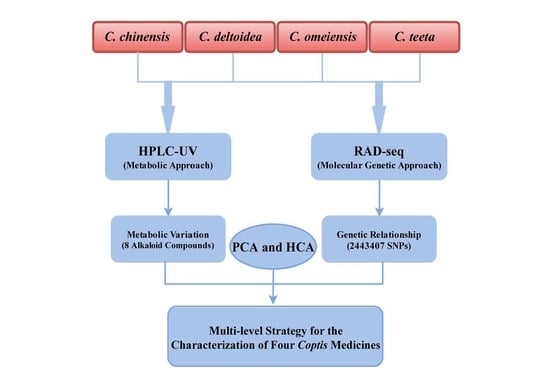
A Multi-Level Strategy Based on Metabolic and Molecular Genetic Approaches for the Characterization of Different Coptis Medicines Using HPLC-UV and RAD-seq Techniques
Abstract
1. Introduction
2. Results
2.1. Results of Validation for HPLC-UV Method
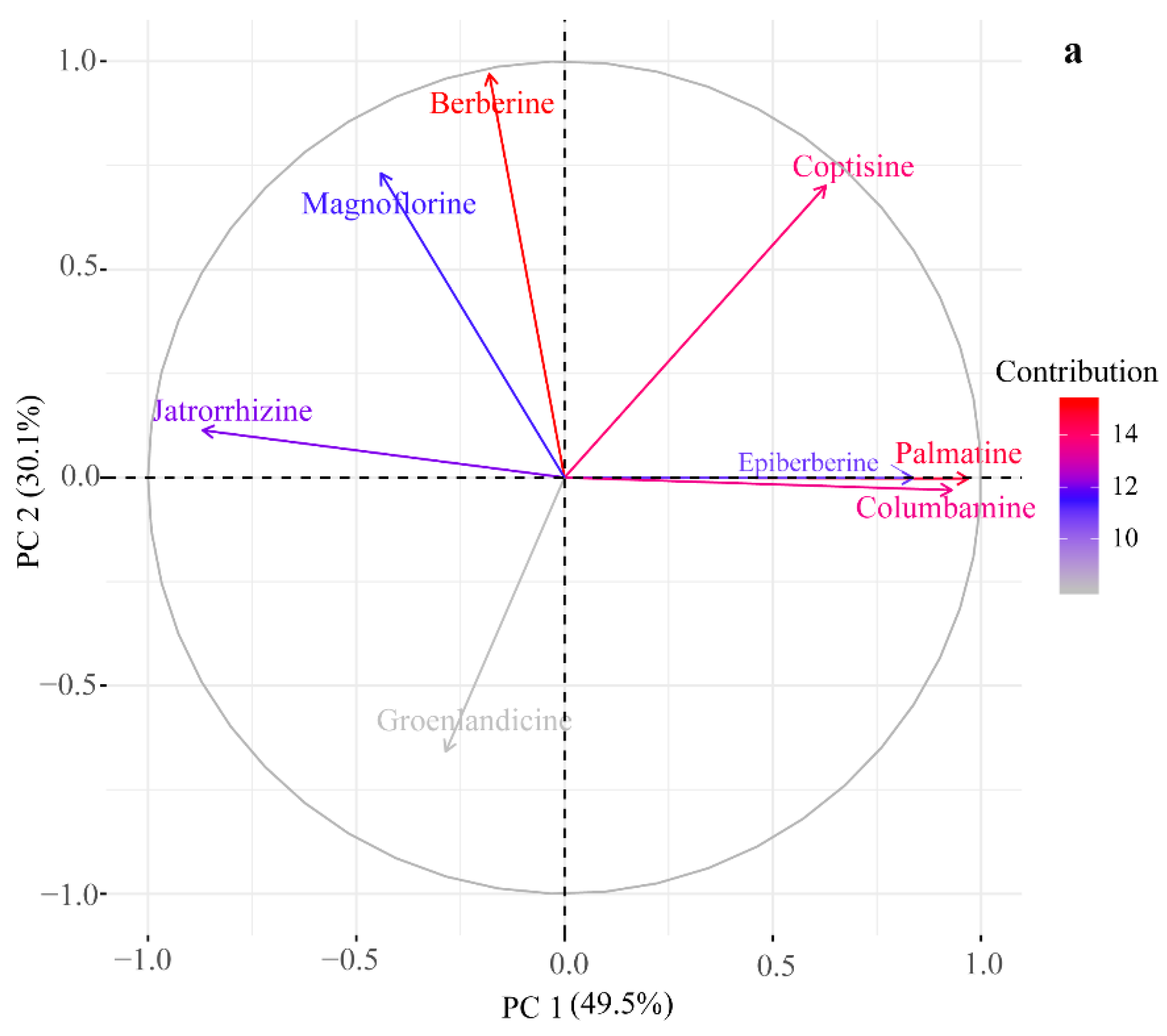
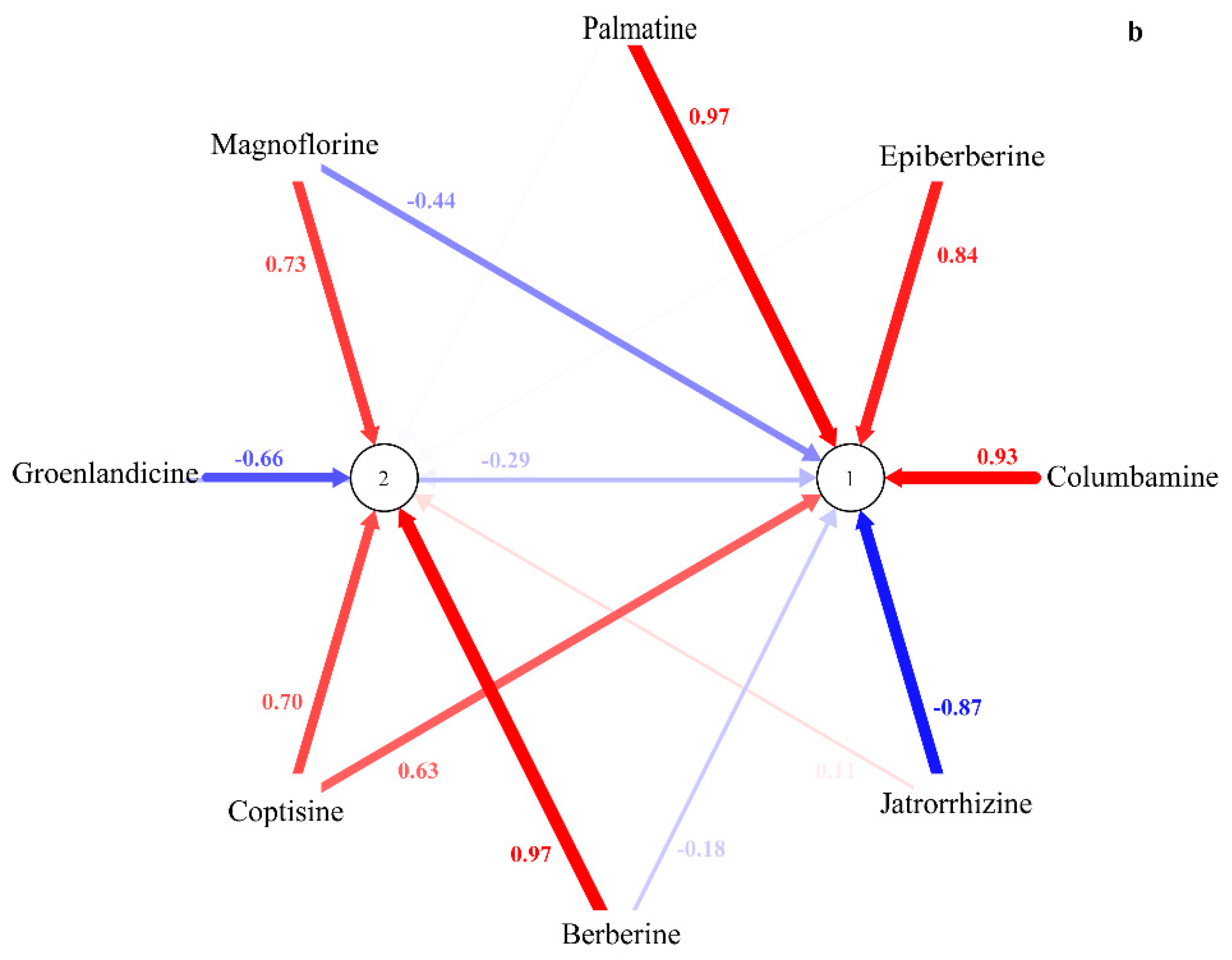
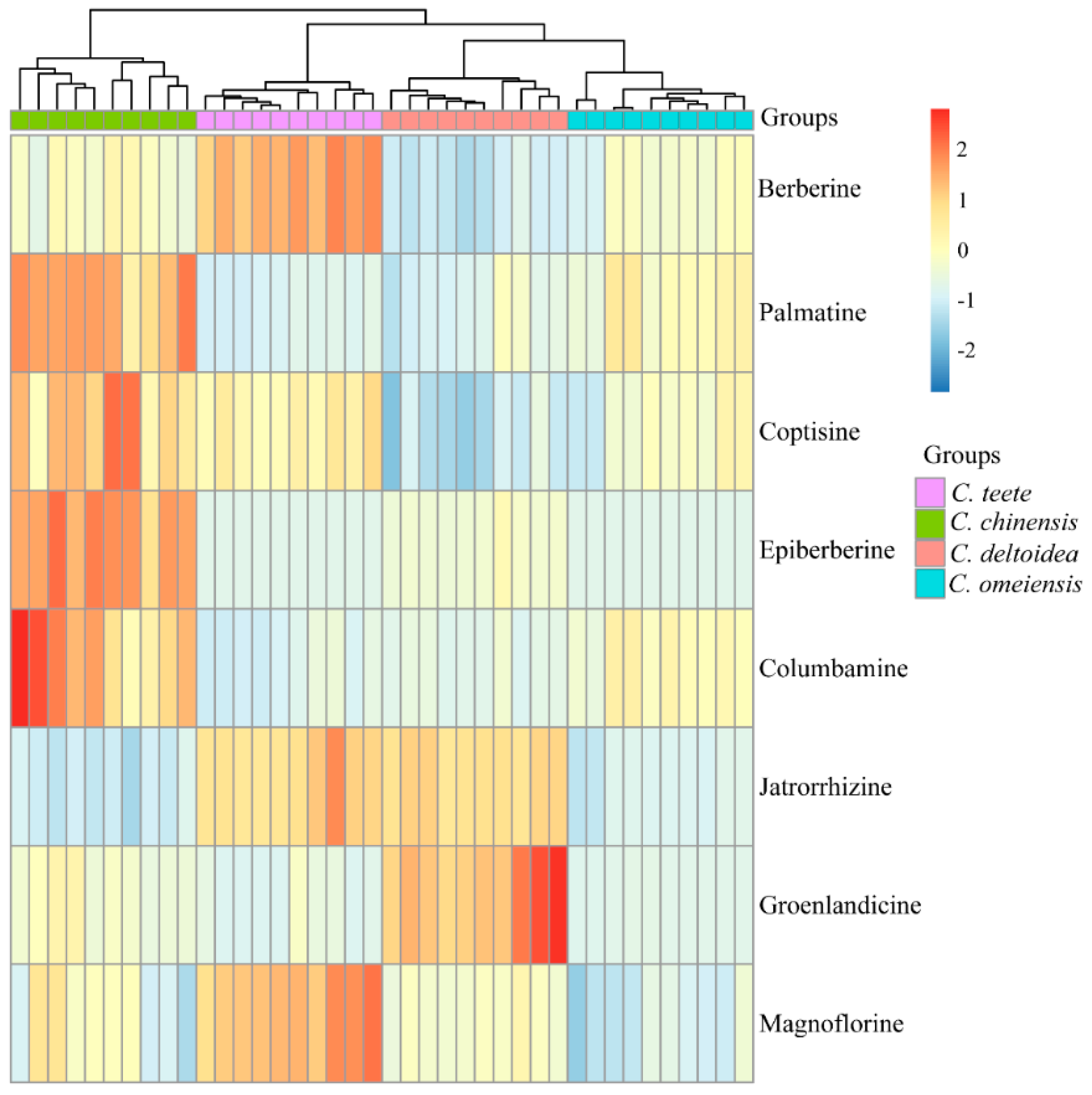
2.2. HPLC-UV Determination of Eight Alkaloids
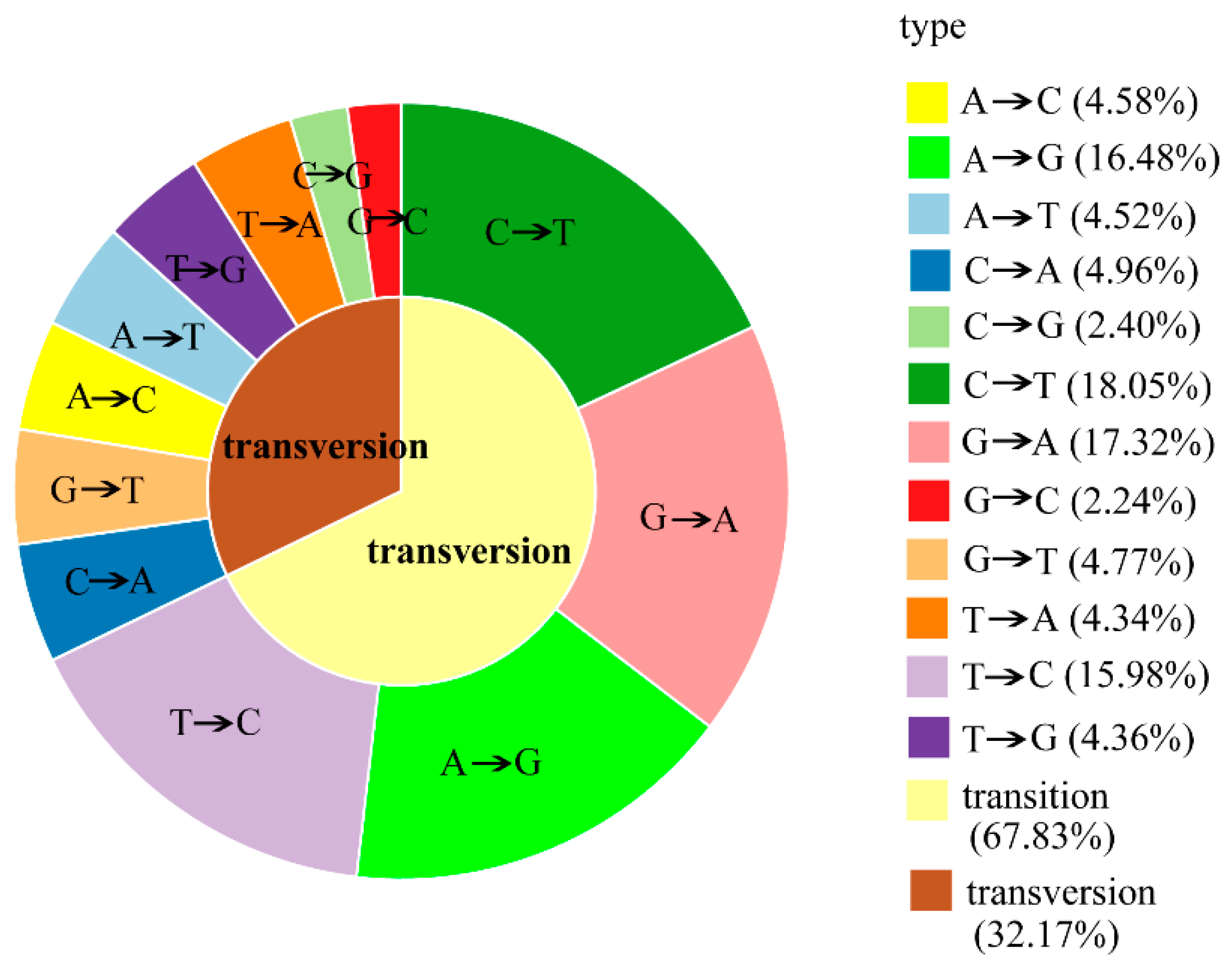
2.3. RAD-seq Data Analysis and SNP Discovery
