Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Qualifying Chemical Probes
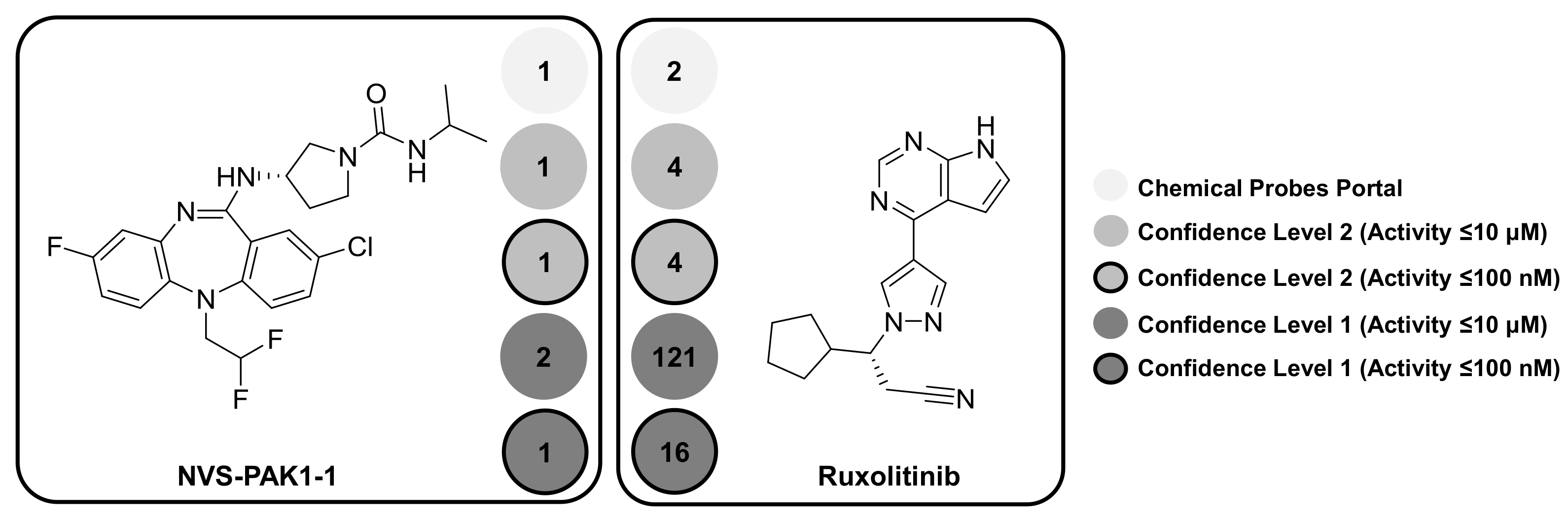
2.2. Selectivity Trends of Chemical Probes
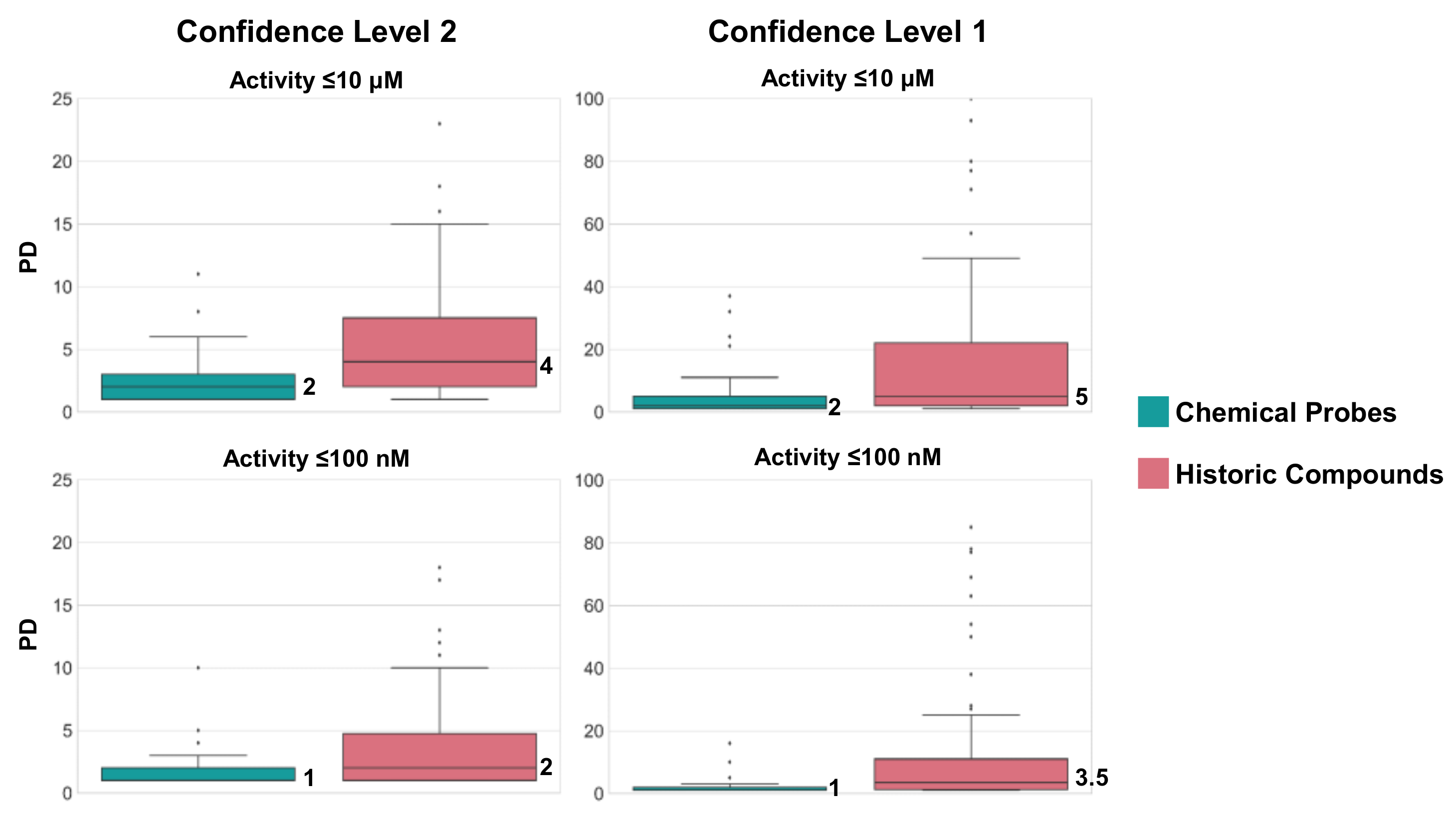
2.3. Chemical Probes and Historic Compounds
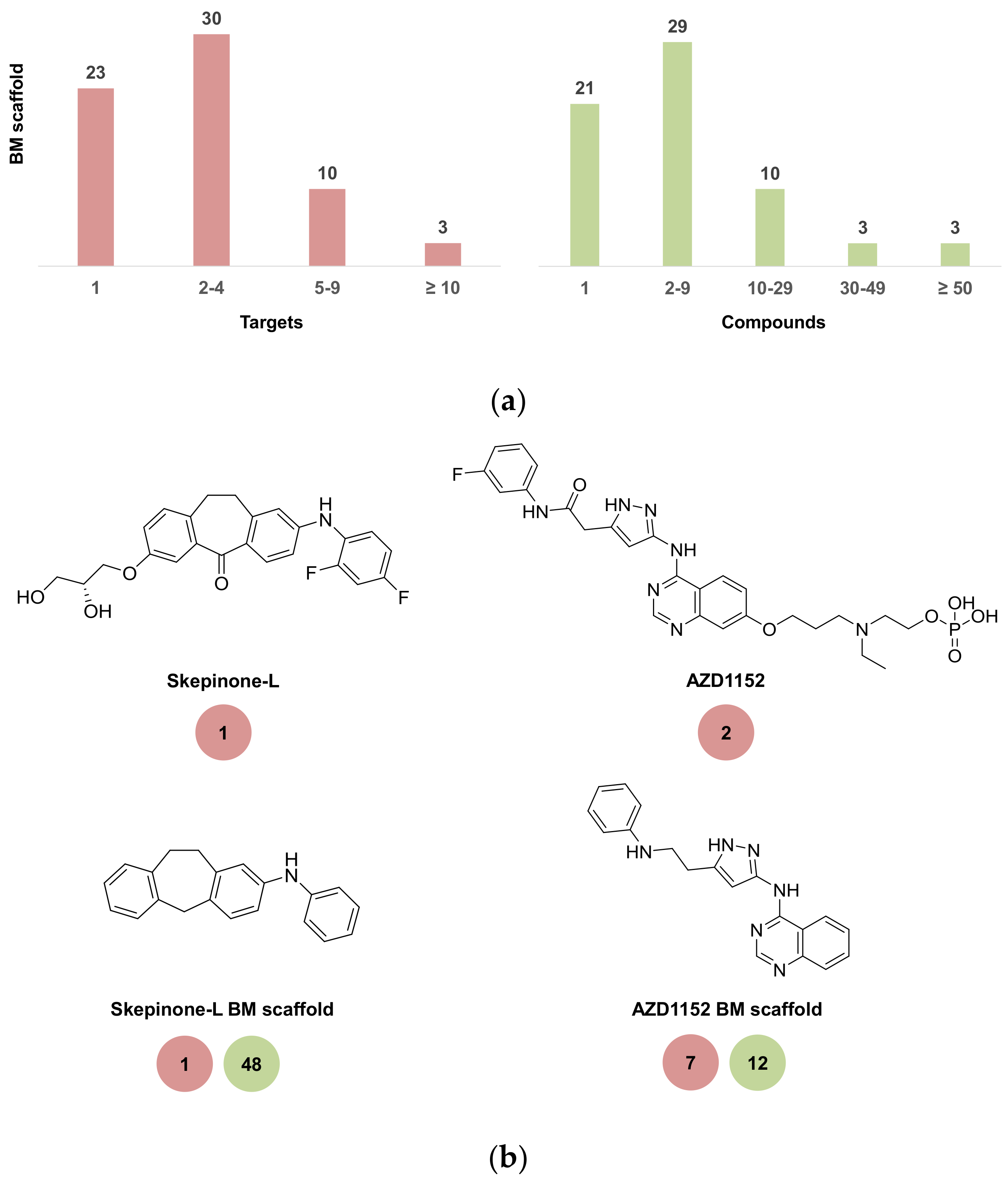
2.4. Scaffold Analysis of Chemical Probes
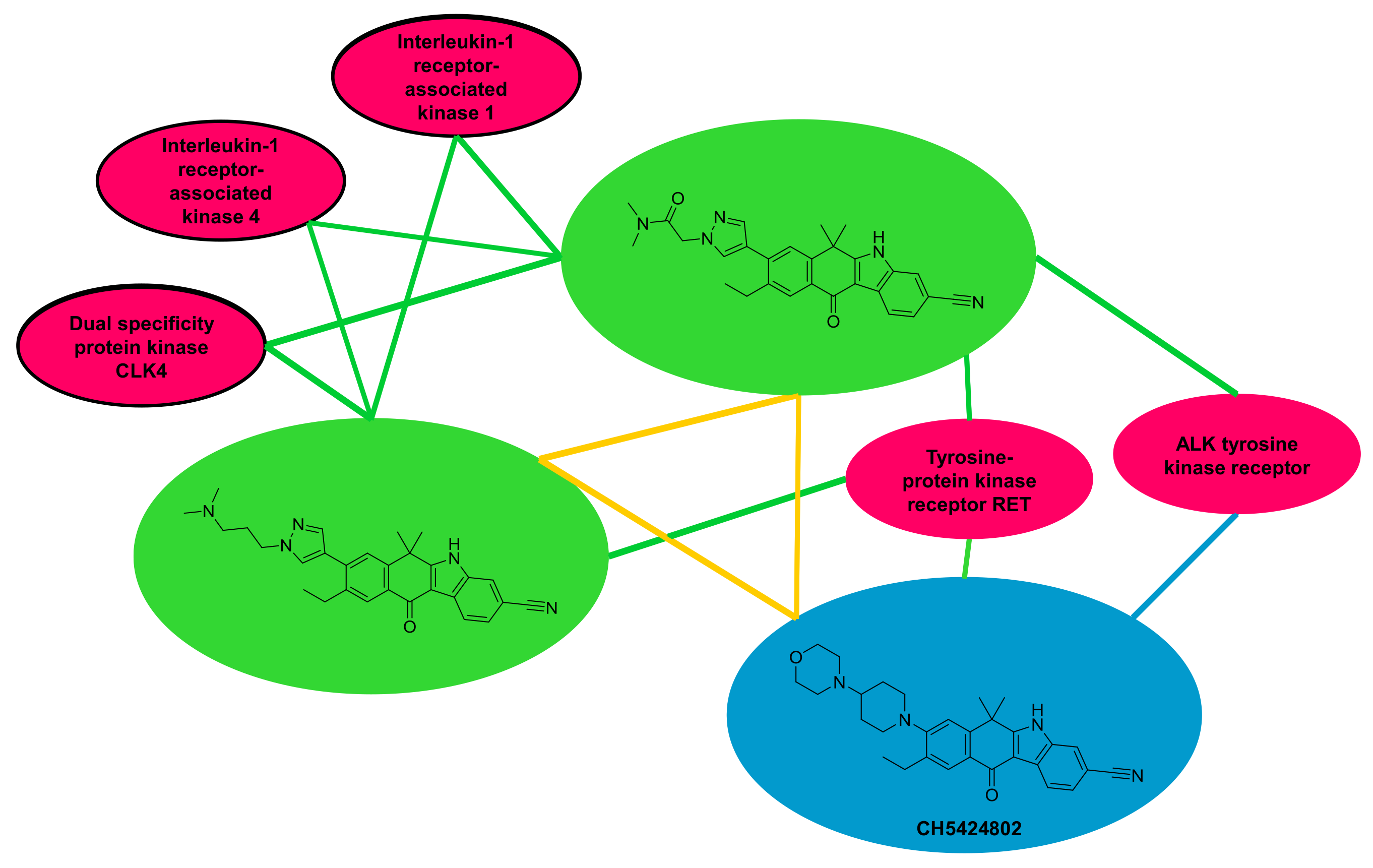
2.5. Off-Target Activity Assessment in Networks
2.6. Summary
3. Materials and Methods
3.1. Chemical Probes
3.2. Activity Data, Confidence Levels, and Historic Compounds
3.3. Bioactive Compounds, Scaffold Analysis, and Off-Target Predictions
Author Contributions
Conflicts of Interest
References
- International Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Finishing the Euchromatic Sequence of the Human Genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Schenone, M.; Dančík, V.; Wagner, B.K.; Clemons, P.A. Target Identification and Mechanism of Action in Chemical Biology and Drug Discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Cornish, P.V.; Ha, T. A Survey of Single-Molecule Techniques in Chemical Biology. ACS Chem. Biol. 2007, 2, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Knowles, J.; Gromo, G. Target Selection in Drug Discovery. Nat. Rev. Drug Discov. 