Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides
Abstract
1. Introduction
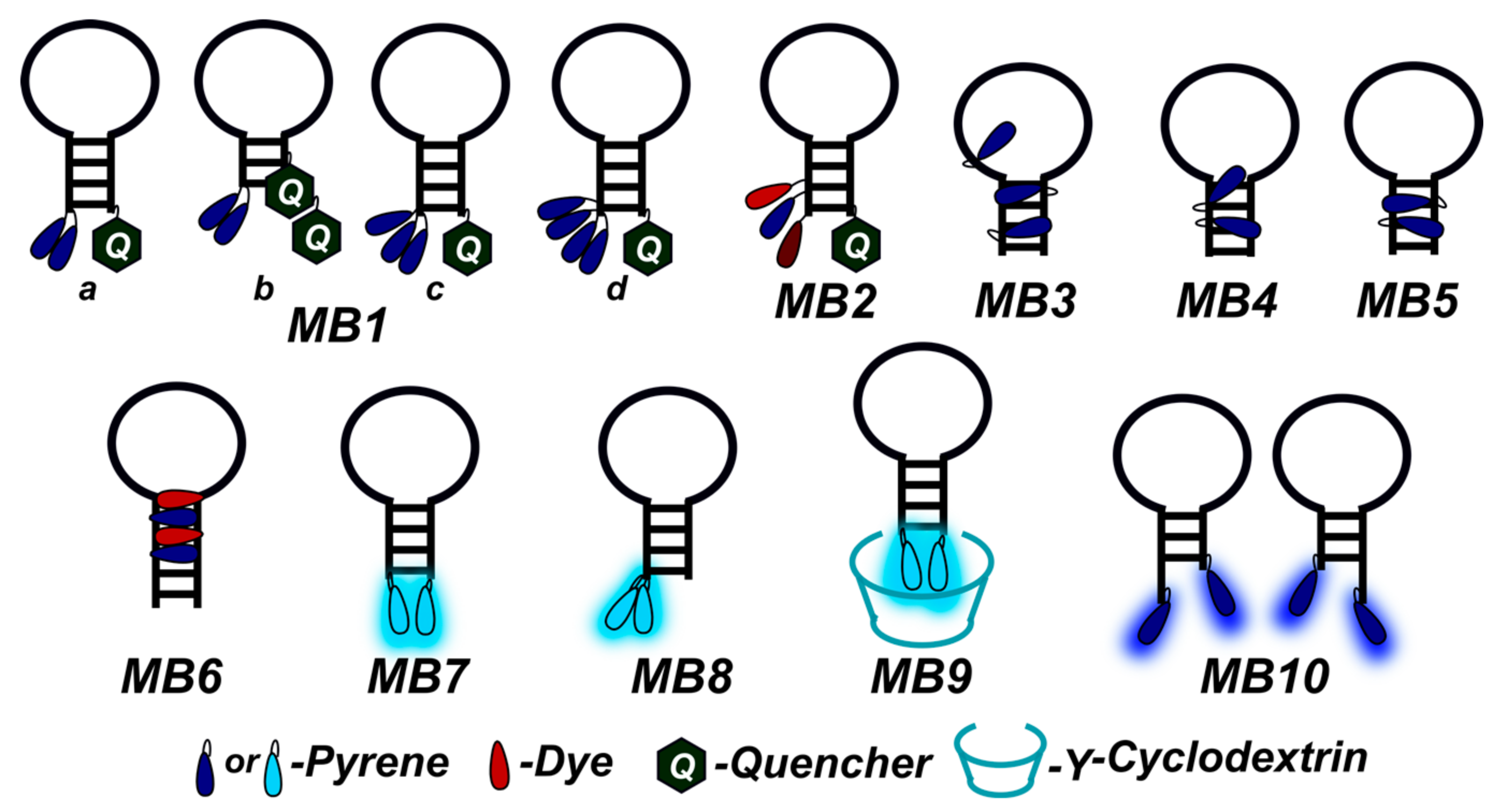
