SistematX, an Online Web-Based Cheminformatics Tool for Data Management of Secondary Metabolites
Abstract
1. Introduction
2. Results and Discussion
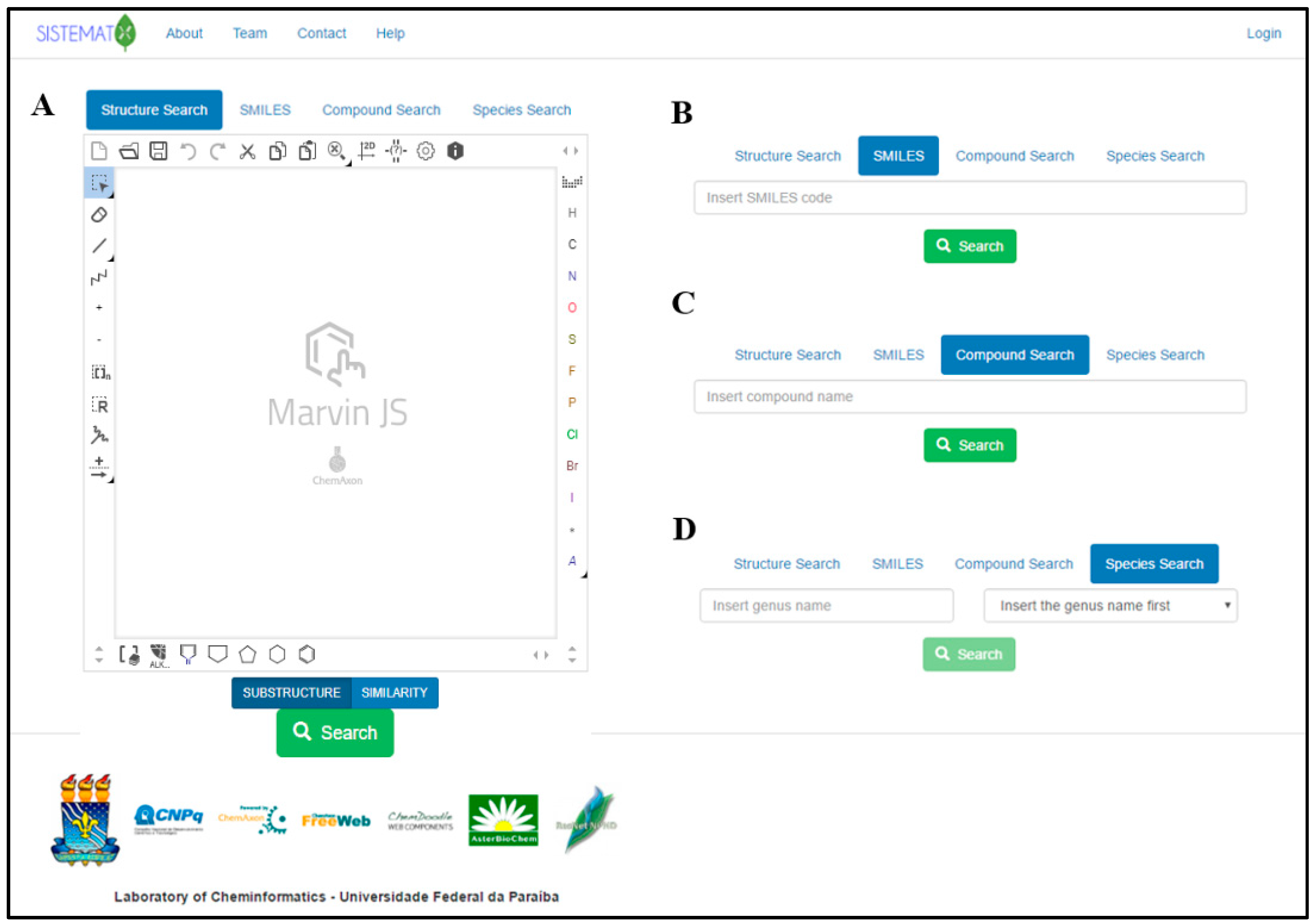
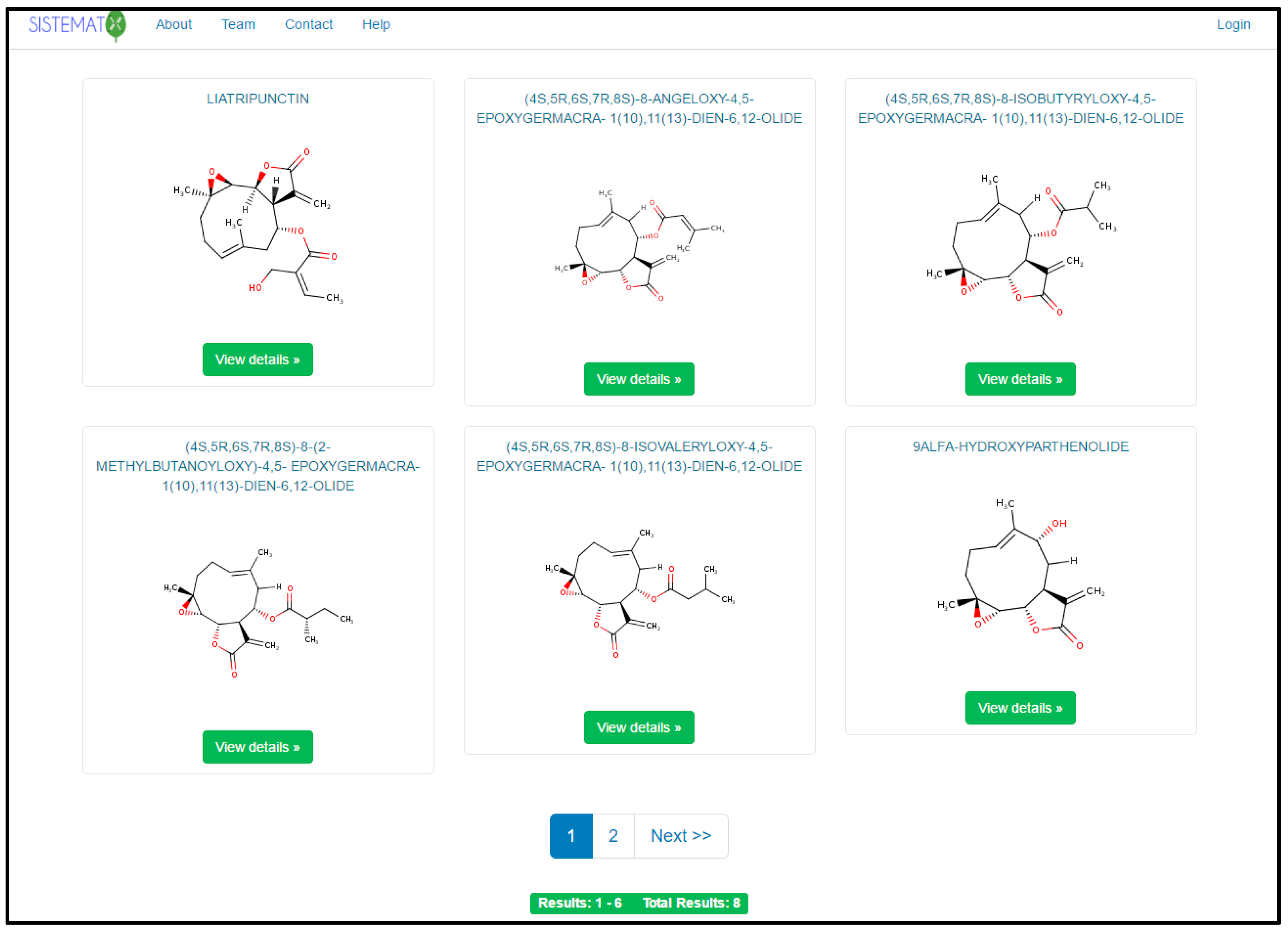
2.1. Utility and Discussion
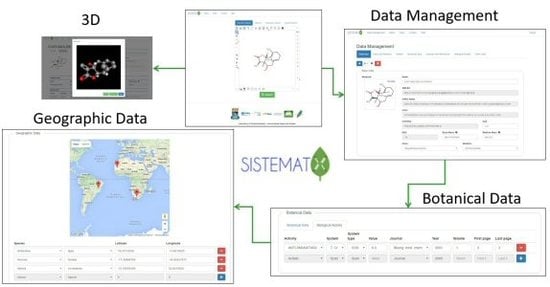
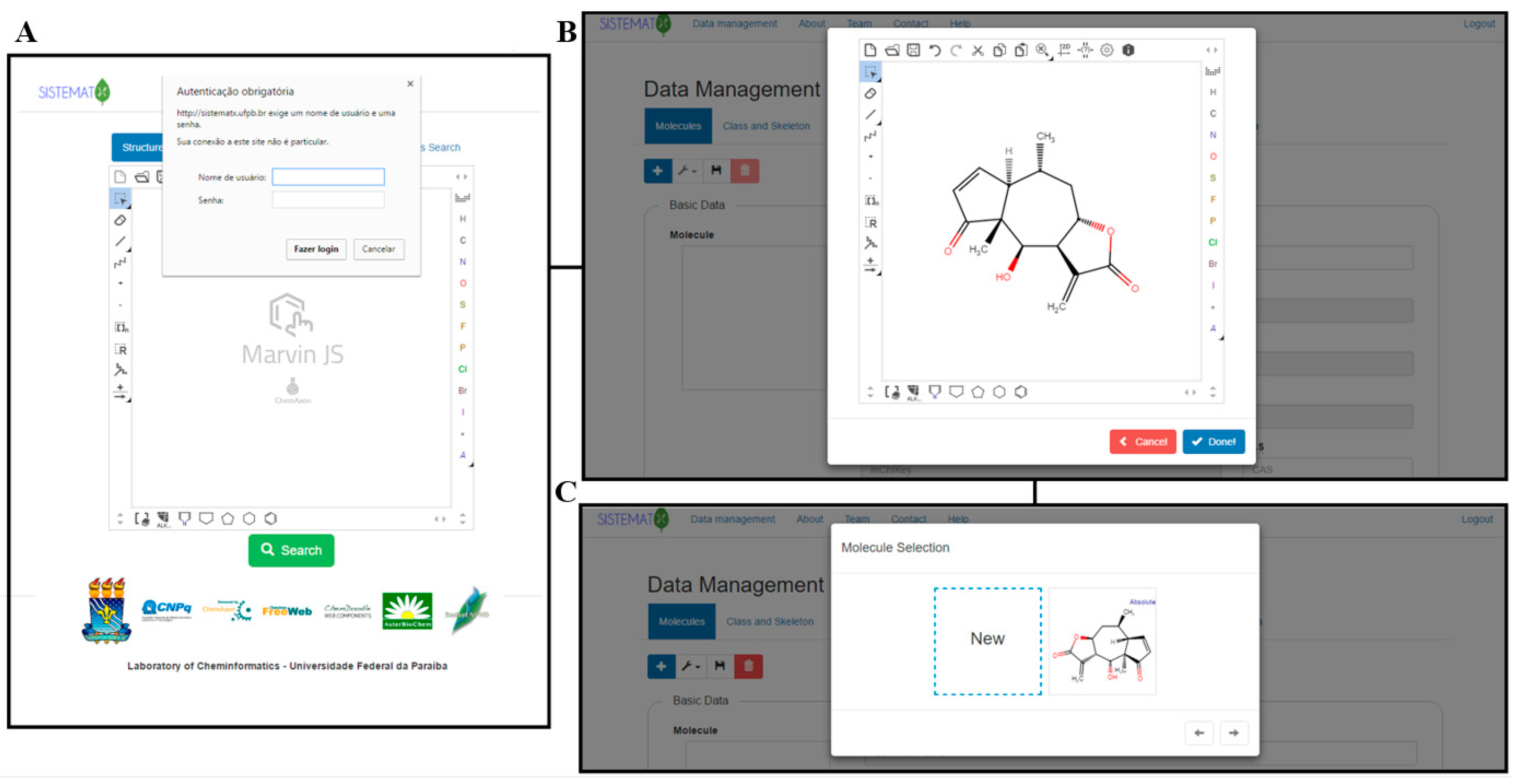
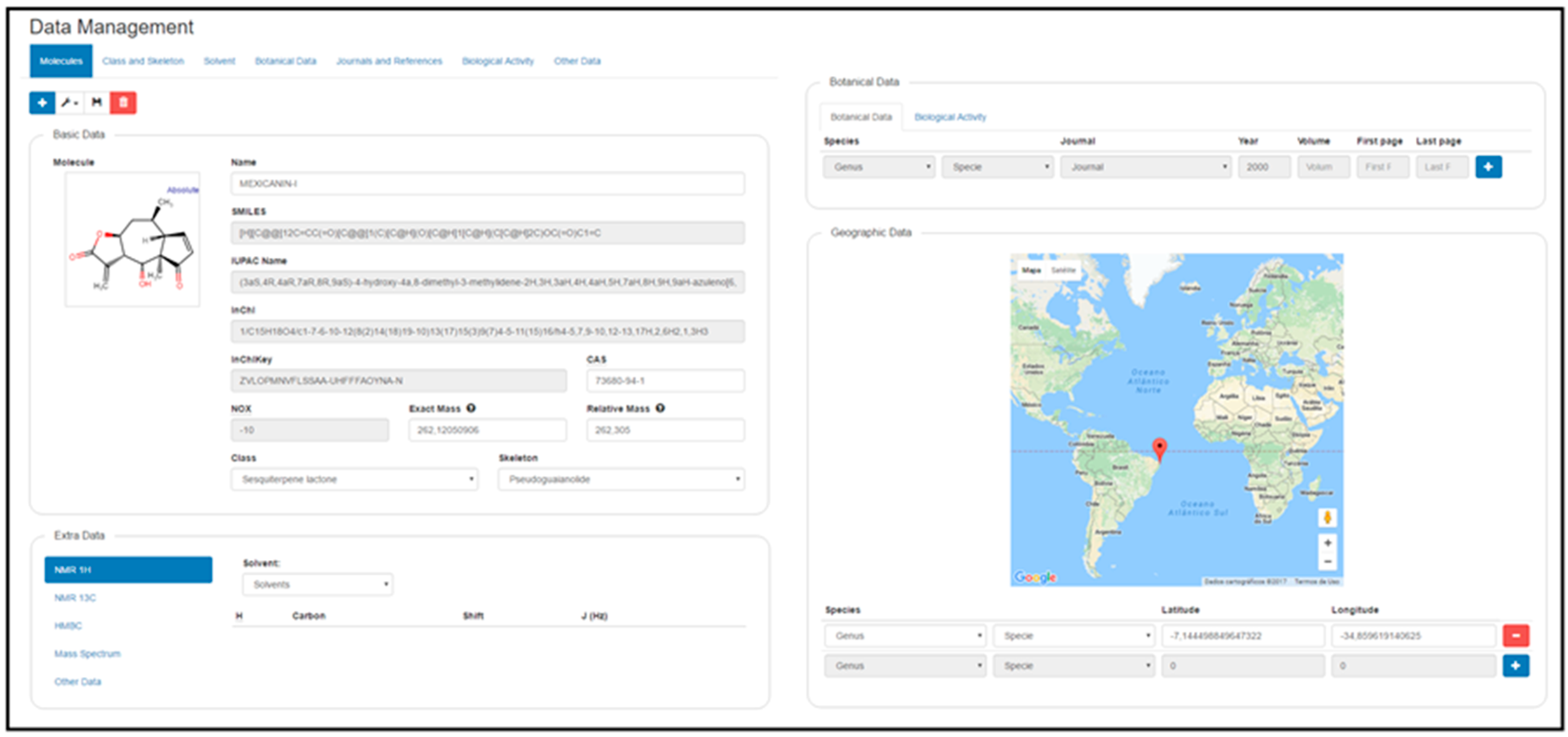
2.2. Data Management
3. Materials and Methods
