Nitrogen-Fixing Bacteria Associated with Peltigera Cyanolichens and Cladonia Chlorolichens
Abstract
1. Introduction
2. Results
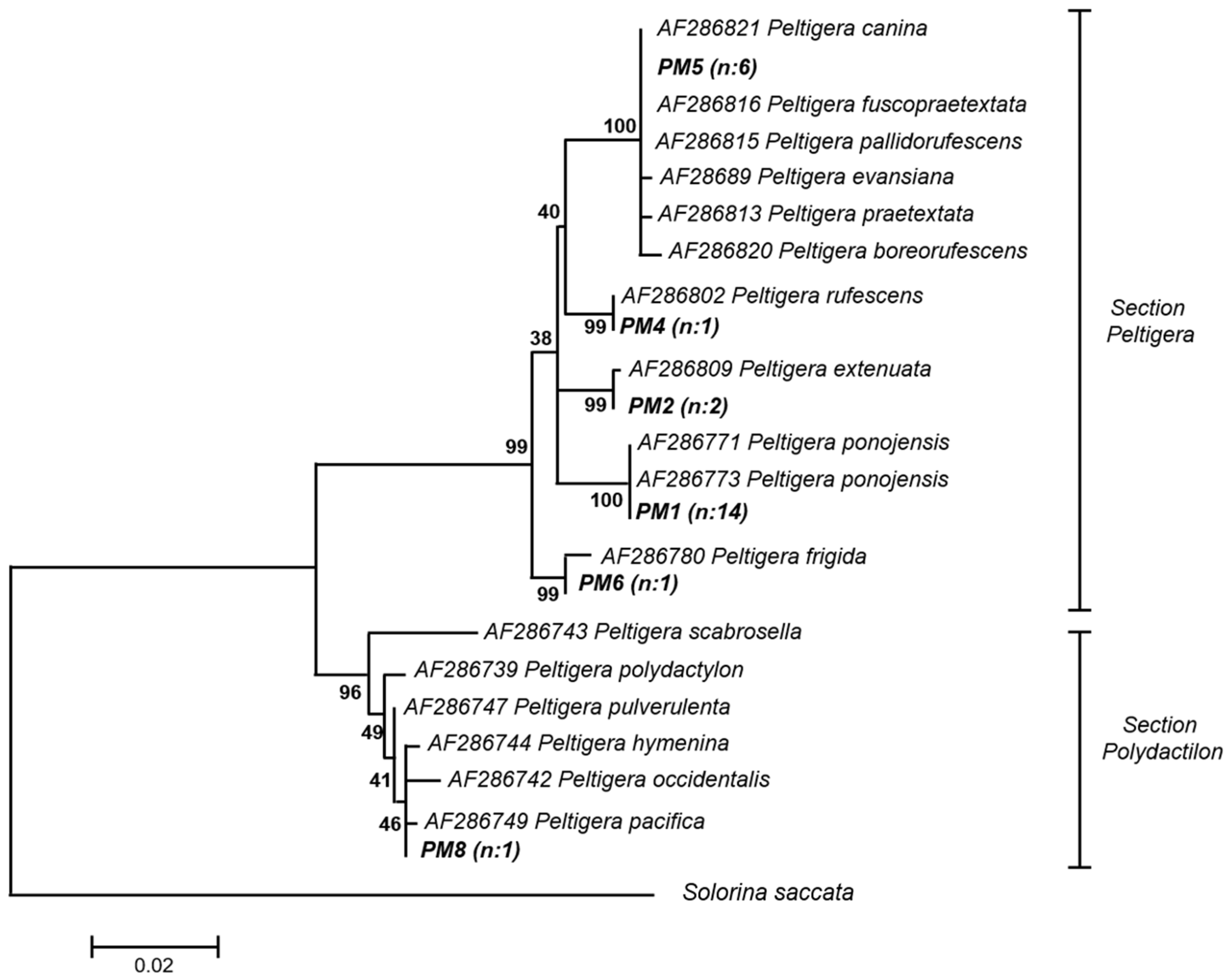
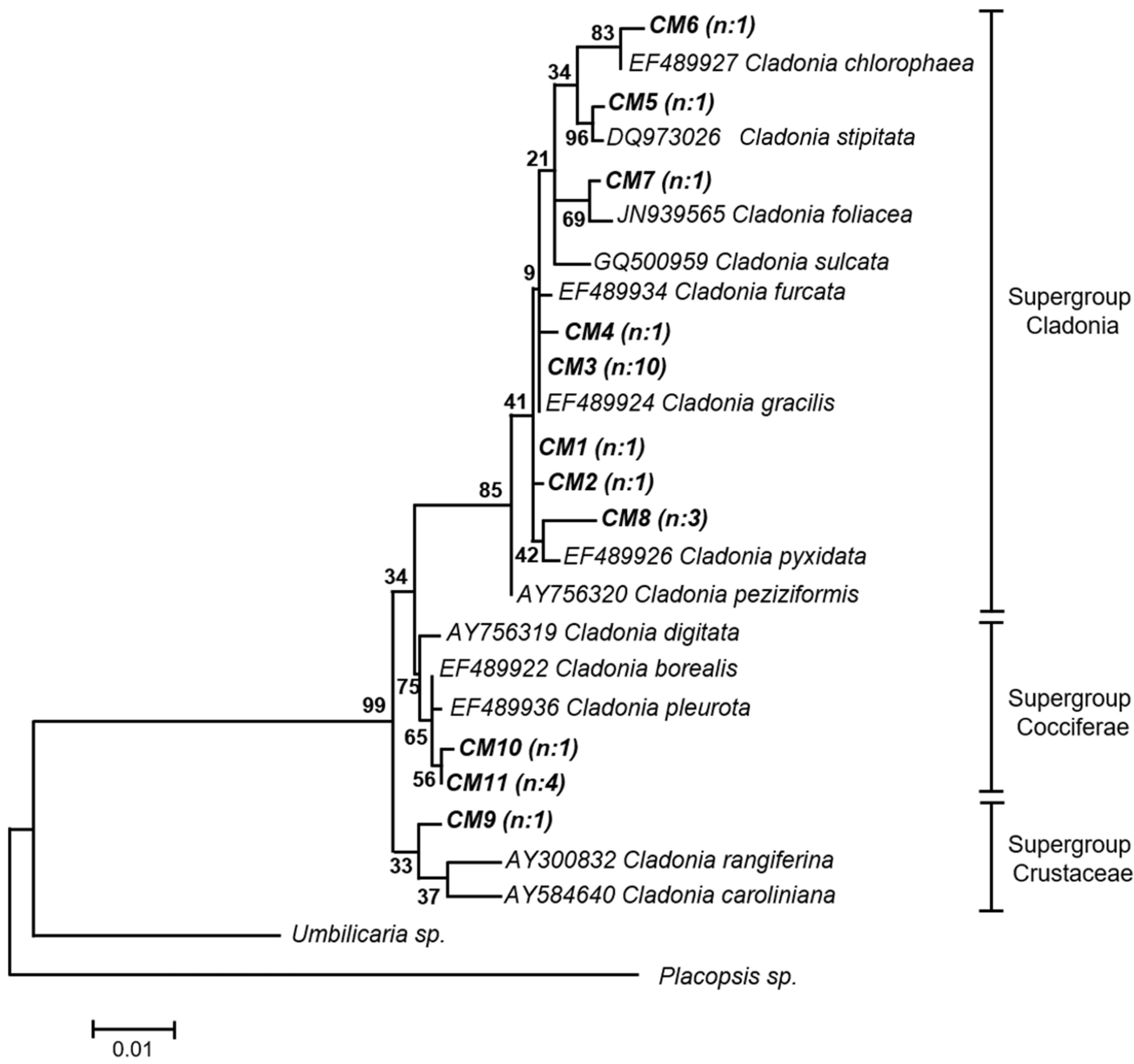
2.1. Molecular Identification of Mycobionts
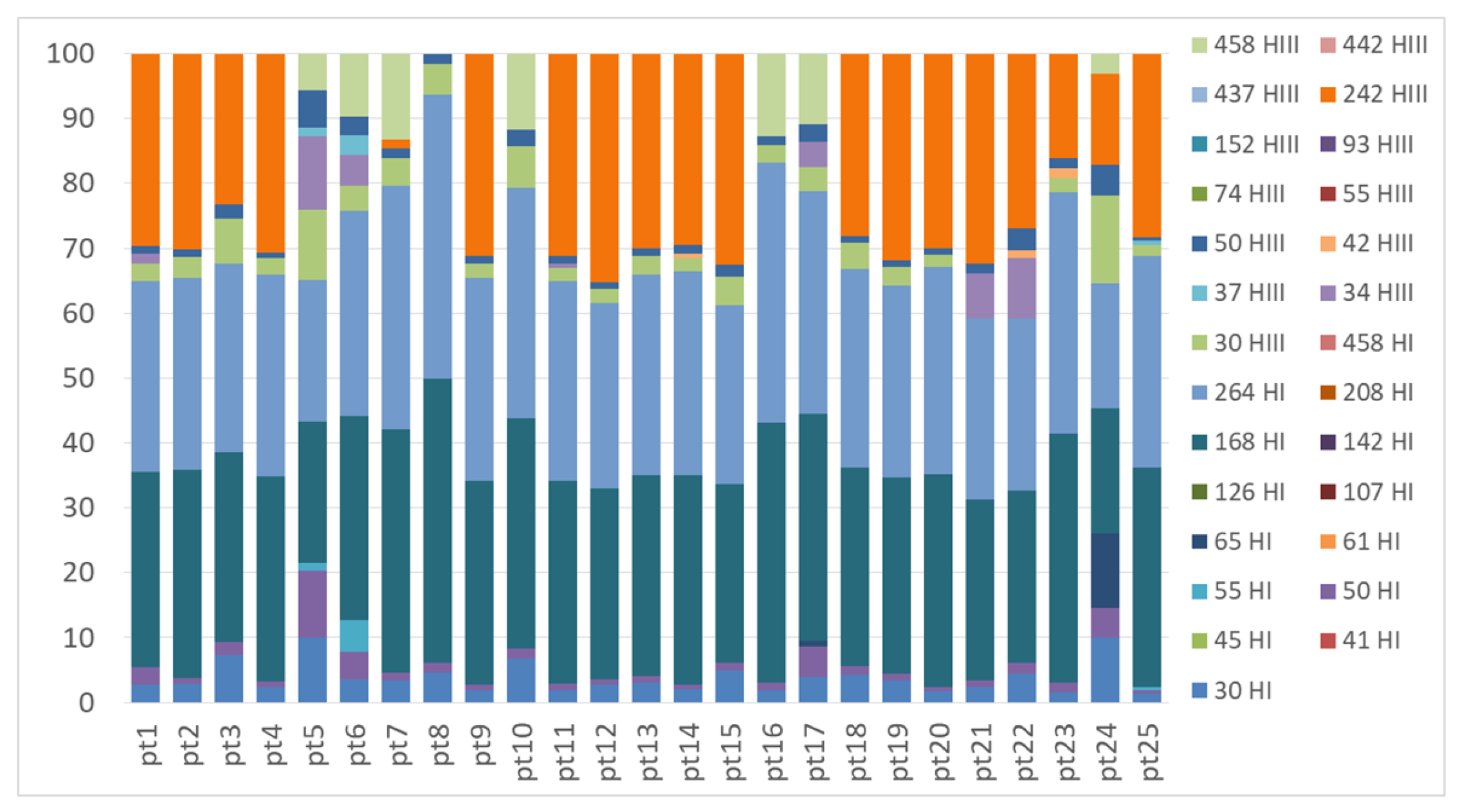
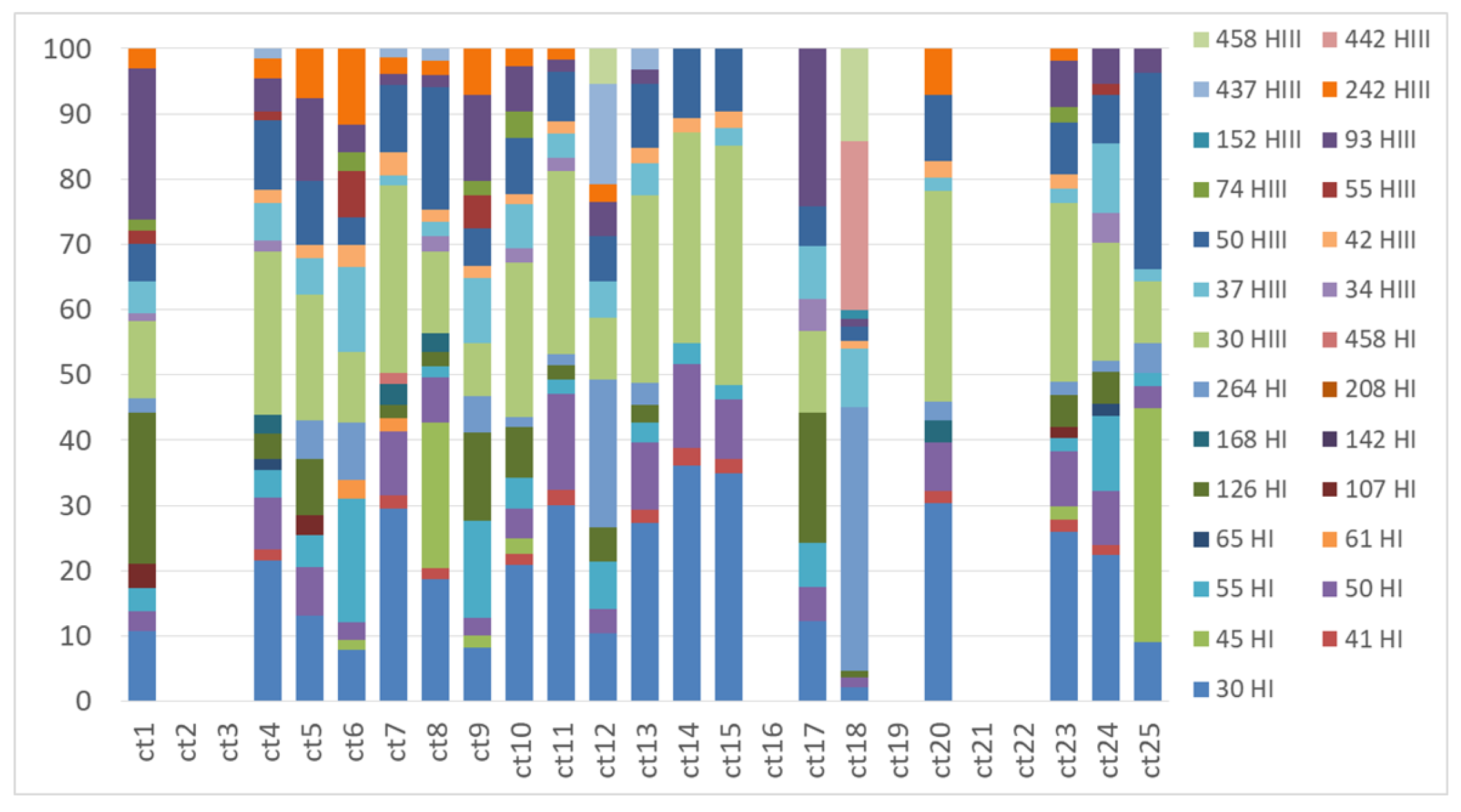
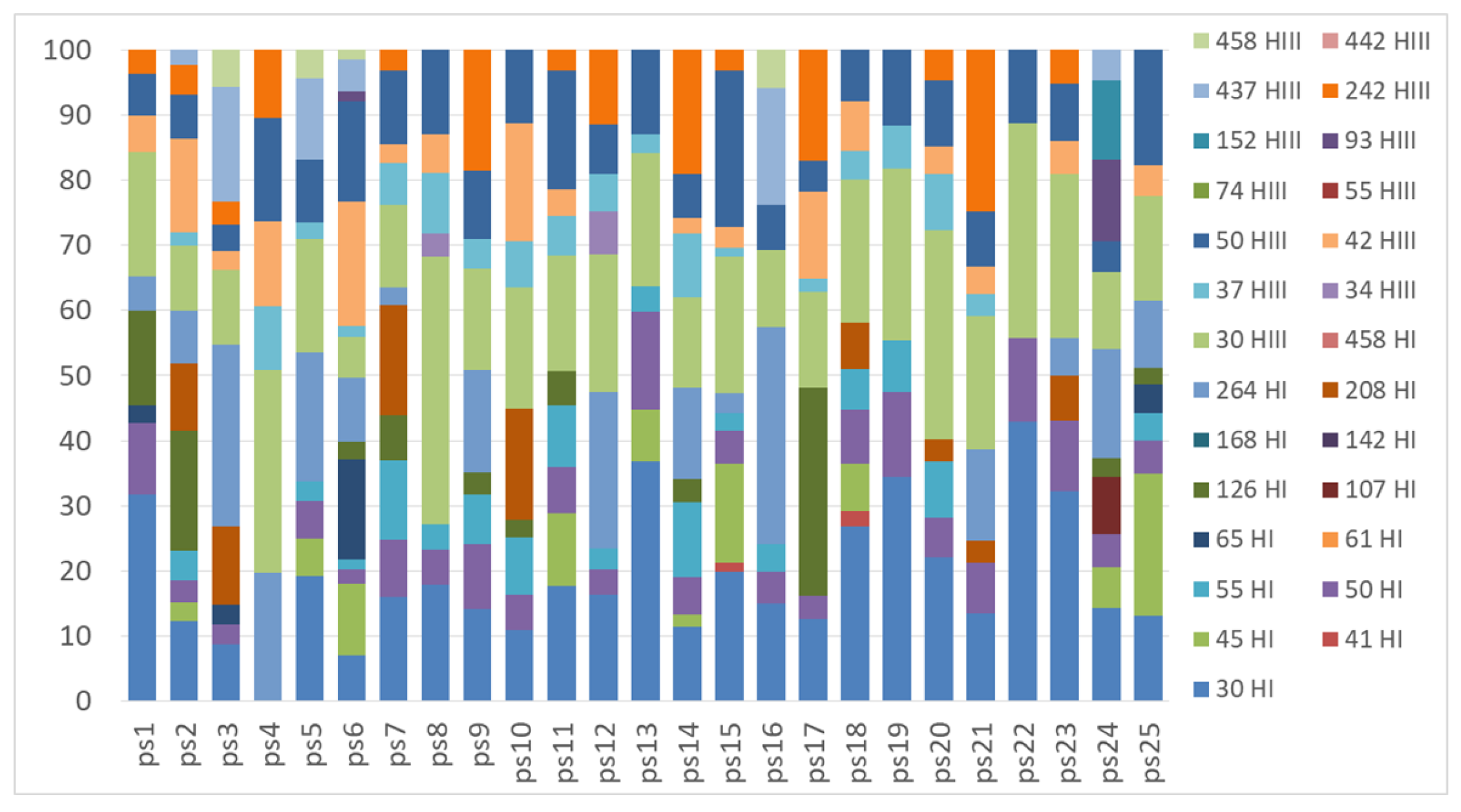
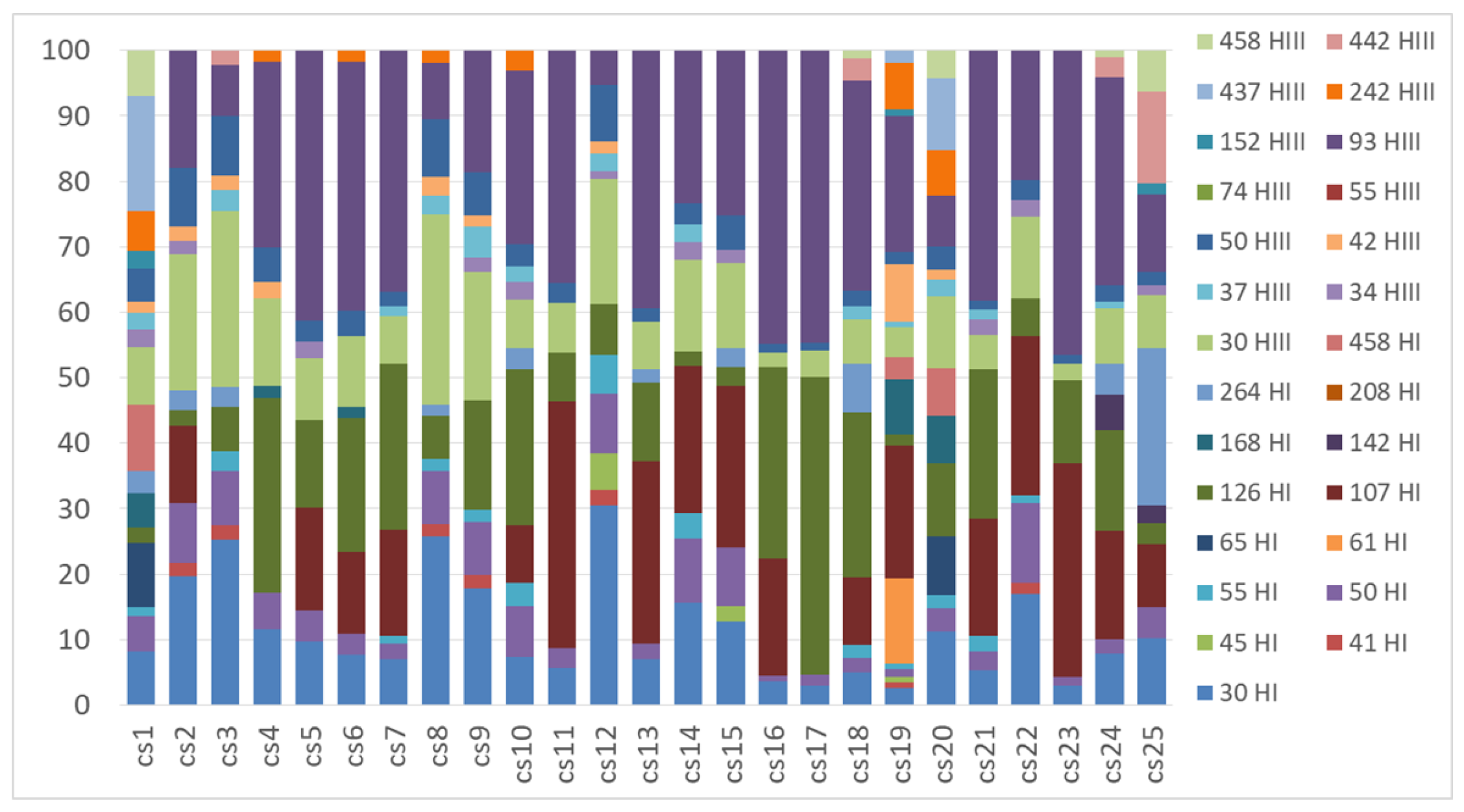
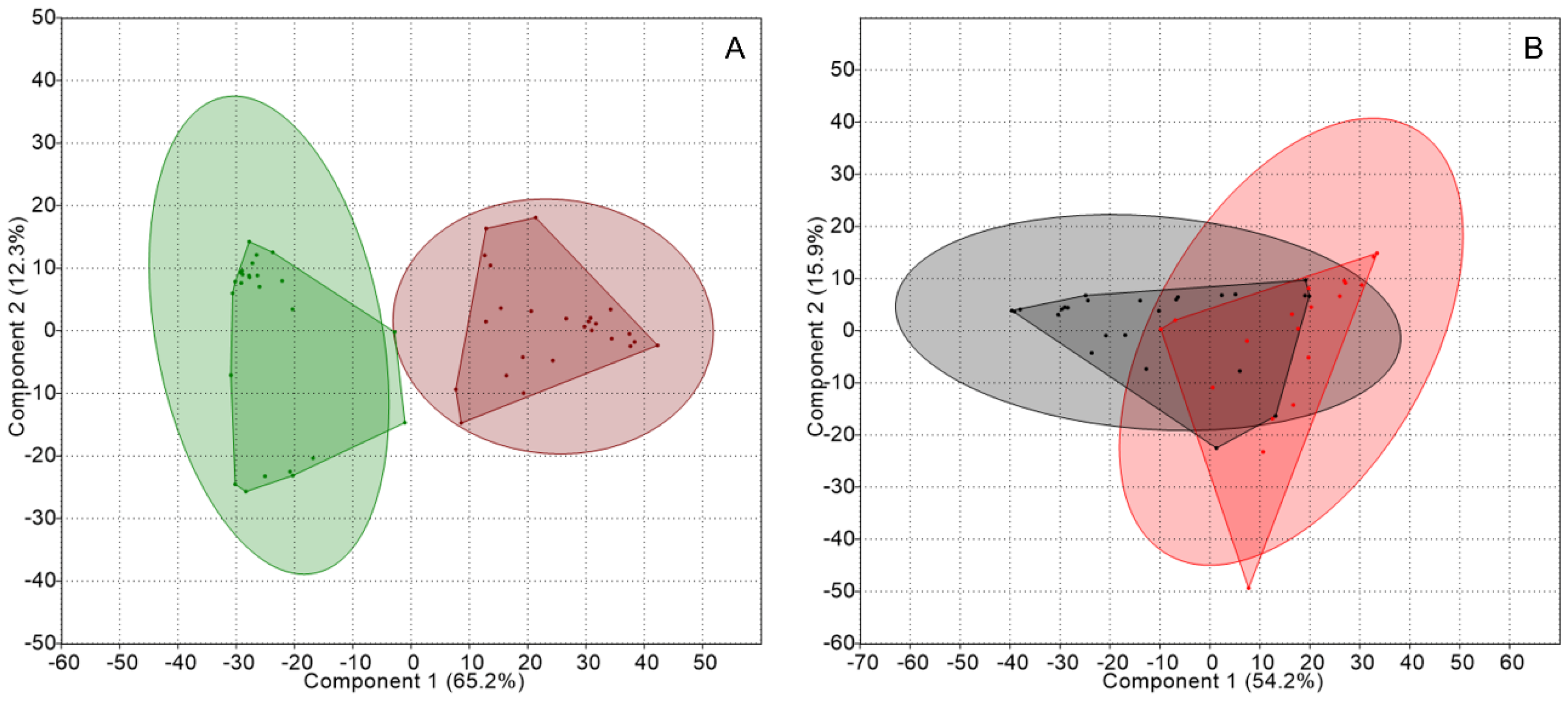
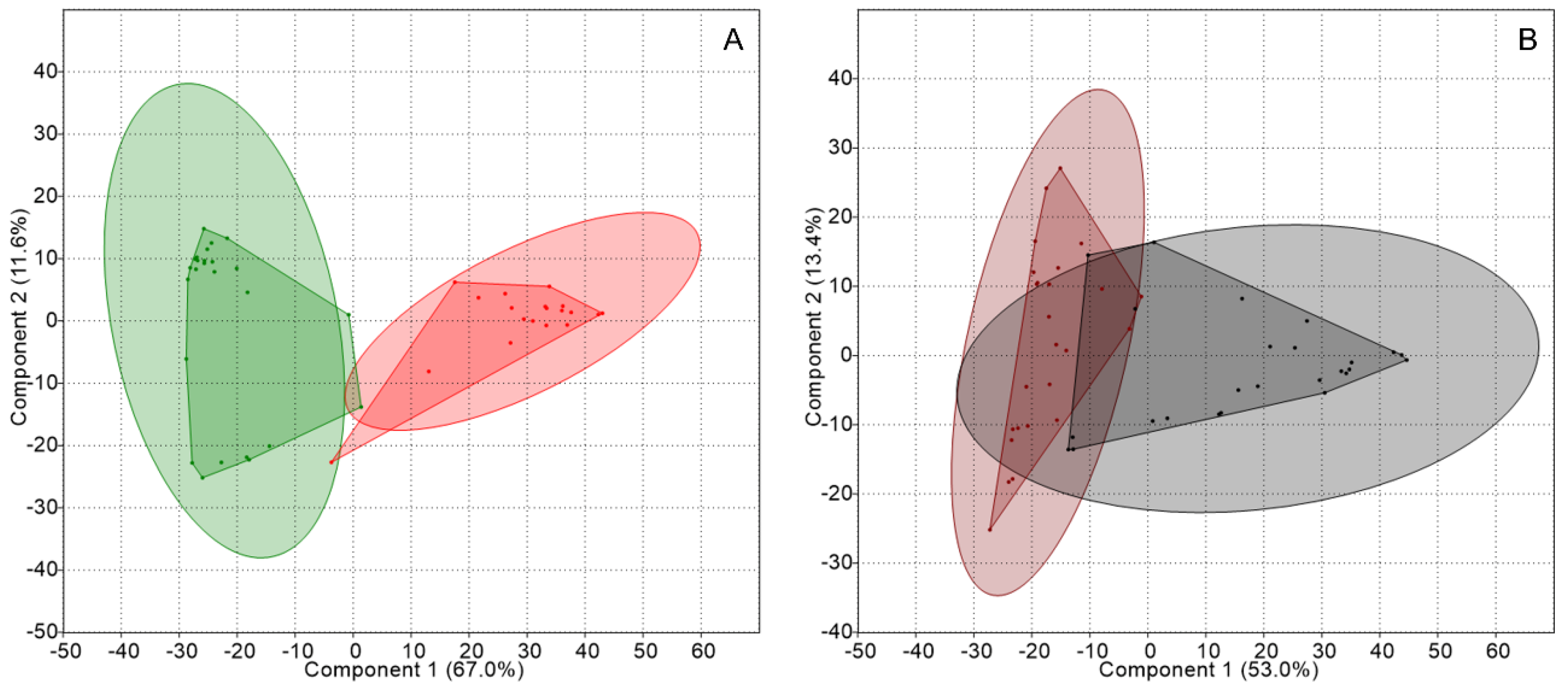
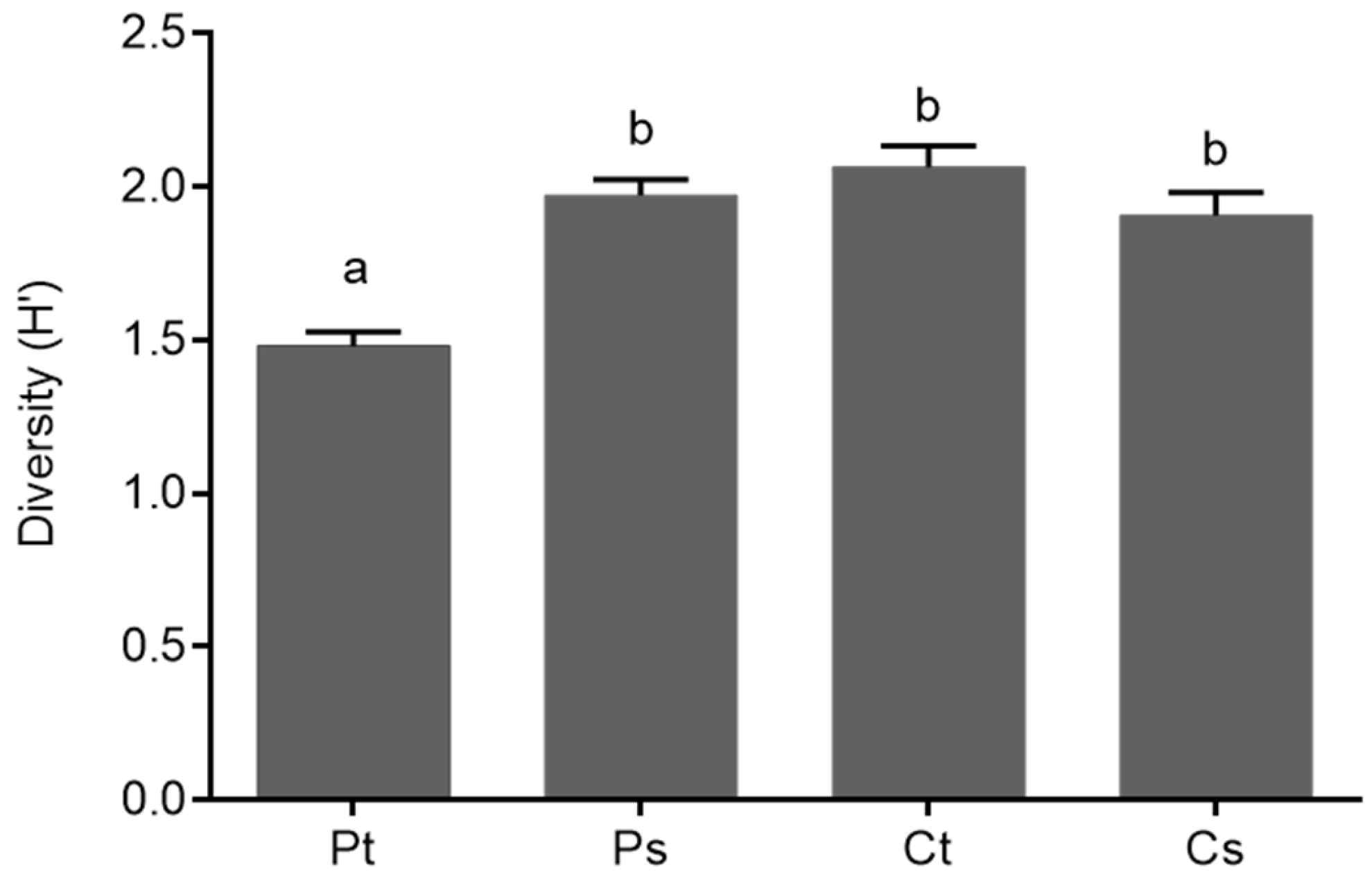
2.2. Genetic Structure of Nitrogen-Fixing Bacteria Associated with Peltigera and Cladonia
3. Discussion
4. Materials and Methods
4.1. Study Site and Sampling
4.2. Pre-Treatment of Samples and DNA Extraction
4.3. Molecular Identification of Mycobionts
4.4. Terminal Restriction Fragment Length Polymorphism
4.5. Data Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Nash, T.H. Lichen Biology, 2nd ed.; Nash, T.H., Ed.; Cambridge University Press: Cambridge, UK, 2008; ISBN 9780521871624. [Google Scholar]
- Petrini, O.; Hake, U.; Dreyfuss, M.M. An analysis of fungal communities isolated from fruticose lichens. Mycologia 1990, 82, 444–451. [Google Scholar] [CrossRef]
- Cornejo, C.; Scheidegger, C. Morphological aspects associated with repair and regeneration in Lobaria pulmonaria and L. amplissima (Peltigerales, Ascomycota). Lichenologist 2013, 45, 285–289. [Google Scholar] [CrossRef]
- Belnap, J. Factors Influencing Nitrogen Fixation. In Biological Soil Crust: Structure, Function, and Management; Belnap, J., Lange, O., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; Volume 150, pp. 241–261. [Google Scholar]
- Rai, A.N. Cyanolichens: Nitrogen metabolism. In Cyanobacterian in Symbiosis; Rai, A.N., Bergman, B., Rasmussen, U., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2002; pp. 97–115. [Google Scholar]
- Grube, M.; Berg, G. Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol. Rev. 2009, 23, 72–85. [Google Scholar] [CrossRef]
- Panosyan, A.; Nikogosyan, G. The presence of Azotobacter in lichens. Biol. Zhurnal Armen. 1966, 19, 3–11. [Google Scholar]
- Henkel, P.; Plotnikova, T. Nitrogen-fixing bacteria in lichens. Izv. Akad. Nauk Seriya Biol. 1973, 807–813. [Google Scholar]