2. Fluorescent Biosensors
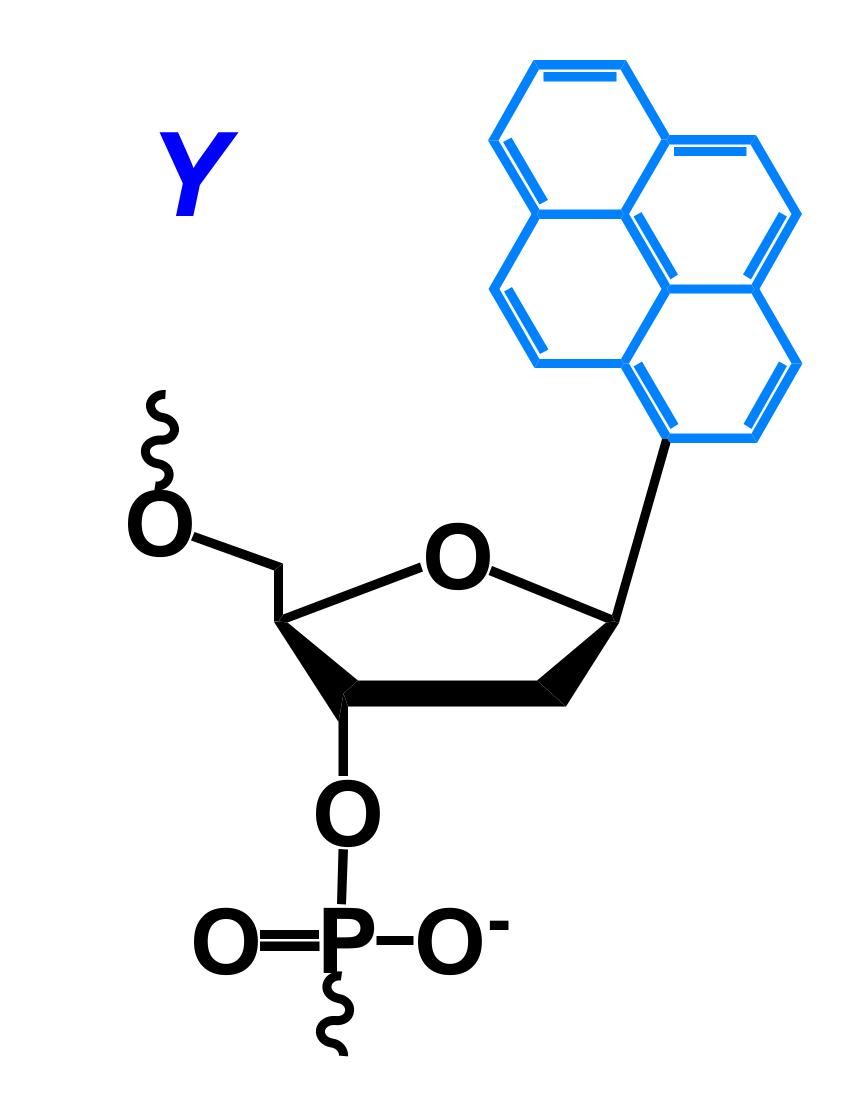
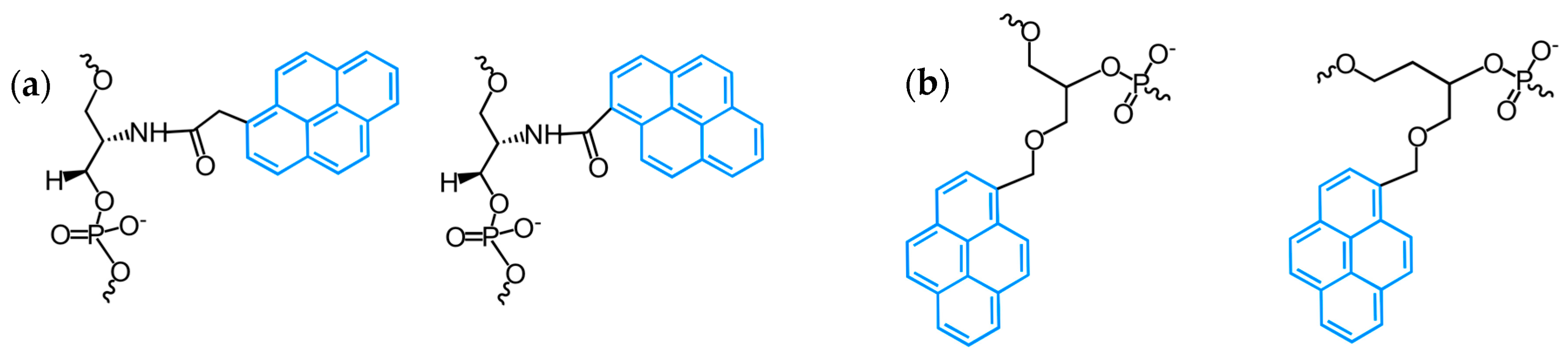