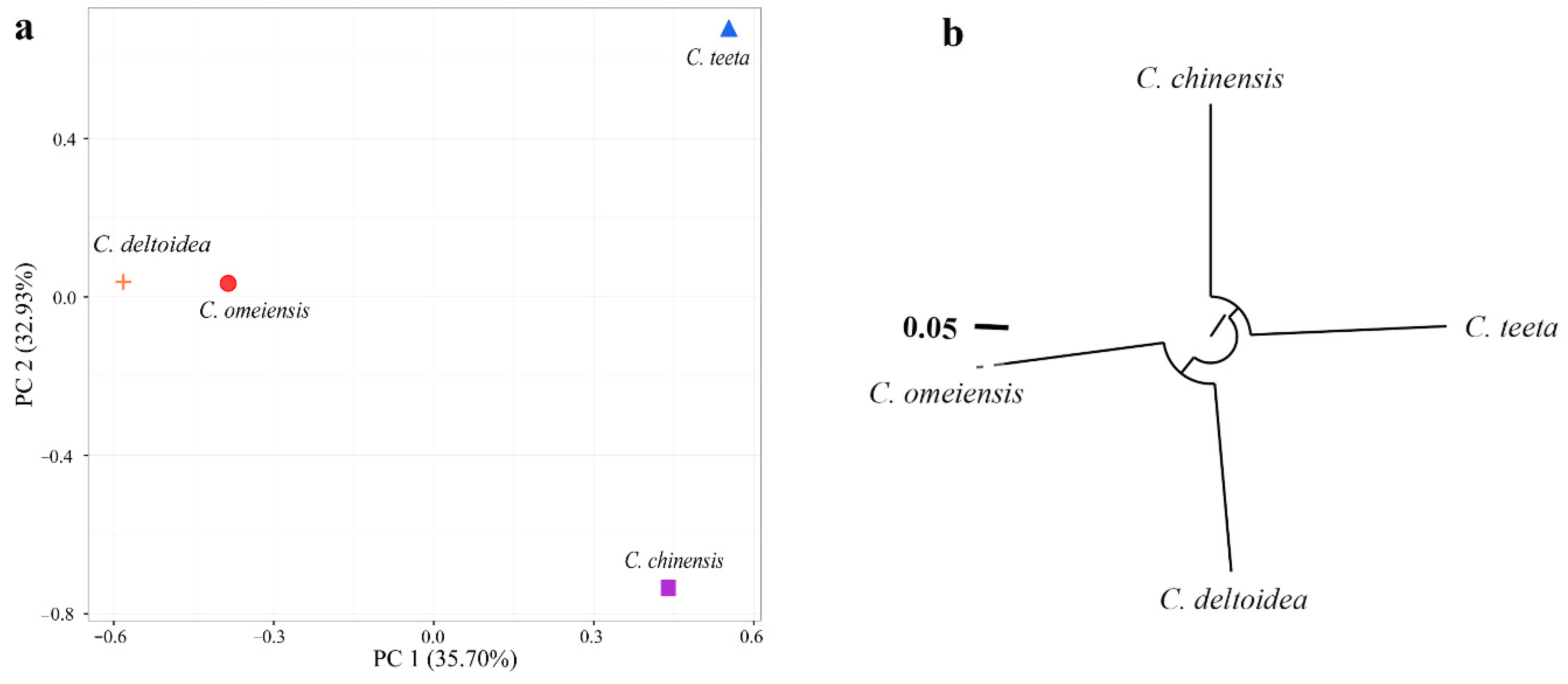
2.4. Genetic Relationship among Four Coptis Plants
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plant Material and Sample Preparation
3.3. HPLC-UV Analysis
3.4. Validation of HPLC–UV Method
3.5. DNA Isolation, RAD Library Construction and Sequencing
3.6. De Novo Assembly of RAD Tags and SNP Identification
- (1)
- After removing the MID and the restriction site, the information of individual stacks was built by sorting and clustering similar first reads from each sample.