- Zook, P.D. A Study of the Role of Bacteria in Lichens; Clark University: Worcester, MA, USA, 1983. [Google Scholar]
- González, I.; Ayuso-Sacido, A.; Anderson, A.; Genilloud, O. Actinomycetes isolated from lichens: Evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 2005, 54, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Cardinale, M.; Viera de Castro, J.V., Jr.; Müller, H.; Berg, G. Species-specific structural and functional diversity of bacterial communities in lichen symbioses. ISME J. 2009, 3, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Kim, E.H.; Lee, H.K.; Hong, S.G. Biodiversity and physiological characteristics of Antarctic and Arctic lichens-associated bacteria. World J. Microbiol. Biotechnol. 2014, 30, 2711–2721. [Google Scholar] [CrossRef] [PubMed]
- Erlacher, A.; Cernava, T.; Cardinale, M.; Soh, J.; Sensen, C.W.; Grube, M.; Berg, G. Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Front. Microbiol. 2015, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, I.A.; Cernava, T.; Berg, G.; Grube, M. Understanding microbial multi-species symbioses. Front. Microbiol. 2016, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Hodkinson, B.P.; Gottel, N.R.; Schadt, C.W.; Lutzoni, F. Photoautotrophic symbiont and geography are major factors affecting highly structured and diverse bacterial communities in the lichen microbiome. Environ. Microbiol. 2012, 14, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, I.A.; Cardinale, M.; Berg, G.; Grube, M. Microbial cargo: Do bacteria on symbiotic propagules reinforce the microbiome of lichens? Environ. Microbiol. 2014, 16, 3743–3752. [Google Scholar] [CrossRef] [PubMed]
- Leiva, D.; Clavero-León, C.; Carú, M.; Orlando, J. Intrinsic factors of Peltigera lichens influence the structure of the associated soil bacterial microbiota. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, I.A.; Cernava, T.; Erlacher, A.; Berg, G.; Grube, M. Differential sharing and distinct co-occurrence networks among spatially close bacterial microbiota of bark, mosses and lichens. Mol. Ecol. 2017, 26, 2826–2838. [Google Scholar] [CrossRef] [PubMed]