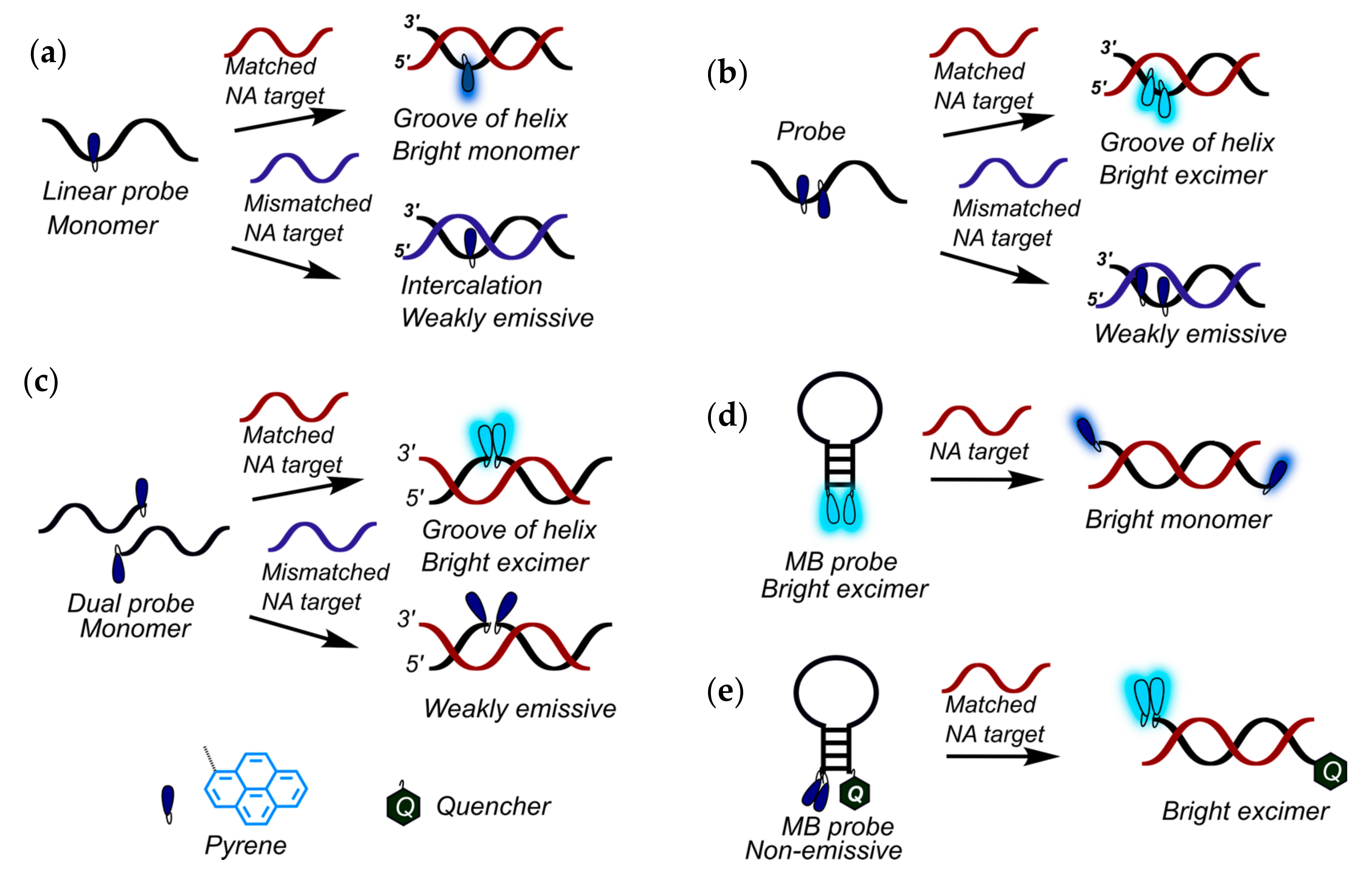
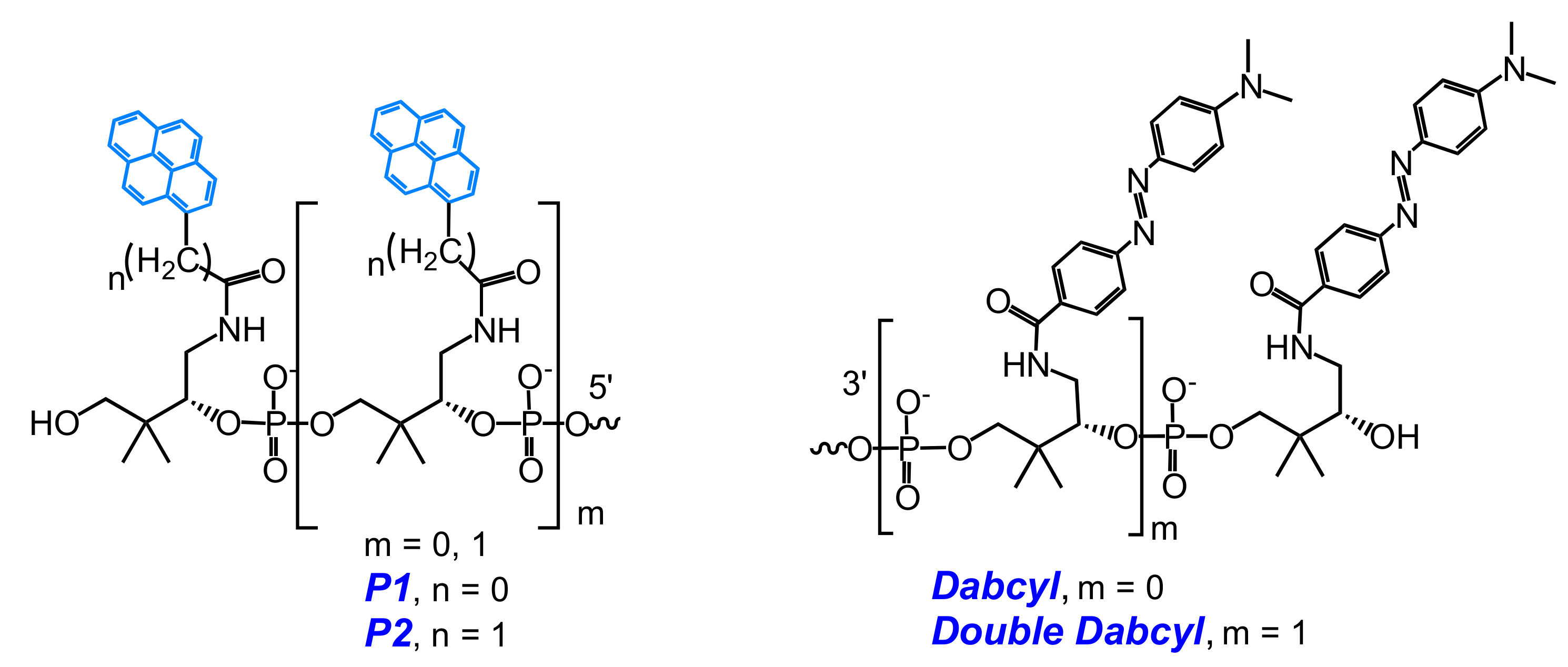
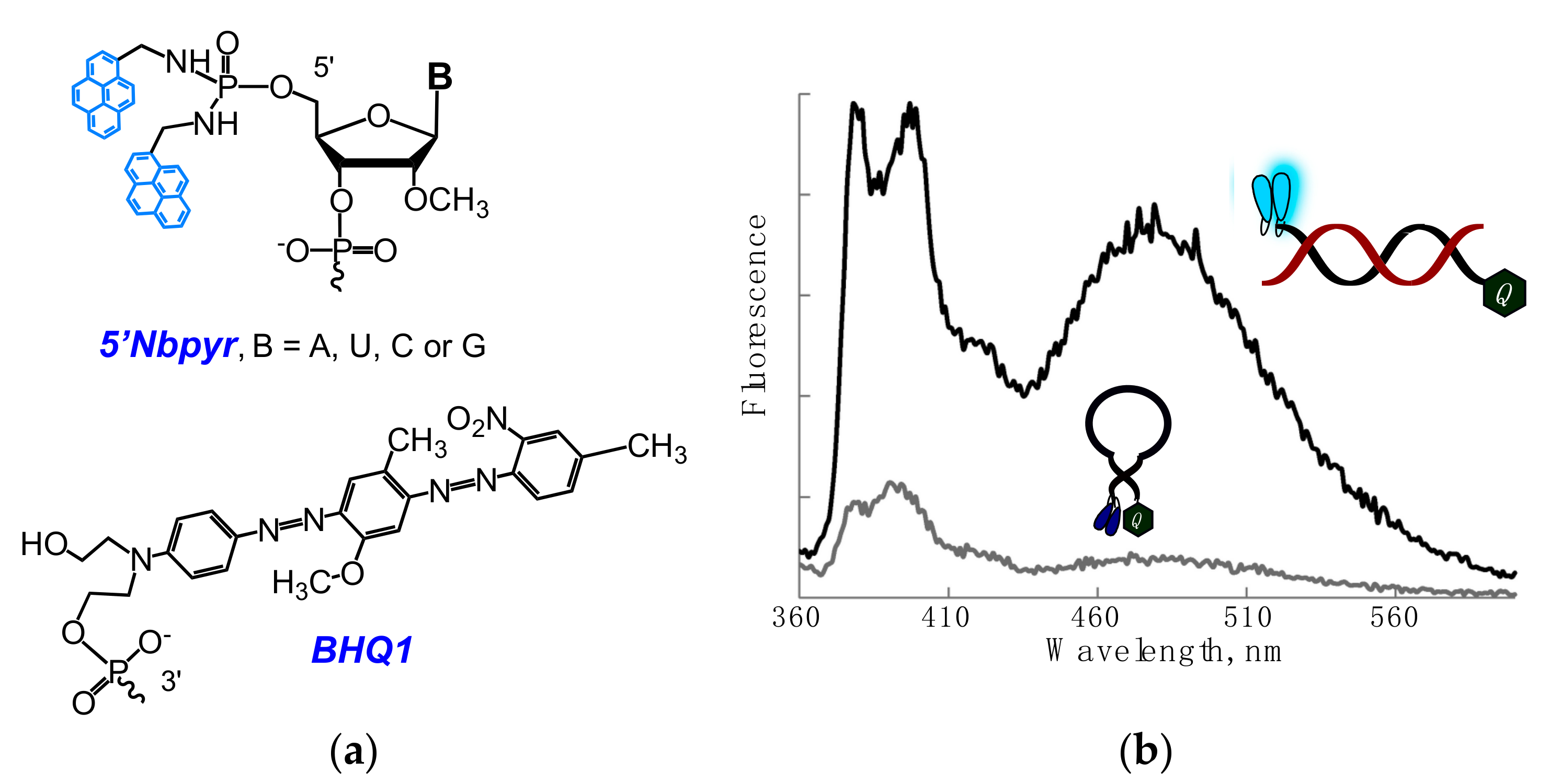