Implementation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blunt, J.W.; Munro, M.H.G. Is There an Ideal Database for Natural Products Research? In Natural Products: Discourse, Diversity, and Design; Osbourn, A., Goss, R.J., Carter, G.T., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; pp. 413–431. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Corley, D.G.; Durley, R.C. Strategies for Database Dereplication of Natural Products. J. Nat. Prod. 1994, 57, 1484–1490. [Google Scholar] [CrossRef]
- Oliveira, T.; Chagas-Paula, D.; Rosa, A.; Gobbo-Neto, L.; Schmidt, T.J.; Da Costa, F.B. Temporal characteristics of a natural products in-house database. Planta Med. 2013, 79, 1113–1114. [Google Scholar] [CrossRef]
- Pence, H.; Williams, A. ChemSpider: An online chemical information resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Bolton, E.E.; Wang, Y.; Thiessen, P.A.; Bryant, S.H. Chapter 12 PubChem: Integrated Platform of Small Molecules and Biological Activities. Annu. Rep. Comput. Chem. 2008, 4, 217–241. [Google Scholar] [CrossRef]
- Degtyarenko, K.; de Matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; McNaught, A.; Alcántara, R.; Darsow, M.; Guedj, M.; Ashburner, M. ChEBI: A database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008, 36, D344–D350. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.B.; O’Neil-Johnson, M.; Williams, A.J.; Wheeler, P.; Pol, R.; Moser, A. Dereplication of natural products using minimal NMR data inputs. Org. Biomol. Chem. 2015, 13, 9957–9962. [Google Scholar] [CrossRef] [PubMed]
- Eugster, P.J.; Boccard, J.; Debrus, B.; Bréant, L.; Wolfender, J.L.; Martel, S.; Carrupt, P.A. Retention time prediction for dereplication of natural products (CxHyOz) in LC–MS metabolite profiling. Phytochemistry 2014, 108, 196–207. [Google Scholar] [CrossRef] [PubMed]
- DNP Database, Dictionary of Natural Products. CRC Press. Available online: http://dnp.chemnetbase.com/ (accessed on 4 March 2017).