- Almendras, K.; Leiva, D.; Carú, M.; Orlando, J. Carbon consumption patterns of microbial communities associated with Peltigera lichens from a Chilean temperate forest. Molecules 2018, 23, 2746. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Fernández, L.; Zúñiga, C.; Carú, M.; Orlando, J. Environmental context shapes the bacterial community structure associated to Peltigera cyanolichens growing in Tierra del Fuego, Chile. World J. Microbiol. Biotechnol. 2014, 30, 1141–1144. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, C.; Leiva, D.; Carú, M.; Orlando, J. Substrates of Peltigera lichens as a potential source of cyanobionts. Microb. Ecol. 2017, 74, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.A.; Dodsworth, J.A. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 2007, 61, 349–377. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, C.; Leiva, D.; Ramírez-Fernández, L.; Carú, M.; Yahr, R.; Orlando, J. Phylogenetic diversity of Peltigera cyanolichens and their photobionts in southern Chile and Antarctica. Microbes Environ. 2015, 30, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Magain, N.; Truong, C.; Goward, T.; Niu, D.; Goffinet, B.; Sérusiaux, E.; Vitikainen, O.; Lutzoni, F.; Miadlikowska, J. Species delimitation at a global scale reveals high species richness with complex biogeography and patterns of symbiont association in Peltigerasection Peltigera (lichenized Ascomycota: Lecanoromycetes). Taxon 2018, 67, 836–870. [Google Scholar] [CrossRef]
- Magain, N.; Miadlikowska, J.; Goffinet, B.; Sérusiaux, E.; Lutzoni, F. Macroevolution of specificity in cyanolichens of the genus Peltigera section Polydactylon (Lecanoromycetes, Ascomycota). Syst. Biol. 2016, 20, syw065. [Google Scholar] [CrossRef] [PubMed]
- Junier, P.; Junier, T.; Witzel, K.-P. TRiFLe, a program for in silico Terminal Restriction Fragment Length Polymorphism analysis with user-defined sequence sets. Appl. Environ. Microbiol. 2008, 74, 6452–6456. [Google Scholar] [CrossRef] [PubMed]
- Miadlikowska, J.; Lutzoni, F.F. Phylogenetic revision of the genus Peltigera (lichen-forming Ascomycota) based on morphological, chemical, and large subunit nuclear ribosomal DNA data. Int. J. Plant Sci. 2000, 161, 925–958. [Google Scholar] [CrossRef]
