke, A.C.; Lamb, D.J.; Wollin, S.L.; Erb, K.J.; Boiarina, E.; Movassagh, H.; Scheffel, J.; Doehn, J.M.; et al. Spleen tyrosine kinase inhibition blocks airway constriction and protects from Th2-induced airway inflammation and remodeling. Allergy 2017, 72, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Singh, M.; Silakari, O. Oxindole-based SYK and JAK3 dual inhibitors for rheumatoid arthritis: Designing, synthesis and biological evaluation. Future Med. Chem. 2017, 9, 1193–1211. [Google Scholar] [CrossRef] [PubMed]
- Bussel, J.; Arnold, D.M.; Grossbard, E.; Mayer, J.; Trelinski, J.; Homenda, W.; Hellmann, A.; Windyga, J.; Sivcheva, L.; Khalafallah, A.A.; et al. Fostamatinib for the treatment of adult persistent and chronic immune thrombocytopenia: Results of two phase 3, randomized, placebo-controlled trials. Am. J. Hematol. 2018, 93, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, R. Novel small-molecular therapeutics for rheumatoid arthritis. Curr. Opin. Rheumatol. 2012, 24, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A.; Phyo, A.P.; Woodrow, C.J. Malaria. Lancet 2018, 391, 1608–1621. [Google Scholar] [CrossRef]
- Zhuang, W.; Li, T.; Wang, C.J.; Shi, X.; Li, Y.L.; Zhang, S.L.; Zhao, Z.Q.; Dong, H.Y.; Qiao, Y.H. Berberine exerts antioxidant effects via protection of spiral ganglion cells against cytomegalovirus-induced apoptosis. Free Radical. Biol. Med. 2018, 121, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.B.; Wang, Z.Z.; Fan, Q.R.; Guo, J.; Galli, G.; Du, G.H.; Wang, X.; Xiao, W. Ginkgolide K protects the heart against endoplasmic reticulum stress injury by activating the inositol-requiring enzyme 1/X box-binding protein-1 pathway. Br. J. Pharmacol. 2016, 173, 2402–2418. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Chang, Y.K.; Bian, Y.; Bae, O.N.; Lim, K.M.; Chung, J.H. Pro-Coagulant and Pro-Thrombotic Effects of Paclitaxel Mediated by Red Blood Cells. Thromb. Haemostasis 2018, 118, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Currie, K.S.; Kropf, J.E.; Lee, T.; Blomgren, P.; Xu, J.; Zhao, Z.; Gallion, S.; Whitney, J.A.; Maclin, D.; Lansdon, E.B.; et al. Discovery of GS-9973, a selective and orally efficacious inhibitor of spleen tyrosine kinase. J. Med. Chem. 2014, 57, 3856–3873. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Deana, A.D.; Ruzzene, M.; Brunati, A.M.; Cesaro, L.; Guerra, B.; Meyer, T.; Mett, H.; Fabbro, D.; Furet, P.; et al. Different Susceptibility of Protein-Kinases to Staurosporine Inhibition-Kinetic-Studies and Molecular-Bases for the Resistance of Protein-Kinase Ck2. Eur. J. Biochem. 1995, 234, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Z.; Su, X. Antipruritic activity of extracts of Humulus scandens, the combinations of bioactive flavonoids. Fitoterapia 2010, 81, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.M.; Altman, M.D.; Bass, A.; Butcher, J.W.; Byford, A.J.; Donofrio, A.; Galloway, S.; Haidle, A.M.; Jewell, J.; Kelly, N.; et al. Overcoming mutagenicity and ion channel activity: Optimization of selective spleen tyrosine kinase inhibitors. J. Med. Chem. 2015, 58, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.Z.; Li, L.L.; Ren, J.X.; Zou, J.; Yang, L.; Wei, Y.Q.; Yang, S.Y. Pharmacophore modeling study based on known spleen tyrosine kinase inhibitors together with virtual screening for identifying novel inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 1944–1949. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.C.; Goldstein, D.M.; Hermann, J.C.; Kuglstatter, A.; Liu, W.; Luk, K.C.; Padilla, F.; Slade, M.; Villasenor, A.G.; Wanner, J.; et al. Rational design of highly selective spleen tyrosine kinase inhibitors. J. Med. Chem. 2012, 55, 10414–10423. [Google Scholar] [CrossRef] [PubMed]