2003, 2, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.M.; Isserlin, R.; Bader, G.D.; Frye, S.V.; Willson, T.M.; Yu, F.H. Too Many Roads Not Taken. Nature 2011, 470, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored Therapeutic Opportunities in the Human Genome. Nat. Rev. Drug Discov. 2018, 17, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Bunnage, M.E.; Chekler, E.L.P.; Jones, L.H. Target Validation Using Chemical Probes. Nat. Chem. Biol. 2013, 9, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.H.; Bunnage, M.E. Applications of Chemogenomic Library Screening in Drug Discovery. Nat. Rev. Drug Discov. 2017, 16, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.M.; Niphakis, M.J.; Cravatt, B.F. Determining Target Engagement in Living Systems. Nat. Chem. Biol. 2013, 9, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Frye, S.V. The Art of the Chemical Probe. Nat. Chem. Biol. 2010, 6, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Workman, P.; Collins, I. Probing the Probes: Fitness Factors for Small Molecule Tools. Chem. Biol. 2010, 17, 561–577. [Google Scholar] [CrossRef] [PubMed]
- Arrowsmith, C.H.; Audia, J.E.; Austin, C.; Baell, J.; Bennett, J.; Blagg, J.; Bountra, C.; Brennan, P.E.; Brown, P.J.; Bunnage, M.E.; et al. The Promise and Peril of Chemical Probes. Nat. Chem. Biol. 2015, 11, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Chemical Probes Portal. Available online: http://www.chemicalprobes.org/ (accessed on 14 July 2018).