- Martínez, I.; Burgaz, A.R.; Vitikainen, O.; Escudero, A. Distribution patterns in the genus Peltigera Willd. Lichenologist 2003, 35, 301–323. [Google Scholar] [CrossRef]
- Quilhot, W.; Cuellar, M.; Díaz, R.; Riquelme, F.; Rubio, C. Lichens of Aisen, Southern Chile. Gayana Bot. 2012, 69, 57–87. [Google Scholar] [CrossRef]
- Ramírez-Fernández, L.; Zúñiga, C.; Méndez, M.A.; Carú, M.; Orlando, J. Genetic diversity of terricolous Peltigera cyanolichen communities in different conservation states of native forest from southern Chile. Int. Microbiol. 2013, 16, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Lumbsch, H.T.; Leavitt, S.D. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Divers. 2011, 50, 59–72. [Google Scholar] [CrossRef]
- Miadlikowska, J.; Lutzoni, F.; Goward, T.; Zoller, S.; Posada, D. New approach to an old problem: Incorporating signal from gap-rich regions of ITS and rDNA large subunit into phylogenetic analyses to resolve the Peltigera canina species complex. Mycologia 2003, 95, 1181–1203. [Google Scholar] [CrossRef] [PubMed]
- Lendemer, J.C.; Hodkinsons, B.P. The Wisdom of Fools: New molecular and morphological insights into the North American apodetiate species of Cladonia. Opusc. Philol. 2009, 7, 79–100. [Google Scholar]
- Sgrignani, J.; Franco, D.; Magistrato, A. Theoretical studies of homogeneous catalysts mimicking nitrogenase. Molecules 2011, 16, 442–465. [Google Scholar] [CrossRef] [PubMed]
- Zehr, J.P.; Jenkins, B.D.; Short, S.M.; Steward, G.F. Nitrogenase gene diversity and microbial community structure: A cross-system comparison. Env. Microbiol 2003, 5, 539–554. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE 2012, 7, e93883. [Google Scholar] [CrossRef] [PubMed]
- Hodkinson, B.P.; Lutzoni, F. A microbiotic survey of lichen-associated bacteria reveals a new lineage from the Rhizobiales. Symbiosis 2009, 49, 163–180. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. A comprehensive aligned nifH gene database: A multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Grube, M.; Castro, J.V., Jr.; Müller, H.; Berg, G.; Castro, J.V.; Müller, H.; Berg, G.; Castro, J.V., Jr.; Müller, H.; Berg, G. Bacterial taxa associated with the lung lichen Lobaria pulmonaria are differentially shaped