2.1. Fluorescent Probes for the Detection of RNA and DNA
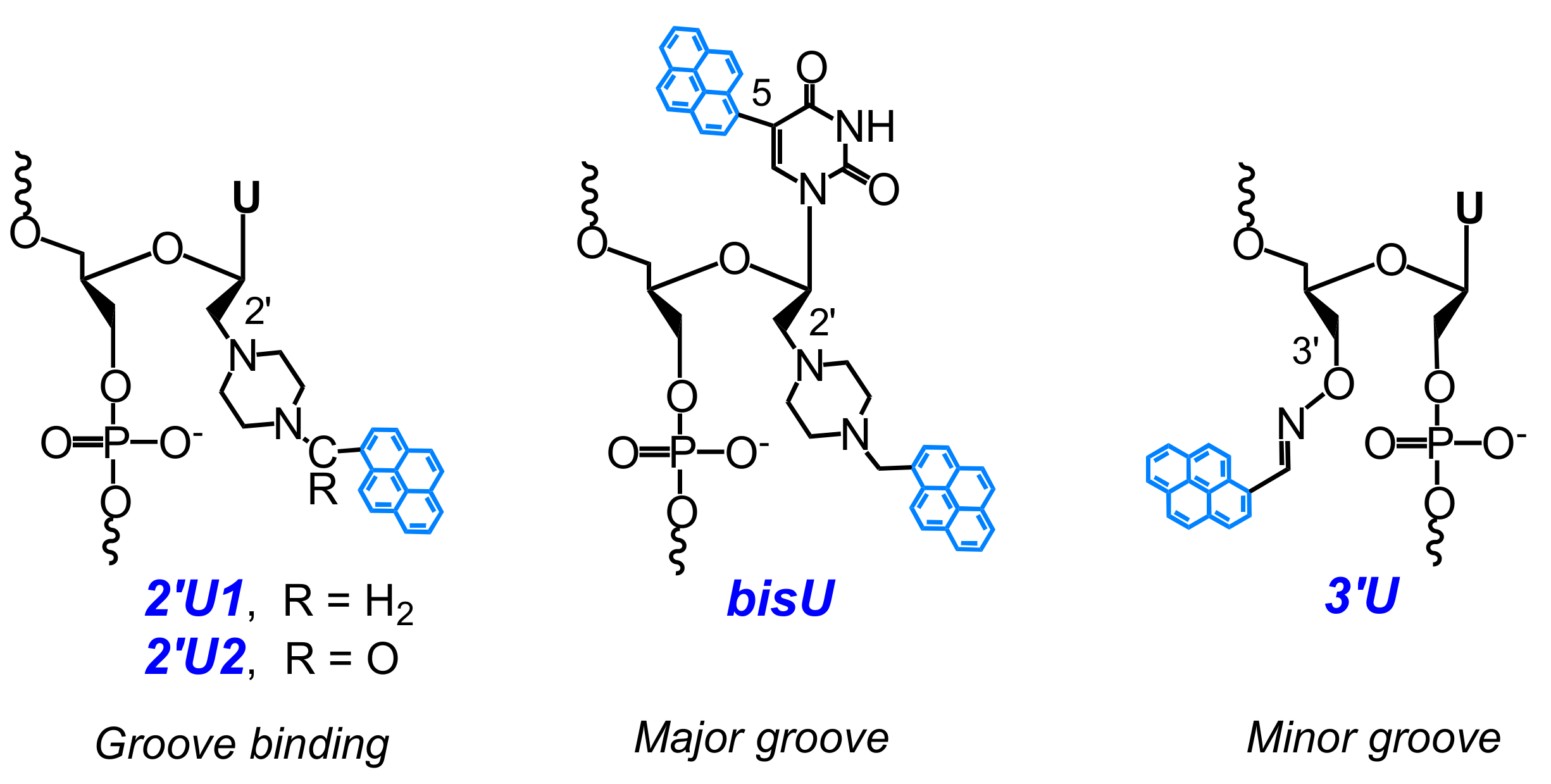
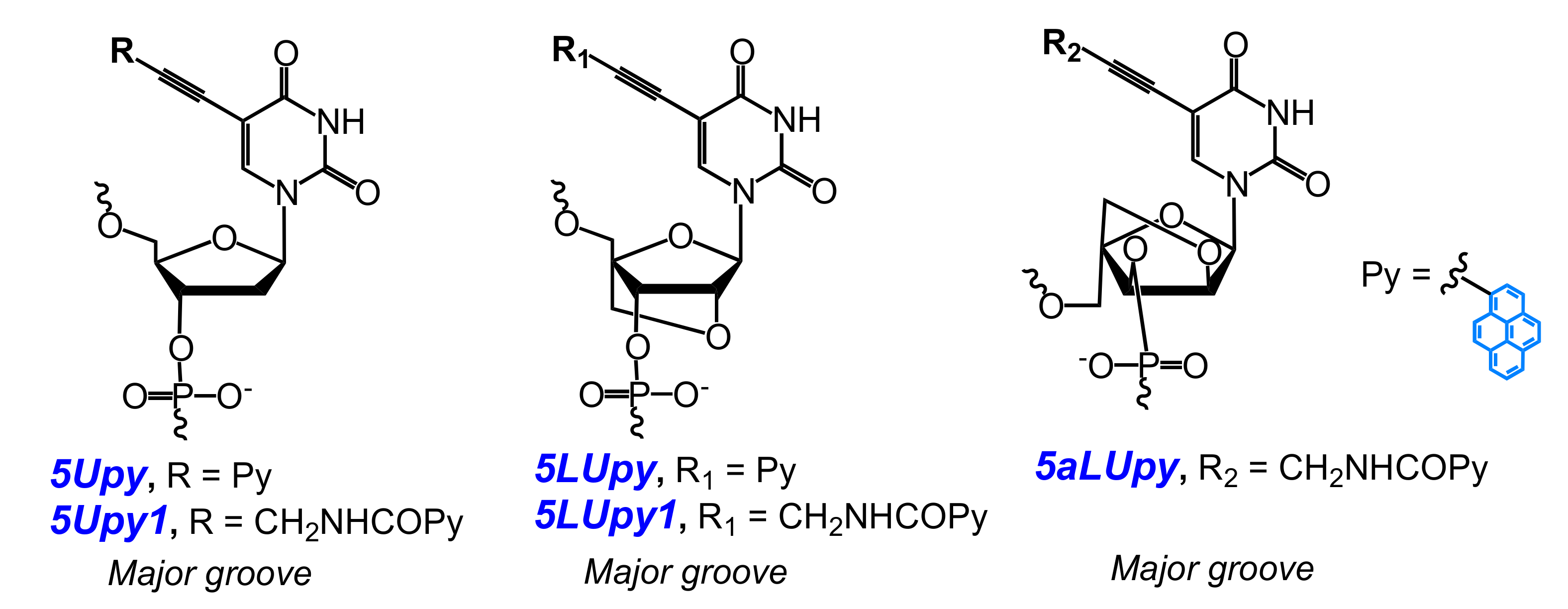