- (2)
- The individual stacks were clustered and the consensus sequences were obtained.
- (3)
- According to the clusters mentioned above, the second reads could be sorted into groups and separately assembled to contigs, which were joined with the first reads consensus sequences. The paired-end reads were used to construct scaffolds serving as a reference for the following analysis.
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Coghlan, M.L.; Maker, G.; Crighton, E.; Haile, J.; Murray, D.C.; White, N.E.; Byard, R.W.; Bellgard, M.I.; Mullaney, I.; Trengove, R.; et al. Combined DNA, toxicological and heavy metal analyses provides an auditing toolkit to improve pharmacovigilance of traditional Chinese medicine (TCM). Sci. Rep. 2015, 5, 17475. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Liu, B.; Ma, K. Traditional Chinese medicine. Lancet 2008, 372, 1938–1940. [Google Scholar] [CrossRef]
- Hao, P.; Jiang, F.; Chen, Y.; Yang, J.; Zhang, K.; Zhang, M.; Zhang, C.; Zhao, Y.; Zhang, Y. Traditional Chinese medication for cardiovascular disease. Nat. Rev. Cardiol. 2015, 12, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Li, W.L.; Zheng, H.C.; Bukuru, J.; de Kimpe, N. Natural medicines used in the traditional Chinese medical system for therapy of diabetes mellitus. J. Ethnopharmacol. 2004, 92, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Luo, Q.; Sun, M.; Corke, H. Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci. 2004, 74, 2157–2184. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, Y.; Lai, Y.; Yang, Q.; Hu, H.; Wang, Y. Sustainable utilization of traditional Chinese medicine resources: Systematic evaluation on different production modes. Evid-Based Compl. Alt. 2015, 2015, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Shaw, D.; Simmonds, M.S.J.; Leon, C.J.; Xu, Q.; Lu, A.; Sutherland, I.; Ignatova, S.; Zhu, Y.; Verpoorte, R.; et al. Good practice in reviewing and publishing studies on herbal medicine, with special emphasis on traditional Chinese medicine and Chinese materia medica. J. Ethnopharmacol. 2012, 140, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jiang, Y.; Cui, B.; Jiang, Y.; Cao, H.; Zhang, W. Determination of the Heavy Metal Levels in Panax notoginseng and the Implications for Human Health Risk Assessment. Hum. Ecol. Risk Assess. 2015, 21, 1218–1229. [Google Scholar] [CrossRef]
- Zhong, X.; Li, D.; Jiang, J. Identification and quality control of Chinese medicine based on the fingerprint techniques. Curr. Med. Chem. 2009, 16, 3064–3075. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Zhang, J.; Zuo, Z.; Zhao, Y.; Wang, Y.; Hang, J. Determination of Iridoids in Gentiana rigescens by Infrared Spectroscopy and Multivariate Analysis. Anal. Lett. 2017, 50, 389–401. [Google Scholar] [CrossRef]
- Chen, C.; Yan, H.; Han, B. Rapid identification of three varieties of Chrysanthemum with near infrared spectroscopy. Rev. Bras. Farmacogn. 2014, 24, 33–37. [Google Scholar] [CrossRef]
- Xiang, K.; Wu, S.; Yu, S.; Liu, Y.; Jabbour, F.; Erst, A.S.; Zhao, L.; Wang, W.; Chen, Z. The first comprehensive phylogeny of Coptis (Ranunculaceae) and its implications for character evolution and classification. PLoS ONE 2016, 11, e153127. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.L.; Sheng, Y.X.; Wang, L.Q.; Zhang, J.L. Development of a rapid resolution liquid chromatographic method for simultaneous analysis of four alkaloids in Rhizoma Coptidis under different cultivation conditions. J. AOAC Int. 2009, 92, 663–671. [Google Scholar] [PubMed]