by geography and habitat. FEMS Microbiol. Lett. 2012, 329, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Vieira de Castro, J.; Müller, H.; Berg, G.; Grube, M. In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol. Ecol. 2008, 66, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Graham, L.E.; Trest, M.T.; Will-Wolf, S.; Miicke, N.S.; Atonio, L.M.; Piotrowski, M.J.; Knack, J.J. Microscopic and metagenomic analyses of Peltigera ponojensis (Peltigerales, Ascomycota). Int. J. Plant Sci. 2018, 179, 241–255. [Google Scholar] [CrossRef]
- Mushegian, A.A.; Peterson, C.N.; Baker, C.C.M.; Pringle, A. Bacterial diversity across individual lichens. Appl. Environ. Microbiol. 2011, 77, 4249–4252. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.T.; Cropsey, G.W.G.; Caporaso, J.G.; Knight, R.; Fierer, N. Bacterial communities associated with the lichen symbiosis. Appl. Environ. Microbiol. 2011, 77, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Cernava, T.; Soh, J.; Fuchs, S.; Aschenbrenner, I.; Lassek, C.; Wegner, U.; Becher, D.; Riedel, K.; Sensen, C.W.; et al. Exploring functional contexts of symbiotic sustain within lichen-associated bacteria by comparative omics. ISME J. 2015, 9, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Eymann, C.; Lassek, C.; Wegner, U.; Bernhardt, J.; Fritsch, O.A.; Fuchs, S.; Otto, A.; Albrecht, D.; Schiefelbein, U.; Cernava, T.; et al. Symbiotic interplay of fungi, algae, and bacteria within the lung lichen Lobaria pulmonaria L. Hoffm. as assessed by state-of-the-art metaproteomics. J. Proteome Res. 2017, 16, 2160–2173. [Google Scholar] [CrossRef] [PubMed]
- Sigurbjörnsdóttir, M.A.; Andrésson, Ó.S.; Vilhelmsson, O.; Auður Sigurbjörnsdóttir, M.; Andrésson, Ó.S.; Vilhelmsson, O.; Sigurbjörnsdóttir, M.A.; Andrésson, Ó.S.; Vilhelmsson, O. Nutrient scavenging activity and antagonistic factors of non-photobiont lichen-associated bacteria: A review. World J. Microbiol. Biotechnol. 2016, 32, 68. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.-F.; Wang, H.-Y.; Si, H.-L.; Zhao, L.; Liu, C.-P.; Zhang, H. Isolation and culture of lichen bacteriobionts. Lichenology 2017, 49, 175–181. [Google Scholar] [CrossRef]