- Structural Genomics Consortium. Available online: https://www.thesgc.org/ (accessed on 22 August 2018).
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A Large-scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [PubMed]
- Bemis, G.W.; Murcko, M.A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef] [PubMed]
- Kunimoto, R.; Bajorath, J. Design of a Tripartite Network for the Prediction of Drug Targets. J. Comput. Aided. Mol. Des. 2018, 32, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Miljković, F.; Bajorath, J. Exploring Selectivity of Multikinase Inhibitors across the Human Kinome. ACS Omega 2018, 3, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Miljković, F.; Bajorath, J. Reconciling Selectivity Trends from a Comprehensive Kinase Inhibitor Profiling Campaign with Known Activity Data. ACS Omega 2018, 3, 3113–3119. [Google Scholar] [CrossRef] [PubMed]
- Miljković, F.; Bajorath, J. Evaluation of Kinase Inhibitor Selectivity Using Cell-Based Profiling Data. Mol. Inform. 2018, 37, 1800024. [Google Scholar] [CrossRef] [PubMed]
- Karpov, A.S.; Amiri, P.; Bellamacina, C.; Bellance, M.-H.; Breitenstein, W.; Daniel, D.; Denay, R.; Fabbro, D.; Fernandez, C.; Galuba, I.; et al. Optimization of a Dibenzodiazepine Hit to a Potent and Selective Allosteric PAK1 Inhibitor. ACS Med. Chem. Lett. 2015, 6, 