- He, Y.; Hou, P.; Fan, G.; Arain, S.; Peng, C. Comprehensive analyses of molecular phylogeny and main alkaloids for Coptis (Ranunculaceae) species identification. Biochem. Syst. Ecol. 2014, 56, 88–94. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, J.; Li, F.; Liu, X.; Deng, M.; Wu, H. Determination of alkaloid contents in various tissues of Coptis Chinensis Franch by reversed phase-high performance liquid chromatography and ultraviolet spectrophotometry. J. Chromatogr. Sci. 2017, 55, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Tjong, Y.; Ip, S.; Lao, L.; Fong, H.H.S.; Sung, J.J.Y.; Berman, B.; Che, C. Analgesic effect of Coptis chinensis rhizomes (Coptidis Rhizoma) extract on rat model of irritable bowel syndrome. J. Ethnopharmacol. 2011, 135, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Piao, X.; Piao, X.; Lu, T.; Wang, D.; Kim, S.W. Preventive effect of Coptis chinensis and berberine on intestinal injury in rats challenged with lipopolysaccharides. Food Chem. Toxicol. 2011, 49, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Liu, M.; Chang, X.; Sun, K. The inhibiting effect of the Coptis chinensis polysaccharide on the type II diabetic mice. Biomed. Pharmacother. 2016, 81, 111–119. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fan, G.; Gao, J.; Peng, C. Correlation analyses between molecular perspective and phytochemical variations in Coptis chinensis Franch. Biochem. Syst. Ecol. 2015, 61, 143–148. [Google Scholar] [CrossRef]
- Gao, X.; Yang, X.W.; Marriott, P.J. Simultaneous analysis of seven alkaloids in Coptis-Evodia herb couple and Zuojin pill by UPLC with accelerated solvent extraction. J. Sep. Sci. 2010, 33, 2714–2722. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Tao, L.H.; Yue, Q.H.; Kuang, T.T.; Tang, C.; Yang, Y.D.; Luo, W.Z.; Zhou, X.D.; Zhang, Y. Metabolic discrimination of Rhizoma Coptidis from different species using 1H-NMR spectroscopy and principal component analysis. Planta Med. 2012, 78, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wei, C.; Liu, H.; Wu, J.; Li, Z.; Zhang, L.; Jian, J.; Li, Y.; Tai, Y.; Zhang, J.; et al. Genetic divergence between Camellia sinensis and its wild relatives revealed via genome-wide SNPs from RAD sequencing. PLoS ONE 2016, 11, e151424. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Pegadaraju, V.; Nipper, R.; Hulke, B.; Qi, L.; Schultz, Q. De novo sequencing of sunflower genome for SNP discovery using RAD (Restriction site Associated DNA) approach. BMC Genom. 2013, 14, 556. [Google Scholar] [CrossRef] [PubMed]
- Karam, M.J.; Lefèvre, F.; Dagher-Kharrat, M.B.; Pinosio, S.; Vendramin, G.G. Genomic exploration and molecular marker development in a large and complex conifer genome using RADseq and mRNAseq. Mol. Ecol. Resour. 2015, 15, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Penjor, T.; Mimura, T.; Kotoda, N.; Matsumoto, R.; Nagano, A.J.; Honjo, M.N.; Kudoh, H.; Yamamoto, M.; Nagano, Y. RAD-Seq analysis of typical and minor Citrus accessions, including Bhutanese varieties. Breeding Sci. 2016, 66, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Arnold, B.; Corbett-Detig, R.B.; Hartl, D.; Bomblies, K. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol. Ecol. 2013, 22, 3179–3190. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.E.; Keller, I.; Wittwer, S.; Selz, O.M.; Mwaiko, S.; Greuter, L.; Sivasundar, A.; Seehausen, O. Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol. Ecol. 2013, 22, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Pfender, W.F.; Saha, M.C.; Johnson, E.A.; Slabaugh, M.B. Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theor. Appl. Genet. 2011, 122, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fan, G.; He, Y. Species evolution and quality evaluation of four Coptis herbal medicinal materials in Southwest China. 3 Biotech 2017, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liang, Y.; Cong, K.; Chen, G.; Zhao, X.; Zhao, Q.; Zhang, J.; Wang, X.; Dong, Y.; Yang, J.; et al. Identification and characterization of genes involved in benzylisoquinoline alkaloid biosynthesis in Coptis Species. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Deng, C.; Nie, H.; Fan, G.; He, Y. Transcriptome analyses provide insights into the difference of alkaloids biosynthesis in the Chinese goldthread (Coptis chinensis Franch.) from different biotopes. PeerJ. 2017, 5, e3303. [Google Scholar] [CrossRef] [PubMed]
- Parameswara, R.V.; Subhashis, C.; Sanjay, S. Berberine: A potential phytochemical with multispectrum therapeutic activities. Expert Opin. Investig. Drugs 2010, 19, 1297–1307. [Google Scholar]
- Lv, X.; Li, Y.; Tang, C.; Zhang, Y.; Zhang, J.; Fan, G. Integration of HPLC-based fingerprint and quantitative analyses for differentiating botanical species and geographical growing origins of Rhizoma Coptidis. Pharm. Biol. 2016, 54, 3264–3271. [Google Scholar] [CrossRef] [PubMed]
- Scaglione, D.; Acquadro, A.; Portis, E.; Tirone, M.; Knapp, S.J.; Lanteri, S. RAD tag sequencing as a source of SNP markers in Cynara cardunculus L. BMC Genom. 2012, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Natarajan, S.; Biswas, M.K.; Shirasawa, K.; Isobe, S.; Kim, H.; Park, J.; Seong, C.; Nou, I. SNP discovery of Korean short day onion inbred lines using double digest restriction site-associated DNA sequencing. PLoS ONE 2018, 13, e201229. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, Z.; Wu, J.; Xu, Y.; Yang, X.; Fan, L.; Fang, R.; Zhou, X. Analysis of genetic variation and diversity of Rice stripe virus populations through high-throughput sequencing. Front. Plant Sci. 2015, 6, 176. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Willing, E.; Hoffmann, M.; Klein, J.D.; Weigel, D.; Dreyer, C. Paired-end RAD-seq for de novo assembly and marker design without available reference. Bioinformatics 2011, 27, 2187–2193. [Google Scholar] [CrossRef] [PubMed]
- Anunciação, D.S.; Leao, D.J.; de Jesus, R.M.; Ferreira, S.L.C. Use of multivariate analysis techniques for evaluation of analytical data-determination of the mineral composition of cabbage (Brassica oleracea). Food Anal. Method 2011, 4, 286–292. [Google Scholar] [CrossRef]
- Dos Santos, W.N.L.; da silva Sauthier, M.C.; dos santos, A.M.P.; de Andrade Santana, D.; Almeida Azevedo, R.S.; Da Cruz Caldas, J. Simultaneous determination of 13 phenolic bioactive compounds in guava (Psidium guajava L.) by HPLC-PAD with evaluation using PCA and Neural Network Analysis (NNA). Microchem. J. 2017, 133, 583–592. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Liu, H.; Zhang, B.; Liao, Y.; Zhang, Z. Quantitative and chemical fingerprint analysis for quality evaluation of the dried bark of wild Phellodendron amurense Rupr based on HPLC-DAD-MS combined with chemometrics methods. Anal. Methods 2015, 7, 2041–2049. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
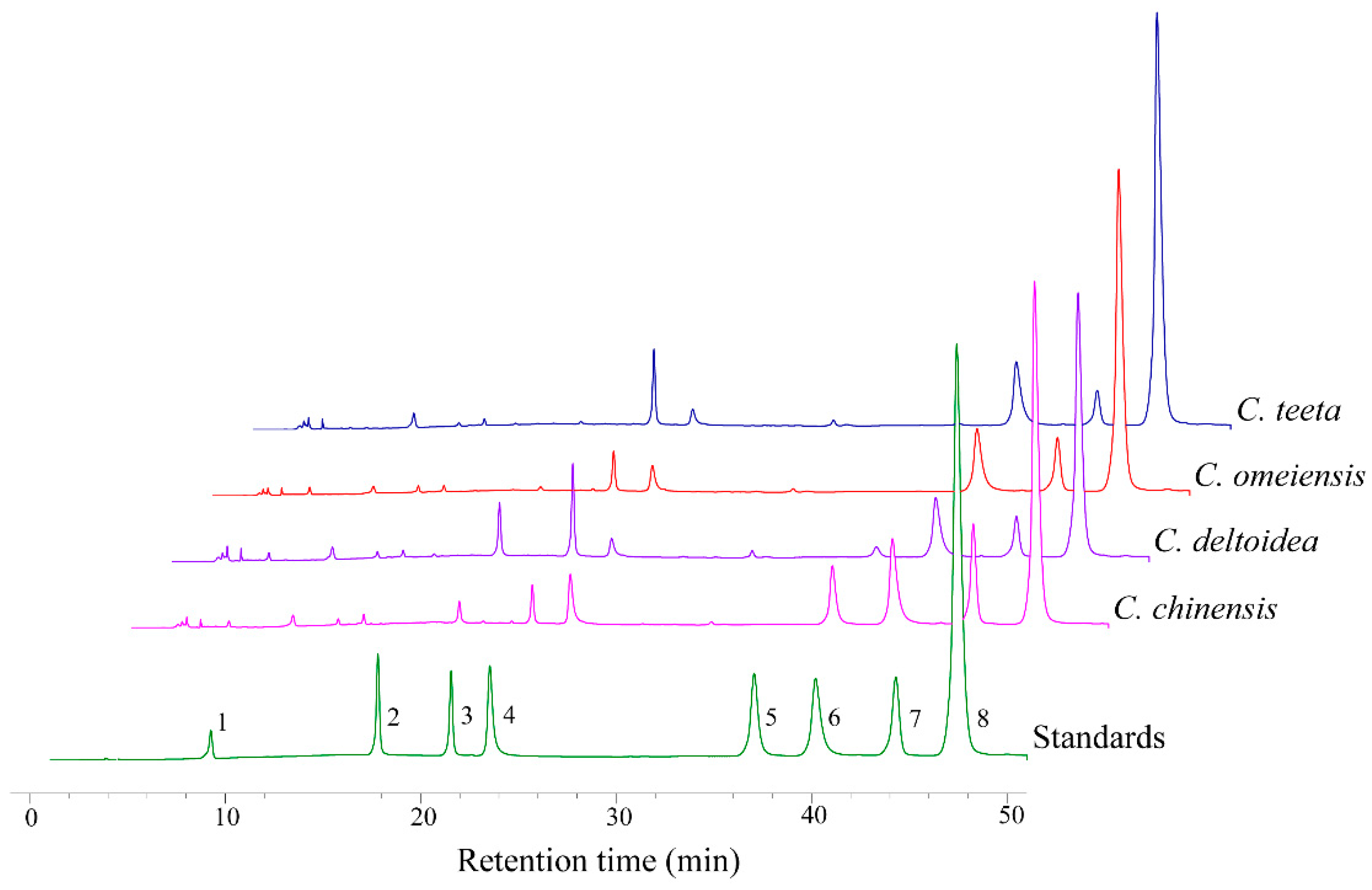