2.2. Base-Discriminating Probes
2.3. Fluorescent Biosensors for Detection of Miscellaneous Targets
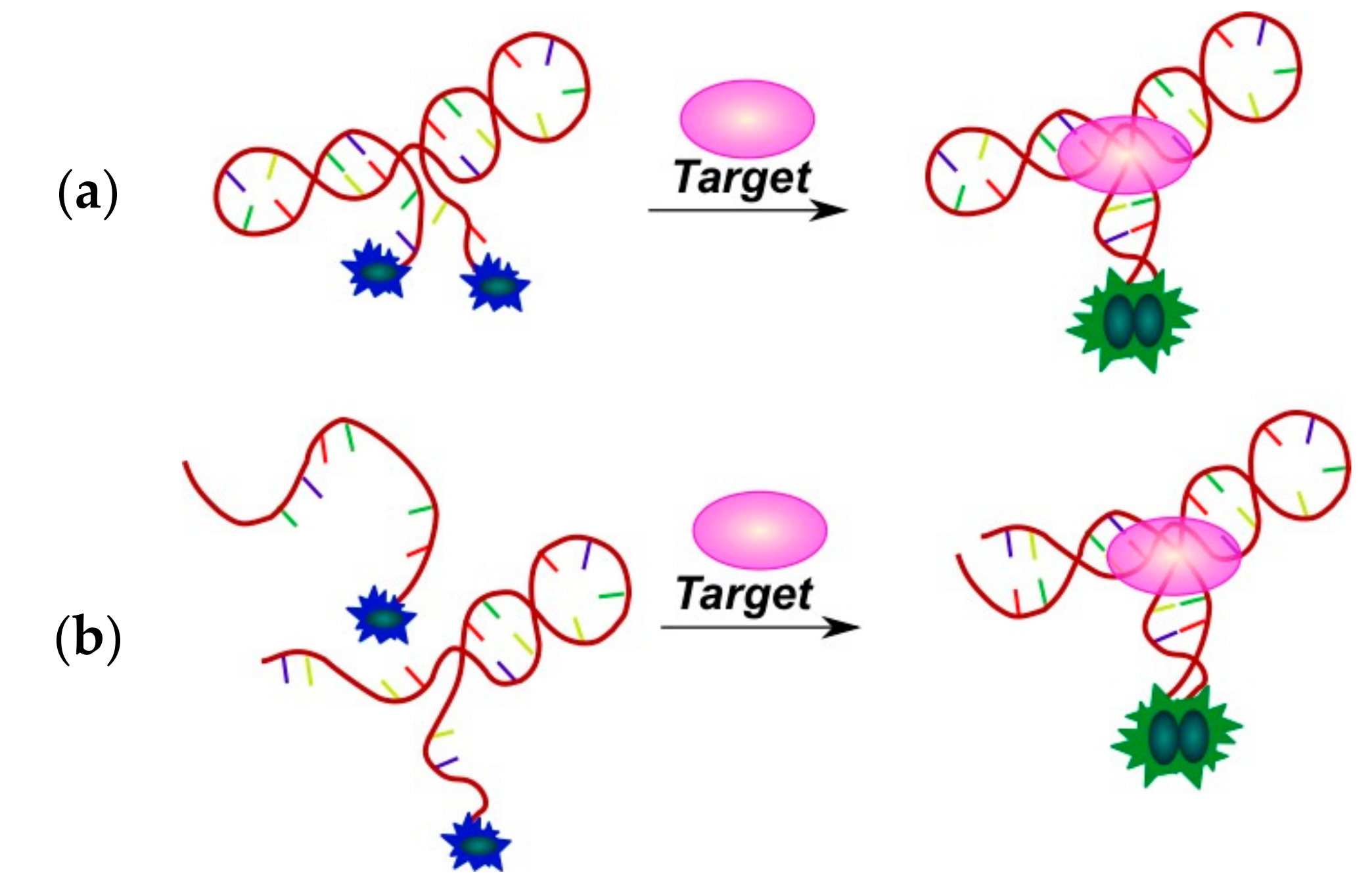
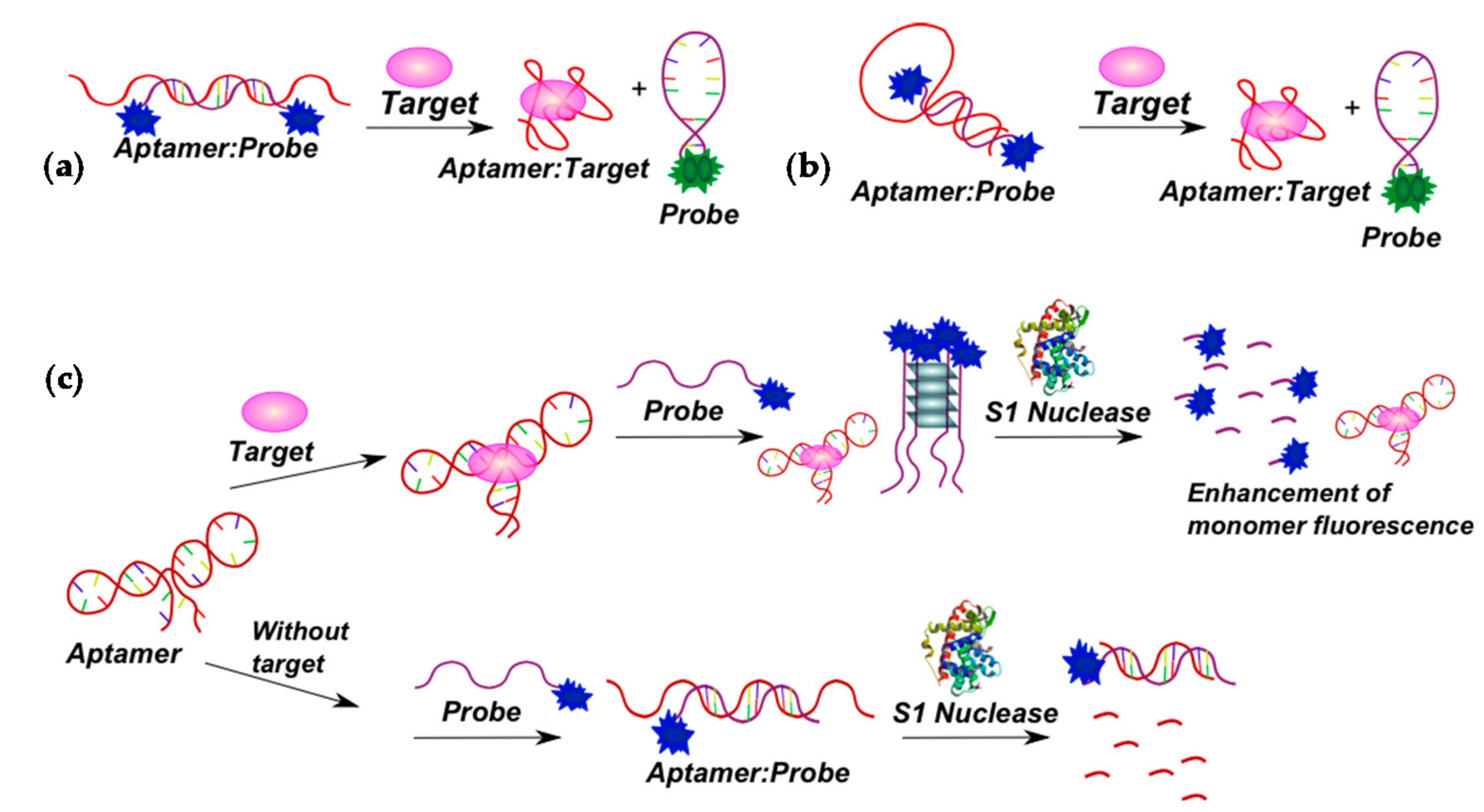
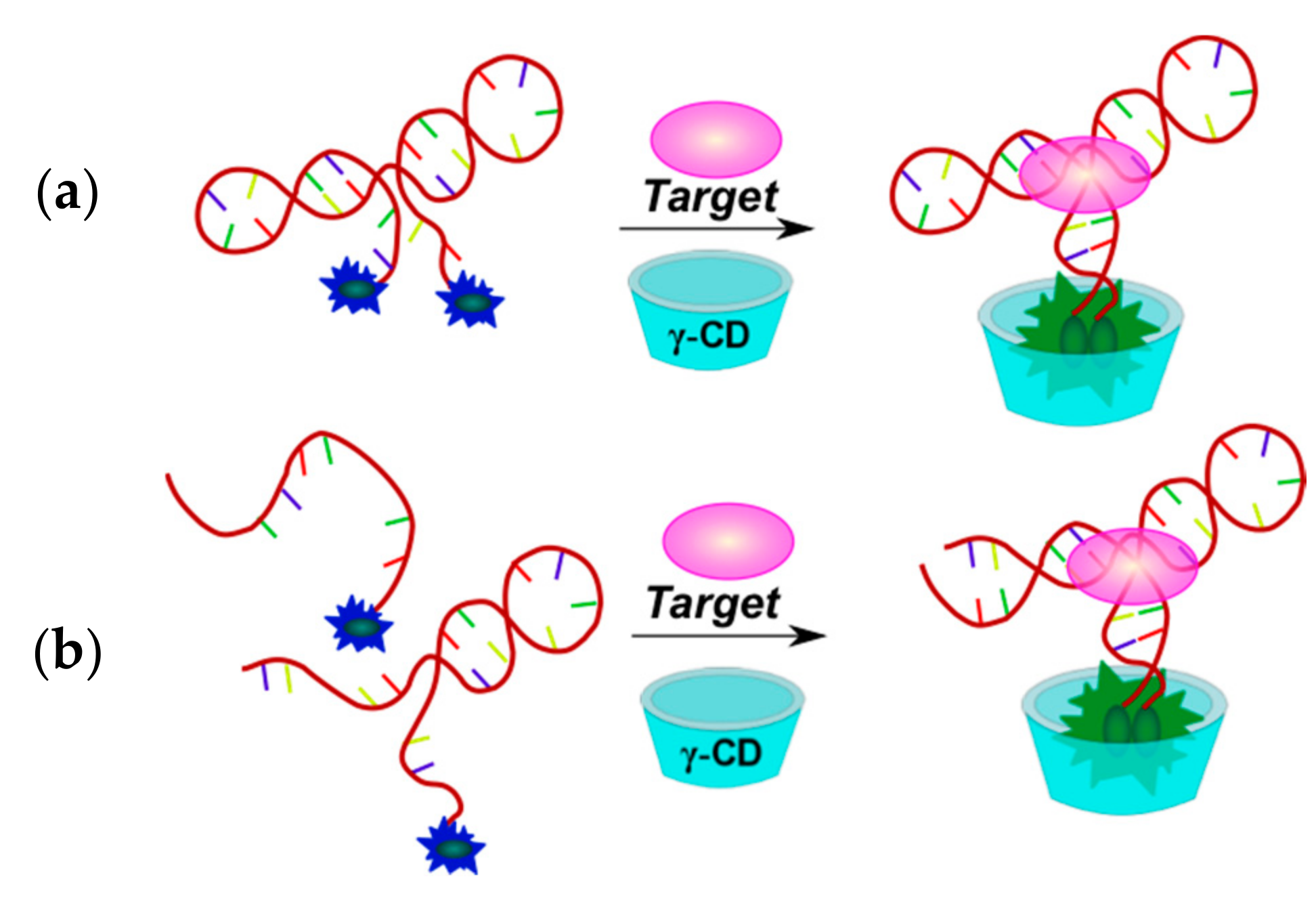