- Thoma, G.; Blanz, J.; Buhlmayer, P.; Druckes, P.; Kittelmann, M.; Smith, A.B.; van Eis, M.; Vangrevelinghe, E.; Zerwes, H.G.; Che, J.W.; et al. Syk inhibitors with high potency in presence of blood. Bioorg. Med. Chem. Lett. 2014, 24, 2278–2282. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.C.; Bhagirath, N.; Chiao, E.; Goldstein, D.M.; Hermann, J.C.; Hsu, P.Y.; Kirchner, S.; Kennedy-Smith, J.J.; Kuglstatter, A.; Lukacs, C.; et al. Using ovality to predict nonmutagenic, orally efficacious pyridazine amides as cell specific spleen tyrosine kinase inhibitors. J. Med. Chem. 2014, 57, 2683–2691. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, X.; Zhang, Z.; Luo, J.; Long, H.; Tu, Z.; Zhou, X.; Ding, K.; Lu, X. 3-aminopyrazolopyrazine derivatives as spleen tyrosine kinase inhibitors. Chem. Biol. Drug Des. 2016, 88, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, O.; Marsden, B.; Pogacic, V.; Rellos, P.; Muller, S.; Bullock, A.N.; Schwaller, J.; Sundstrom, M.; Knapp, S. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. USA 2007, 104, 20523–20528. [Google Scholar] [CrossRef] [PubMed]
- Li, G.B.; Ji, S.; Yang, L.L.; Zhang, R.J.; Chen, K.; Zhong, L.; Ma, S.; Yang, S.Y. LEADOPT: An automatic tool for structure-based lead optimization, and its application in structural optimizations of VEGFR2 and SYK inhibitors. Eur. J. Med. Chem. 2015, 93, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Bonesi, M.; Loizzo, M.R.; Acquaviva, R.; Malfa, G.A.; Aiello, F.; Tundis, R. Anti-inflammatory and Antioxidant Agents from Salvia Genus (Lamiaceae): An Assessment of the Current State of Knowledge. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2017, 16, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Ge, G.; Zhang, Q.; Ma, J.; Qiao, Z.; Huang, J.; Cheng, W.; Wang, H. Protective effect of Salvia miltiorrhiza aqueous extract on myocardium oxidative injury in ischemic-reperfusion rats. Gene 2014, 546, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Park, O.; Cho, J.H.; Chen, B.H.; Kim, I.H.; Ahn, J.H.; Lee, J.C.; Yan, B.C.; Yoo, K.Y.; Lee, C.H.; et al. Anti-inflammatory effect of tanshinone I in neuroprotection against cerebral ischemia-reperfusion injury in the gerbil hippocampus. Neurochem. Res. 2014, 39, 1300–1312. [Google Scholar] [CrossRef] [PubMed]
- Trinh, H.T.; Chae, S.J.; Joh, E.H.; Son, K.H.; Jeon, S.J.; Kim, D.H. Tanshinones isolated from the rhizome of Salvia miltiorrhiza inhibit passive cutaneous anaphylaxis reaction in mice. J. Ethnopharmacol. 2010, 132, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Kim, K.M. Tanshinones inhibit mast cell degranulation by interfering with IgE receptor-mediated tyrosine phosphorylation of PLCgamma2 and MAPK. Planta Med. 2004, 70, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Chiarelli, L.R.; Mori, M.; Barlocco, D.; Beretta, G.; Gelain, A.; Pini, E.; Porcino, M.; Mori, G.; Stelitano, G.; Costantino, L.; et al. Discovery and development of novel salicylate synthase (MbtI) furanic inhibitors as antitubercular agents. Eur. J. Med. Chem. 2018, 155, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; He, Y.; Yang, J.; Han, W.; Zhai, X.; Zhao, Y.; Li, Y. Molecular Modeling Study for the Design of Novel Peroxisome Proliferator-Activated Receptor Gamma Agonists using 3D-QSAR and Molecular Docking. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Wang, J.; Wang, R.; Lin, Y.; Hu, Y.; Wang, Y.; Shu, M.; Lin, Z. Combined pharmacophore modeling, 3D-QSAR, homology modeling and docking studies on CYP11B1 inhibitors. Molecules 2015, 20, 1014–1030. [Google Scholar] [CrossRef] [PubMed]
- Empereur-mot, C.; Guillemain, H.; Latouche, A.; Zagury, J.F.; Viallon, V.; Montes, M. Predictiveness curves in virtual screening. J. Cheminf. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, H.; Wang, Q.; Deng, S.H.; Huang, M.; Ma, X.H.; Song, P.; Du, J.W.; Huang, Y.; Wen, Y.Z.; et al. Inhibitory Effect of Kurarinone on Growth of Human Non-small Cell Lung Cancer: An Experimental Study Both in Vitro and in Vivo Studies. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Qin, C.; Sin, J.E.; Yang, X.; Tao, L.; Zeng, X.; Zhang, P.; Gao, C.M.; Jiang, Y.Y.; Zhang, C.; et al. Discovery of novel dual VEGFR2 and Src inhibitors using a multistep virtual screening approach. Future Med. Chem. 2017, 9, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.; Arikawa, Y.; Cramlett, J.; Dong, Q.; de Jong, R.; Feher, V.; Grimshaw, C.E.; Farrell, P.J.; Hoffman, I.D.; Jennings, A.; et al. Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK). Bioorg. Med. Chem. Lett. 2016, 26, 5947–5950. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des. 2007, 21, 281–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Yang, Y.; Wu, X.; Fan, H.; Qiao, Y. Identification of berberine as a direct thrombin inhibitor from traditional Chinese medicine through structural, functional and binding studies. Sci. Rep. 2017, 7, 44040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Wang, X.; Li, X.; Peng, S.; Wang, S.F.; Huang, C.Z.; Huang, C.Z.; Zhang, Q.; Li, D.; Jiang, J.; et al. Identification of a specific agonist of human TAS2R14 from Radix Bupleuri through virtual screening, functional evaluation and binding studies. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, R.; Jain, A.N. Surflex-Dock: Docking benchmarks and real-world application. J. Comput. Aided Mol. Des. 2012, 26, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.M.; Yang, J.H.; Kim, Y.S.; Yang, H.J.; Cho, W.K.; Ma, J.Y. Inhibitory Effects of Viscum coloratum Extract on IgE/Antigen-Activated Mast Cells and Mast Cell-Derived Inflammatory Mediator-Activated Chondrocytes. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Park, K.I.; Kim, D.G.; Yoo, J.M.; Ma, J.Y. The Herbal Medicine KIOM-MA128 Inhibits the Antigen/IgE-Mediated Allergic Response in Vitro and in Vivo. Molecules 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Shinmoto, H.; Tsushida, T. Coumarin and flavone derivatives from estragon and thyme as inhibitors of chemical mediator release from RBL-2H3 Cells. Biosci. Biotechnol. Biochem. 2005, 69, 1–6. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

| ID | Total Score | Name | CAS No. | Sources |
|---|---|---|---|---|
| 1 | 9.61 | 10-Gingerol | 23513-15-7 | Zingiber officinale |
| 2 | 8.02 | Calceolarioside B | 105471-98-5 | Akebia quinata |
| 3 | 7.71 | 6-Gingerol | 23513-14-6 | Zingiber officinale |
| 4 | 7.59 | Corylifolinin | 20784-50-3 | Glycyrrhiza uralensis |
| 5 | 7.48 | Arbutin | 497-76-7 | Saussurea lappa |
| 6 | 7.37 | Piceid | 27208-80-6 | Lilium brownii var. viridulum |
| 7 | 7.07 | Ostruthin | 148-83-4 | Angelica decursiva |
| 8 | 7.01 | Wogonoside | 51059-44-0 | Scutellaria baicalensis |
| 9 | 6.91 | Mulberroside A | 102841-42-9 | Morus alba |
| 10 | 6.88 | Anisodamine hydrobromide | 55449-49-5 | Anisodustanguticus (Maxim.) Pascher |
| 11 | 6.84 | Curcumin | 458-37-7 | Curcuma zedoaria |
| 12 | 6.55 | Dehydrodiisoeugenol | 2680-81-1 | Syzygium aromaticum |
| 13 | 6.30 | Tanshinone I | 568-73-0 | Salvia miltiorrhiza |
| 14 | 6.25 | l-Arctigenin | 7770-78-7 | Arctium lappa |
| 15 | 6.21 | Mulberroside C | 102841-43-0 | Morus alba |
| 16 | 6.18 | 6-Shogaol | 555-66-8 | Zingiber officinale |
| 17 | 6.15 | Propyl gallate | 121-79-9 | Rheum officinale |
| 18 | 6.03 | Sesamolin | 526-07-8 | Sesamum indicum |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Guo, J.; Ning, Z.; Wu, X. Discovery of a Natural Syk Inhibitor from Chinese Medicine through a Docking-Based Virtual Screening and Biological Assay Study. Molecules 2018, 23, 3114. https://doi.org/10.3390/molecules23123114
Wang X, Guo J, Ning Z, Wu X. Discovery of a Natural Syk Inhibitor from Chinese Medicine through a Docking-Based Virtual Screening and Biological Assay Study. Molecules. 2018; 23(12):3114. https://doi.org/10.3390/molecules23123114
Chicago/Turabian StyleWang, Xing, Junfang Guo, Zhongqi Ning, and Xia Wu. 2018. "Discovery of a Natural Syk Inhibitor from Chinese Medicine through a Docking-Based Virtual Screening and Biological Assay Study" Molecules 23, no. 12: 3114. https://doi.org/10.3390/molecules23123114
APA StyleWang, X., Guo, J., Ning, Z., & Wu, X. (2018). Discovery of a Natural Syk Inhibitor from Chinese Medicine through a Docking-Based Virtual Screening and Biological Assay Study. Molecules, 23(12), 3114. https://doi.org/10.3390/molecules23123114