776–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintás-Cardama, A.; Vaddi, K.; Liu, P.; Manshouri, T.; Li, J.; Scherle, P.A.; Caulder, E.; Wen, X.; Li, Y.; Waeltz, P.; et al. Preclinical Characterization of the Selective JAK1/2 Inhibitor INCB018424: Therapeutic Implications for the Treatment of Myeloproliferative Neoplasms. Blood 2010, 115, 3109–3117. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, S.C.; Fischer, S.; Schollmeyer, D.; Schattel, V.; Grütter, C.; Rauh, D.; Laufer, S.A. Design, Synthesis, and Biological Evaluation of Novel Disubstituted Dibenzosuberones as Highly Potent and Selective Inhibitors of p38 Mitogen Activated Protein Kinase. J. Med. Chem. 2012, 55, 5868–5877. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, S.C.; Romir, J.; Fischer, S.; Koeberle, A.; Schattel, V.; Albrecht, W.; Grütter, C.; Werz, O.; Rauh, D.; Stehle, T.; et al. Skepinone-L is a Selective p38 Mitogen-activated Protein Kinase Inhibitor. Nat. Chem. Biol. 2012, 8, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Mortlock, A.A.; Foote, K.M.; Heron, N.M.; Jung, F.H.; Pasquet, G.; Lohmann, J.-J.M.; Warin, N.; Renaud, F.; De Savi, C.; Roberts, N.J.; et al. Discovery, Synthesis, and In Vivo Activity of a New Class of Pyrazoloquinazolines as Selective Inhibitors of Aurora B Kinase. J. Med. Chem. 2007, 50, 2213–2224. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ikezoe, T.; Nishioka, C.; Tasaka, T.; Taniguchi, A.; Kuwayama, Y.; Komatsu, N.; Bandobashi, K.; Togitani, K.; Koeffler, H.P.; et al. AZD1152, a Novel and Selective Aurora B Kinase Inhibitor, Induces Growth Arrest, Apoptosis, and Sensitization for Tubulin Depolymerizing Agent or Topoisomerase II Inhibitor in Human Acute Leukemia Cells In Vitro and In Vivo. Blood 2007, 110, 2034–2040. [Google Scholar] [CrossRef] [PubMed]
- Kenny, P.W.; Sadowski, J. Structure Modification in Chemical Databases. In Chemoinformatics in Drug Discovery; Wiley-Blackwell: Hoboken, NJ, USA, 2005; pp. 271–285. [Google Scholar]
- Stumpfe, D.; Dimova, D.; Bajorath, J. Computational Method for the Systematic Identification of Analog Series and Key Compounds Representing Series and Their Biological Activity Profiles. J. Med. Chem. 2016, 59, 7667–7676. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Tsukaguchi, T.; Hiroshima, S.; Kodama, T.; Kobayashi, T.; Fukami, T.A.; Oikawa, N.; Tsukuda, T.; Ishii, N.; Aoki, Y. CH5424802, a Selective ALK Inhibitor Capable of Blocking the Resistant Gatekeeper Mutant. Cancer Cell 2011, 19, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New Features for Data Integration and Network Visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Target Class | Chemical Probes | Target-Based Categories |
|---|---|---|