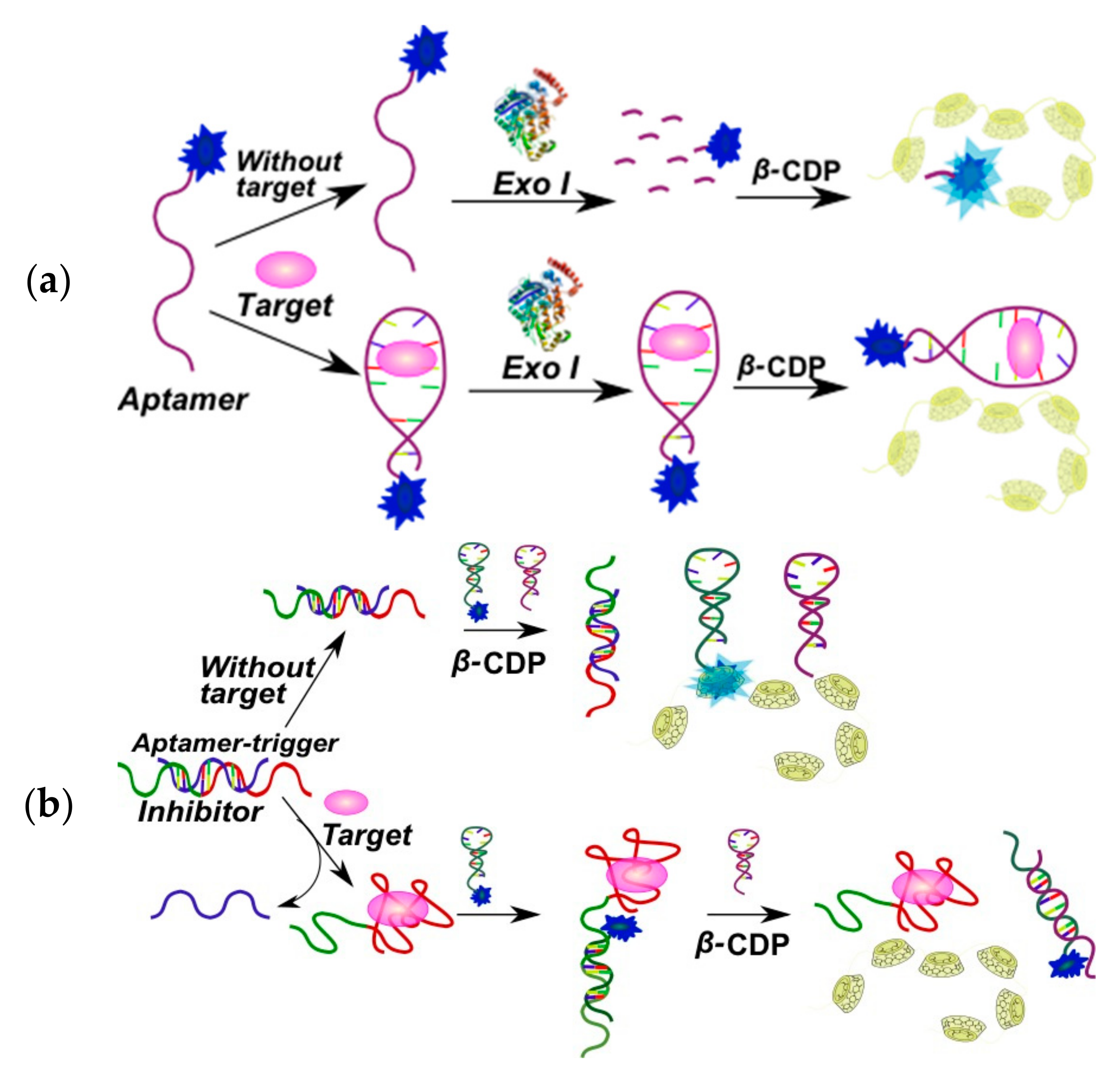
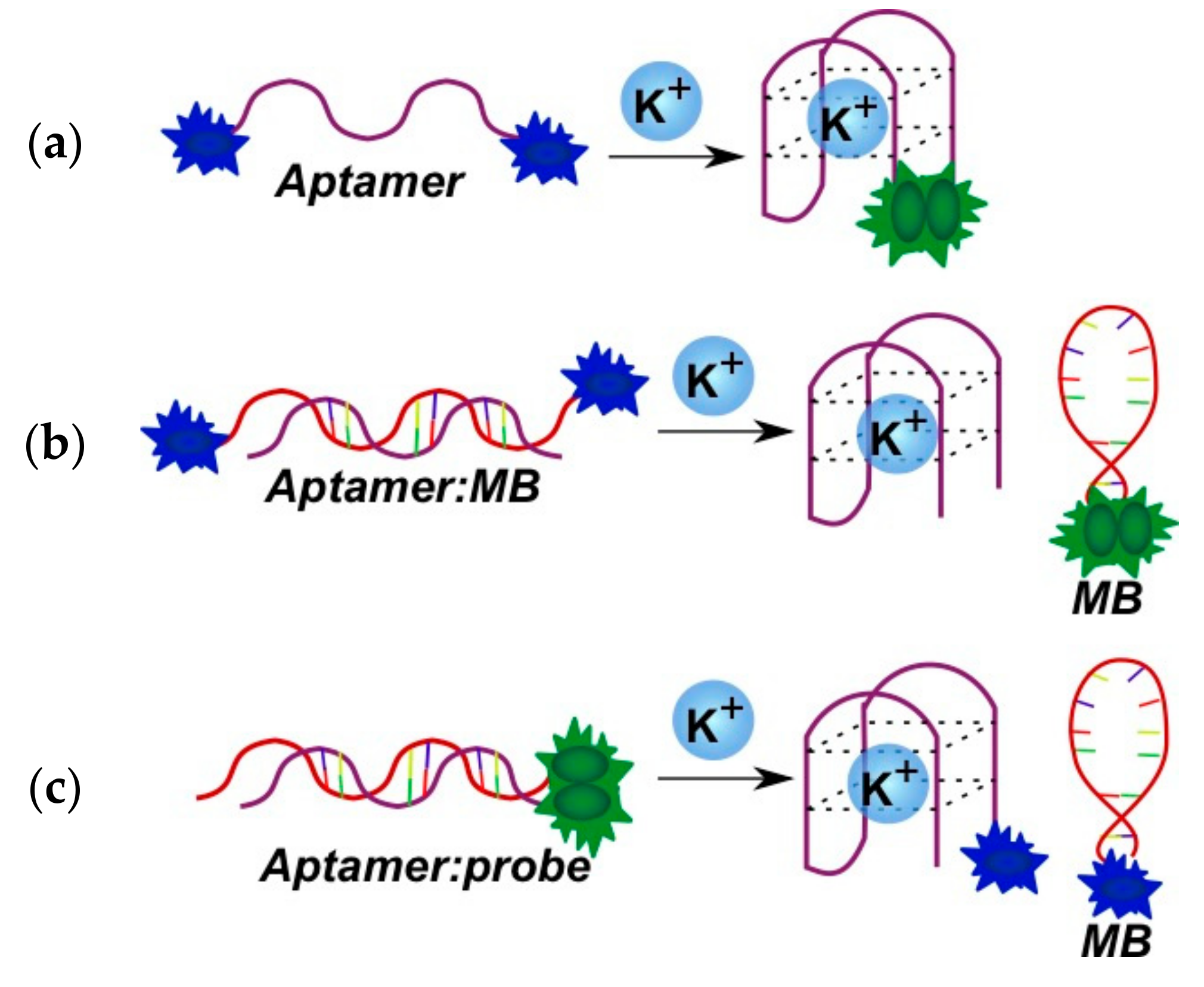
2.3.1. Fluorescent Aptasensors Containing Pyrene Residues