- Salgado, F.; Albornoz, L.; Cortéz, C.; Stashenko, E.; Urrea-Vallejo, K.; Nagles, E.; Galicia-Virviescas, C.; Cornejo, A.; Ardiles, A.; Simirgiotis, M.; et al. Secondary metabolite profiling of species of the genus Usnea by UHPLC-ESI-OT-MS-MS. Molecules 2018, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Parrot, D.; Legrave, N.; Delmail, D.; Grube, M.; Suzuki, M.; Tomasi, S. Review—Lichen-associated bacteria as a hot spot of chemodiversity: Focus on uncialamycin, a promising compound for future medicinal applications. Planta Med. 2016, 82, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Boustie, J.; Grube, M. Lichens-a promising source of bioactive secondary metabolites. Plant Genet. Resour. Charact. Util. 2005, 3, 273–287. [Google Scholar] [CrossRef]
- White, P.A.S.; Oliveira, R.C.M.; Oliveira, A.P.; Serafini, M.R.; Araújo, A.A.S.; Gelain, D.P.; Moreira, J.C.F.; Almeida, J.R.G.S.; Quintans, J.S.S.; Quintans-Junior, L.J.; et al. V Antioxidant activity and mechanisms of action of natural compounds isolated from lichens: A systematic review. Molecules 2014, 19, 14496–14527. [Google Scholar] [CrossRef] [PubMed]
- Legouin, B.; Lohézic-Le Dévéhat, F.; Ferron, S.; Rouaud, I.; Le Pogam, P.; Cornevin, L.; Bertrand, M.; Boustie, J. Specialized metabolites of the lichen Vulpicida pinastri act as photoprotective agents. Molecules 2017, 22, 1162. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.S.; Kim, T.K.; Kim, J.E.; Hong, J.M.; Nguyen, T.T.T.; Han, S.J.; Youn, U.J.; Yim, J.H.; Kim, I.C. Anticancer activity of ramalin, a secondary metabolite from the antarctic lichen Ramalina terebrata, against colorectal cancer cells. Molecules 2017, 22, 1361. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.M.; Suh, S.S.; Kim, T.K.; Kim, J.E.; Han, S.J.; Youn, U.J.; Yim, J.H.; Kim, I.C. Anti-cancer activity of lobaric acid and lobarstin extracted from the antarctic lichen Stereocaulon alpnum. Molecules 2018, 23, 658. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.P.; Wright, A.J. Environmental filtering, niche construction, and trait variability: The missing discussion. Trends Ecol. Evol. 2017, 32, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, M.; Steinová, J.; Rabensteiner, J.; Berg, G.; Grube, M. Age, sun and substrate: Triggers of bacterial communities in lichens. Environ. Microbiol. Rep. 2012, 4, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Printzen, C.; Fernández-Mendoza, F.; Muggia, L.; Berg, G.; Grube, M. Alphaproteobacterial communities in geographically distant populations of the lichen Cetraria aculeata. FEMS Microbiol. Ecol. 2012, 82, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Lambright, D.; Kapustka, A. The association of N2-fixing bacteria with Dermatocarpon miniatum and Lepraria sp. Bot. Soc. Am. Misc. 1981, 160, 5. [Google Scholar]
- Liba, C.M.; Ferrara, F.I.S.; Manfio, G.P.; Fantinatti-Garboggini, F.; Albuquerque, R.C.; Pavan, C.; Ramos, P.L.; Moreira-Filho, C.A.; Barbosa, H.R. Nitrogen-fixing chemo-organotrophic bacteria isolated from cyanobacteria-deprived lichens and their ability to solubilize phosphate and to release amino acids and phytohormones. J. Appl. Microbiol. 2006, 101, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Till-Bottraud, I.; Fajardo, A.; Rioux, D. Multi-stemmed trees of Nothofagus pumilio second-growth forest in Patagonia are formed by highly related individuals. Ann. Bot. 2012, 110, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Soc. Study Evol. 1985, 39, 1–15. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2011, 39, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Rösch, C.; Mergel, A.; Bothe, H.; Ro, C. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 2002, 68, 3818–3829. [Google Scholar] [CrossRef] [PubMed]
- Almasia, R.; Carú, M.; Handford, M.; Orlando, J. Environmental conditions shape soil bacterial community structure in a fragmented landscape. Soil Biol. Biochem. 2016, 103, 39–45. [Google Scholar] [CrossRef]
- Dunbar, J.; Ticknor, L.O.; Kuske, C.R. Phylogenetic specificity and reproducibility and new method for analysis of Terminal Restriction Fragment Profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 2001, 67, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kaplan, C.W.; Kitts, C.L. Variation between observed and true Terminal Restriction Fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 2003, 54, 121–125. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9–18. [Google Scholar] [CrossRef]