- Graham, J.G.; Farnsworth, N.R. The NAPRALERT database as an aid for discovery of novel bioactive compounds. In Comprehensive Natural Products II: Chemistry and Biology; Elsevier Ltd.: Amsterdam, The Netherlands, 2010; Volume 3, pp. 81–94. ISBN 9780080453828. [Google Scholar]
- Dabb, S.; Blunt, J.; Munro, M. MarinLit: Database and essential tools for the marine natural products community. In Proceedings of the 248th National Meeting of the American-Chemical-Society (ACS), San Francisco, CA, USA, 10–14 August 2014. [Google Scholar]
- Banerjee, P.; Erehman, J.; Gohlke, B.-O.; Wilheim, T.; Preissner, R.; Dunkel, M. Super Natural II—A database of natural products. Nucleic Acids Res. 2015, 43, D935–D939. [Google Scholar] [CrossRef] [PubMed]
- Drewry, D.H.; Macarron, R. Enhancements of screening collections to address areas of unmet medical need: an industry perspective. Curr. Opin. Chem. Biol. 2010, 14, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Valli, M.; dos Santos, R.N.; Figueira, L.D.; Nakajima, C.H.; Castro-Gamboa, I.; Andricopulo, A.D.; Bolzani, V.S. Development of a natural products database from the biodiversity of Brazil. J. Nat. Prod. 2013, 76, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Hatherley, R.; Brown, D.K.; Musyoka, T.M.; Penkler, D.L.; Faya, N.; Lobb, K.A.; Tastan Bishop, Ö. SANCDB: A South African natural compound database. J. Cheminform. 2015, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Nam, S.; Jang, H.; Kim, A.; Lee, J.J. TM-MC: A database of medicinal materials and chemical compounds in Northeast Asian traditional medicine. BMC Complement. Altern. Med. 2015, 15, 218. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.-C. TCM Database@Taiwan: The world’s largest traditional Chinese medicine database for drug screening in silico. PLoS ONE 2011, 6, e15939. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Telukunta, K.K.; Döring, K.; Simoben, C.V.; Moumbock, A.F.A.; Malange, Y.I.; Njume, L.E.; Yong, J.N.; Sippl, W.; Günther, S. NANPDB: A Resource for Natural Products from Northern African Sources. J. Nat. Prod. 2017, 80, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Fang, Z.; Zhang, M.; Yi, Z.; Wen, C.; Shi, T. TCMID: Traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013, 41, D1089–D1095. [Google Scholar] [CrossRef] [PubMed]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK Family Databases: Integrated Metabolite-Plant Species Databases for Multifaceted Plant Research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.W.; Lin, Y.C.; Chang, H.S.; Wang, C.C.; Chen, I.S.; Jheng, J.L.; Li, J.H. TIPdb-3D: The three-dimensional structure database of phytochemicals from Taiwan indigenous plants. Database (Oxford) 2014. [Google Scholar] [CrossRef] [PubMed]
- AsterDB, AsterBioChem in House Database. Available online: http://www.asterbiochem.org/asterdb (accessed on 4 March 2017).
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 29265. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.J.; Rzeppa, S.; Kaiser, M.; Brun, R. Larrea tridentata—Absolute configuration of its epoxylignans and investigations on its antiprotozoal activity. Phytochem. Lett. 2012, 5, 632–638. [Google Scholar] [CrossRef]
- Gaquerel, E.; Kuhl, C.; Neumann, S. Computational annotation of plant metabolomics profiles via a novel network-assisted approach. Metabolomics 2013, 9, 904–918. [Google Scholar] [CrossRef]
- Hendrickson, J.B.; Cram, D.J.; Hammond, G.S. Organic Chemistry, 3rd ed.; McGraw-Hill: New York, NY, USA, 1970; ISBN 07-028150-5. [Google Scholar]
- Gottlieb, O. The role of oxygen in phytochemical evolution towards diversity. Phytochemistry 1989, 28, 2545–2558. [Google Scholar] [CrossRef]
- Züst, T.; Heichinger, C.; Grossniklaus, U.; Harrington, R.; Kliebenstein, D.J.; Turnbull, L.A. Natural enemies drive geographic variation in plant defenses. Science 2012, 338, 116–119. [Google Scholar] [CrossRef] [PubMed]
- MySQL. Available online: http://dev.mysql.com/downloads/mysql/ (accessed on 31 January 2017).
- Scotti, M.; da Silva, R.O., Jr.; Yudi, S.; Brayner, R.; Scotti, L. SISTEMAT X—A web tool to manage databases of secondary metabolites. In Proceedings of the MOL2NET, 5–15 December 2015. [Google Scholar]
- Bootstrap. Available online: http://getbootstrap.com/ (accessed on 10 January 2017).