| Standard | Calibration Curve | Linearity (μg/mL) | r2 | LOD (μg/mL) | LOQ (μg/mL) |
|---|---|---|---|---|---|
| Berberine | y = 18906563.4500 x − 18038.6417 | 55.29–470.00 | 0.9998 | 7.24 | 24.13 |
| Palmatine | y = 19332328.3900 x + 6058.2880 | 4.47–54.32 | 0.9998 | 0.69 | 2.32 |
| Jatrorrhizine | y = 14849036.3600 x + 241.1122 | 2.95–35.84 | 0.9998 | 0.51 | 1.69 |
| Coptisine | y = 19176427.8100 x − 2518.8334 | 5.69–69.16 | 0.9999 | 0.61 | 2.04 |
| Columbamine | y = 31118373.4300 x − 2025.8588 | 2.42–29.40 | 0.9998 | 0.39 | 1.31 |
| Epiberberine | y = 19026602.7700 x + 4470.7873 | 5.01–86.81 | 0.9999 | 1.11 | 3.70 |
| Magnoflorine | y = 4769076.5410 x + 293.8634 | 3.25–56.40 | 0.9998 | 0.82 | 2.73 |
| Groenlandicine | y = 13999875.3500 x − 6591.2541 | 2.63–65.20 | 0.9999 | 0.66 | 2.19 |
| Alkaloid | Species | |||
|---|---|---|---|---|
| C. omeiensis | C. teeta | C. chinensis | C. deltoidea | |
| Ber | 62.17 ± 5.36a | 93.46 ± 5.12b | 64.93 ± 4.97a | 48.66 ± 3.24c |
| Pal | 9.94 ± 1.65a | 6.21 ± 0.53b | 15.39 ± 1.92c | 6.72 ± 1.45b |
| Jat | 4.51 ± 0.56a | 10.06 ± 1.09b | 3.88 ± 0.55a | 9.84 ± 0.44b |
| Cop | 15.07 ± 1.65a | 17.23 ± 1.07b | 19.84 ± 2.40c | 12.02 ± 1.26d |
| Col | 2.37 ± 0.33a | 1.44 ± 0.26b | 3.79 ± 0.92c | 1.64 ± 0.16b |
| Epi | N | N | 11.21 ± 1.75a | 1.95 ± 0.74b |
| Mag | 3.17 ± 0.63a | 7.33 ± 0.67b | 4.40 ± 1.26c | 4.57 ± 0.35c |
| Gro | 0.74 ± 0.09a | 0.95 ± 0.80a | 2.66 ± 1.11b | 9.11 ± 2.36c |
| Species | Raw Reads Number | Clean Reads Number (%) | Read Length | Adapter (%) | Low Quality (%) |
|---|---|---|---|---|---|
| C. omeiensis | 10,872,358 | 10,655,424 (98%) | 150/150 | 114,860 (1.06%) | 98,434 (0.91%) |
| C. teeta | 11,100,218 | 10,874,838 (97.97%) | 150/150 | 125,744 (1.13%) | 95,970 (0.86%) |
| C. chinensis | 11,795,008 | 11,548,500 (97.91%) | 150/150 | 110,340 (0.94%) | 132,294 (1.12%) |
| C. deltoidea | 10,979,432 | 10,793,194 (98.3%) | 150/150 | 64,318 (0.59%) | 118,454 (1.08%) |
| Total | 44,747,016 | 43,871,958 | - | 415,262 | 445,152 |
| Feature | Value |
|---|---|
| Number of assembled reference sequence contigs | 965,140 |
| Total assembly length (bp) | 321,905,653 |
| Minimum contig length (bp) | 162 |
| Maximum contig length (bp) | 135,561 |
| GC% | 38.03 |
| N50 Contig Length (bp) | 440 |
| N90 Contig Length (bp) | 170 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, F.; Shen, C.; Qi, L.; Ma, Y. A Multi-Level Strategy Based on Metabolic and Molecular Genetic Approaches for the Characterization of Different Coptis Medicines Using HPLC-UV and RAD-seq Techniques. Molecules 2018, 23, 3090. https://doi.org/10.3390/molecules23123090
Zhong F, Shen C, Qi L, Ma Y. A Multi-Level Strategy Based on Metabolic and Molecular Genetic Approaches for the Characterization of Different Coptis Medicines Using HPLC-UV and RAD-seq Techniques. Molecules. 2018; 23(12):3090. https://doi.org/10.3390/molecules23123090
Chicago/Turabian StyleZhong, Furong, Chan Shen, Luming Qi, and Yuntong Ma. 2018. "A Multi-Level Strategy Based on Metabolic and Molecular Genetic Approaches for the Characterization of Different Coptis Medicines Using HPLC-UV and RAD-seq Techniques" Molecules 23, no. 12: 3090. https://doi.org/10.3390/molecules23123090
APA StyleZhong, F., Shen, C., Qi, L., & Ma, Y. (2018). A Multi-Level Strategy Based on Metabolic and Molecular Genetic Approaches for the Characterization of Different Coptis Medicines Using HPLC-UV and RAD-seq Techniques. Molecules, 23(12), 3090. https://doi.org/10.3390/molecules23123090