- Stenroos, S.; Hyvönen, J.; Myllys, L.; Thell, A.; Ahti, T. Phylogeny of the genus Cladonia s.lat. (Cladoniaceae, Ascomycetes) inferred from molecular, morphological, and chemical data. Cladistics 2002, 18, 237–278. [Google Scholar] [CrossRef]
Sample Availability: Not Available. |
| HhaI | HindIII | Putative Identification | Pt | Ps | Ct | Cs |
|---|---|---|---|---|---|---|
| 58 (0) | 40 (3) | Alphaproteobacteria; Rhodobacterales | ||||
| 90 (6) | uncultured bacterium | |||||
| 154 (0) | Actinobacteria; Frankiales | |||||
| 465 (7) | Cyanobacteria; Nostocales | |||||
| 144 (3) | 53 (5) | Actinobacteria; Frankiales | ||||
| Alphaproteobacteria; Rhizobiales | ||||||
| Betaproteobacteria; Burkholderiales | ||||||
| Gammaproteobacteria; Pseudomonadales | ||||||
| Firmicutes; Clostridiales | ||||||
| uncultured bacterium | ||||||
| 144 (3–6) | 465 (4–10) | Cyanobacteria; Nostocales | ||||
| 465 (4–24) | 53 (3–5) | Firmicutes; Clostridiales | ||||
| uncultured bacterium |
| Samples | ANOSIM R | ANOSIM p | SIMPER Dissimilarity (%) |
|---|---|---|---|
| TRFLP-Pt | 0.9355 | 0.0001 | 75.6 |
| TRFLP-Ps | |||
| TRFLP-Ct | 0.3871 | 0.0001 | 59.1 |
| TRFLP-Cs | |||
| TRFLP-Pt | 0.9427 | 0.0001 | 80.7 |
| TRFLP-Ct | |||
| TRFLP-Ps | 0.6212 | 0.0001 | 65.6 |
| TRFLP-Cs |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almendras, K.; García, J.; Carú, M.; Orlando, J. Nitrogen-Fixing Bacteria Associated with Peltigera Cyanolichens and Cladonia Chlorolichens. Molecules 2018, 23, 3077. https://doi.org/10.3390/molecules23123077
Almendras K, García J, Carú M, Orlando J. Nitrogen-Fixing Bacteria Associated with Peltigera Cyanolichens and Cladonia Chlorolichens. Molecules. 2018; 23(12):3077. https://doi.org/10.3390/molecules23123077
Chicago/Turabian StyleAlmendras, Katerin, Jaime García, Margarita Carú, and Julieta Orlando. 2018. "Nitrogen-Fixing Bacteria Associated with Peltigera Cyanolichens and Cladonia Chlorolichens" Molecules 23, no. 12: 3077. https://doi.org/10.3390/molecules23123077
APA StyleAlmendras, K., García, J., Carú, M., & Orlando, J. (2018). Nitrogen-Fixing Bacteria Associated with Peltigera Cyanolichens and Cladonia Chlorolichens. Molecules, 23(12), 3077. https://doi.org/10.3390/molecules23123077