- jQuery. Available online: http://jquery.com/ (accessed on 17 January 2017).
- jQuery Autocomplete. Available online: http://jqueryui.com/autocomplete/ (accessed on 20 January 2017).
- ChemAxon. Marvin JS. Available online: http://chemaxon.com/products/marvin/marvin-js/ (accessed on 24 January 2017).
- ChemAxon. Jchem Web Services. Available online: http://chemaxon.com/products/jchem-web-services/ (accessed on 24 January 2017).
- ChemDoodle by iChemLabs. Available online: http://web.chemdoodle.com/ (accessed on 24 January 2017).
- Google Maps by Google, Inc. Available online: http://developers.google.com/maps/ (accessed on 13 January 2017).
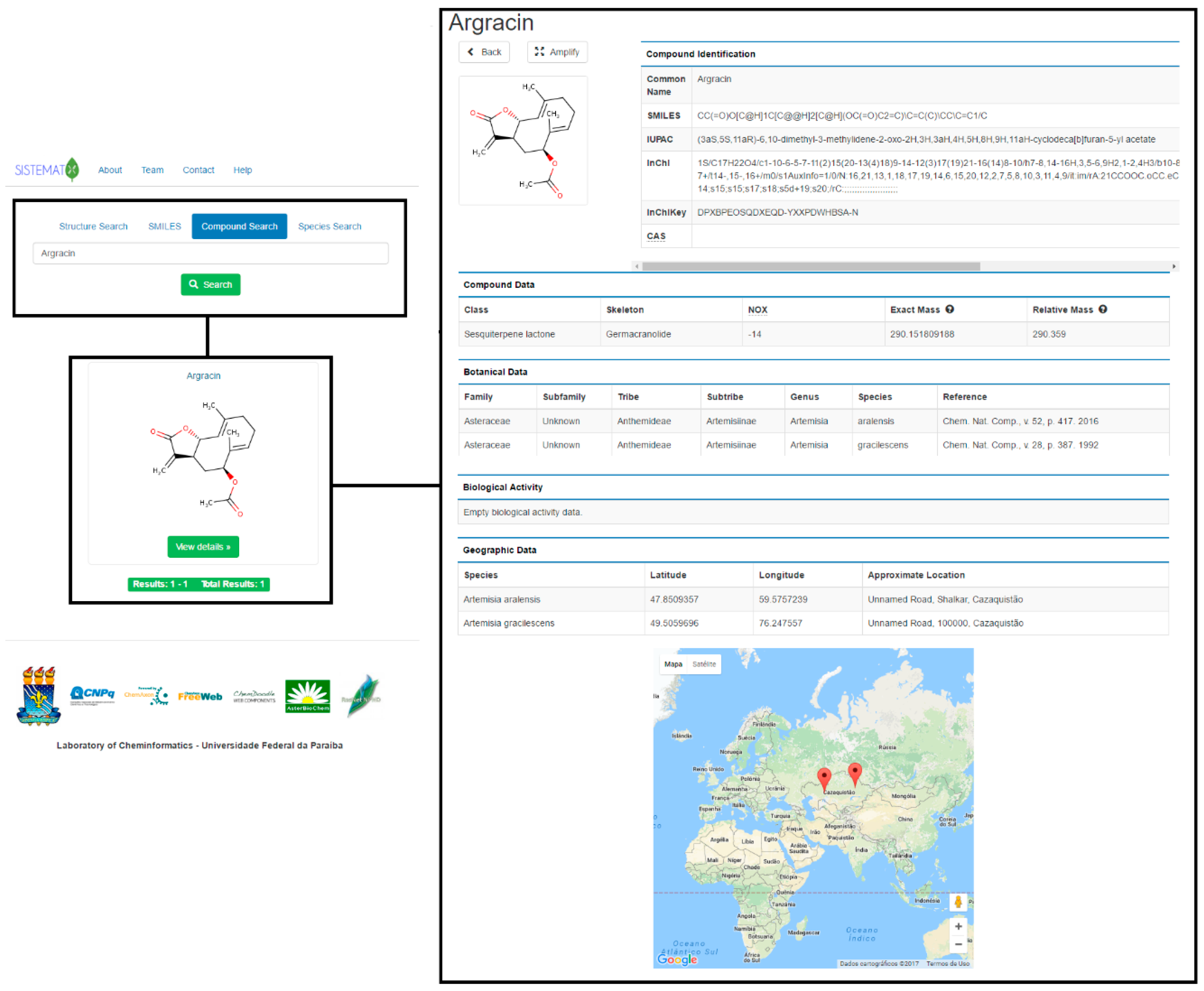
| API | Description | Engine |
|---|---|---|
| 1. Structure | ||
| 2D drawing | Allows drawing and visualization of chemical structures | ChemAxon |
| 3D generator | Uses 2D drawing to generate a 3D representation of the molecule | ChemAxon |
| 3D | Graphical visualization of 3D molecules with JavaScript | ChemDoodl |
| 2. Compound Identification | ||
| SMILES | Simplified Molecular Input Line Entry System | ChemAxon |
| IUPAC | IUPAC Nomenclature | ChemAxon |
| InChI | IUPAC International Chemical Identifier | ChemAxon |
| InChIKey | InChIKey is a compact format of the InChI code | ChemAxon |
| CAS | Chemical Abstracts Service Registry Number | ChemAxon |
| 3. Compound Data | ||
| NOX | Oxidation number (NOX) of an organic compound | ChemAxon |
| Exact Mass | Uses the mass of the most abundant isotope of each element | ChemAxon |
| Relative Mass | Uses the average atomic mass of each element | ChemAxon |
| 4. Geographic data | ||
| Latitude | Can be inserted by the administrator or appears by clicking in the world map | Google Inc. |