| Protein kinases | 33 | Chemical probes for kinase targets |
| Lipid kinases | 1 | |
| Epigenetics | 16 | Chemical probes for non-kinase targets |
| Other post-translation modification proteins | 13 | |
| Other proteins | 3 | |
| Structural proteins | 1 |
| Chemical Probe | Confidence Level 2 | Confidence Level 1 | ||
|---|---|---|---|---|
| ≤10 μM | ≤100 nM | ≤10 μM | ≤100 nM | |
| BMS-911543 | 6 (2) 1 | 3 | 6 (2) | 3 |
| BYL-719 | 4 (2) | 1 | 8 (3) | 1 |
| CCT244747 | 2 (1) | 1 | 2 (1) | 1 |
| CCT251545 | 4 (1) | 2 (1) | 4 (1) | 2 (1) |
| P505-15 | 6 (1) | 3 | 6 (1) | 3 |
| PF3644022 | 2 (1) | 1 | 2 (1) | 1 |
| Rapamycin | 5 (4) | 2 (1) | 7 (6) | 3 (2) |
| Ruxolitinib | 4 | 4 | 121 (1) | 16 |
| SGX-523 | 1 | 1 | 8 (1) | 1 |
| VX-745 | 3 (2) | 1 | 24 (2) | 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miljković, F.; Bajorath, J. Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes. Molecules 2018, 23, 2434. https://doi.org/10.3390/molecules23102434
Miljković F, Bajorath J. Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes. Molecules. 2018; 23(10):2434. https://doi.org/10.3390/molecules23102434
Chicago/Turabian StyleMiljković, Filip, and Jürgen Bajorath. 2018. "Data-Driven Exploration of Selectivity and Off-Target Activities of Designated Chemical Probes" Molecules 23, no. 10: 2434. https://doi.org/10.3390/molecules23102434