| Longitude | Can be inserted by the administrator or appears by clicking in the world map | Google Inc. |
| Approximate | Using the latitude and longitude, appears an an approximate location of the specie | Google Inc. |
| Visualization | Uses the world map to possible to visualize the localization of the species | Google Inc. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scotti, M.T.; Herrera-Acevedo, C.; Oliveira, T.B.; Costa, R.P.O.; Santos, S.Y.K.d.O.; Rodrigues, R.P.; Scotti, L.; Da-Costa, F.B. SistematX, an Online Web-Based Cheminformatics Tool for Data Management of Secondary Metabolites. Molecules 2018, 23, 103. https://doi.org/10.3390/molecules23010103
Scotti MT, Herrera-Acevedo C, Oliveira TB, Costa RPO, Santos SYKdO, Rodrigues RP, Scotti L, Da-Costa FB. SistematX, an Online Web-Based Cheminformatics Tool for Data Management of Secondary Metabolites. Molecules. 2018; 23(1):103. https://doi.org/10.3390/molecules23010103
Chicago/Turabian StyleScotti, Marcus Tullius, Chonny Herrera-Acevedo, Tiago Branquinho Oliveira, Renan Paiva Oliveira Costa, Silas Yudi Konno de Oliveira Santos, Ricardo Pereira Rodrigues, Luciana Scotti, and Fernando Batista Da-Costa. 2018. "SistematX, an Online Web-Based Cheminformatics Tool for Data Management of Secondary Metabolites" Molecules 23, no. 1: 103. https://doi.org/10.3390/molecules23010103
APA StyleScotti, M. T., Herrera-Acevedo, C., Oliveira, T. B., Costa, R. P. O., Santos, S. Y. K. d. O., Rodrigues, R. P., Scotti, L., & Da-Costa, F. B. (2018). SistematX, an Online Web-Based Cheminformatics Tool for Data Management of Secondary Metabolites. Molecules, 23(1), 103. https://doi.org/10.3390/molecules23010103