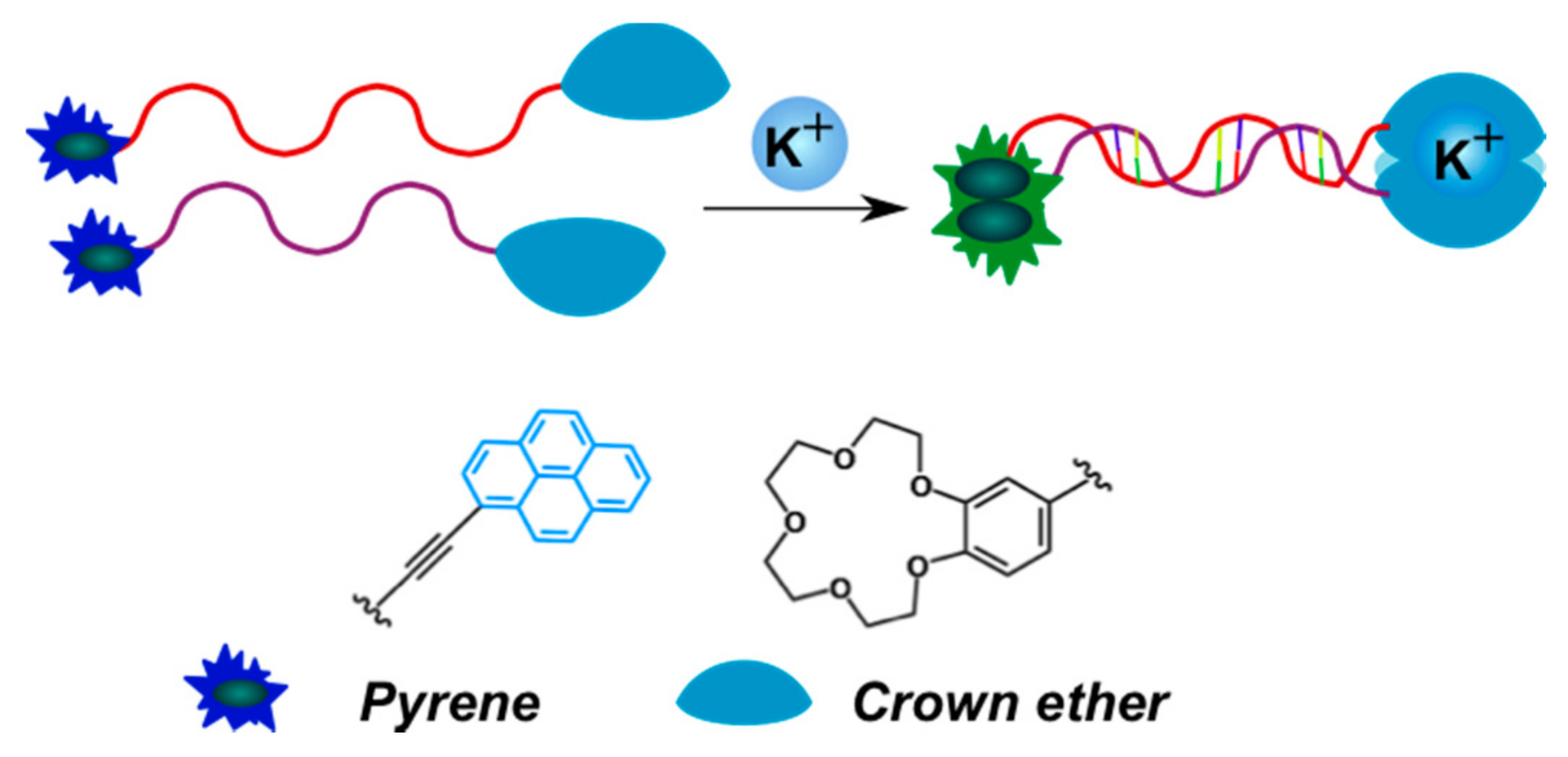
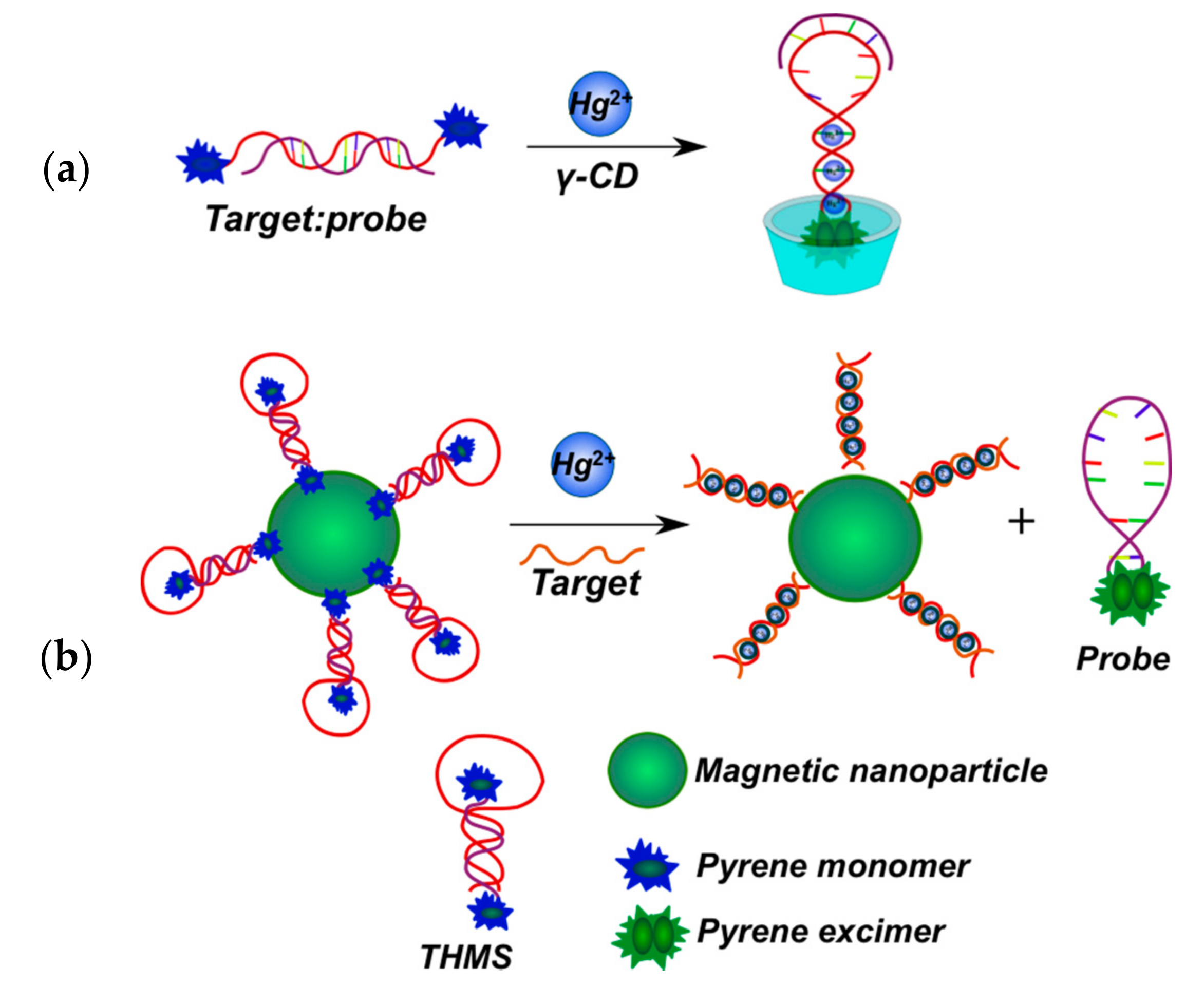
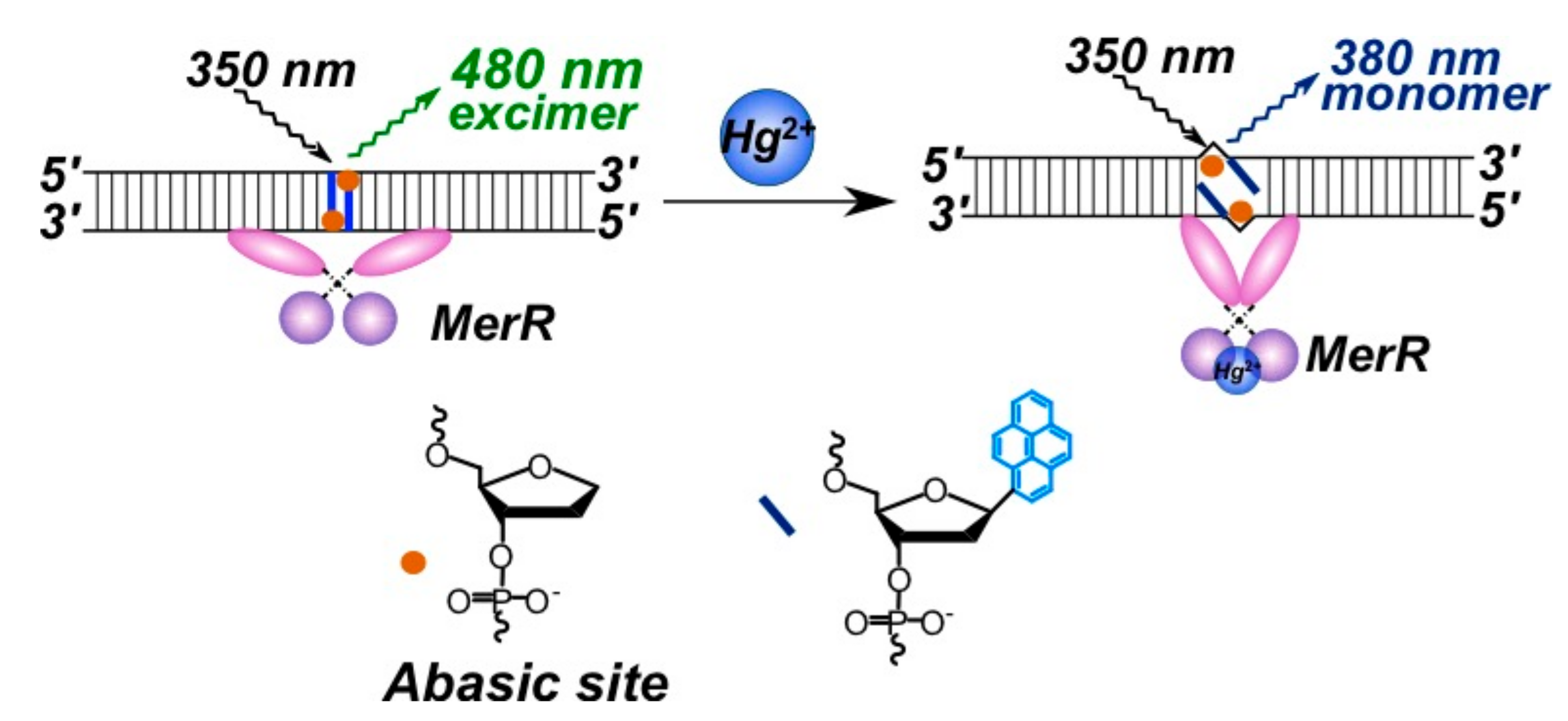
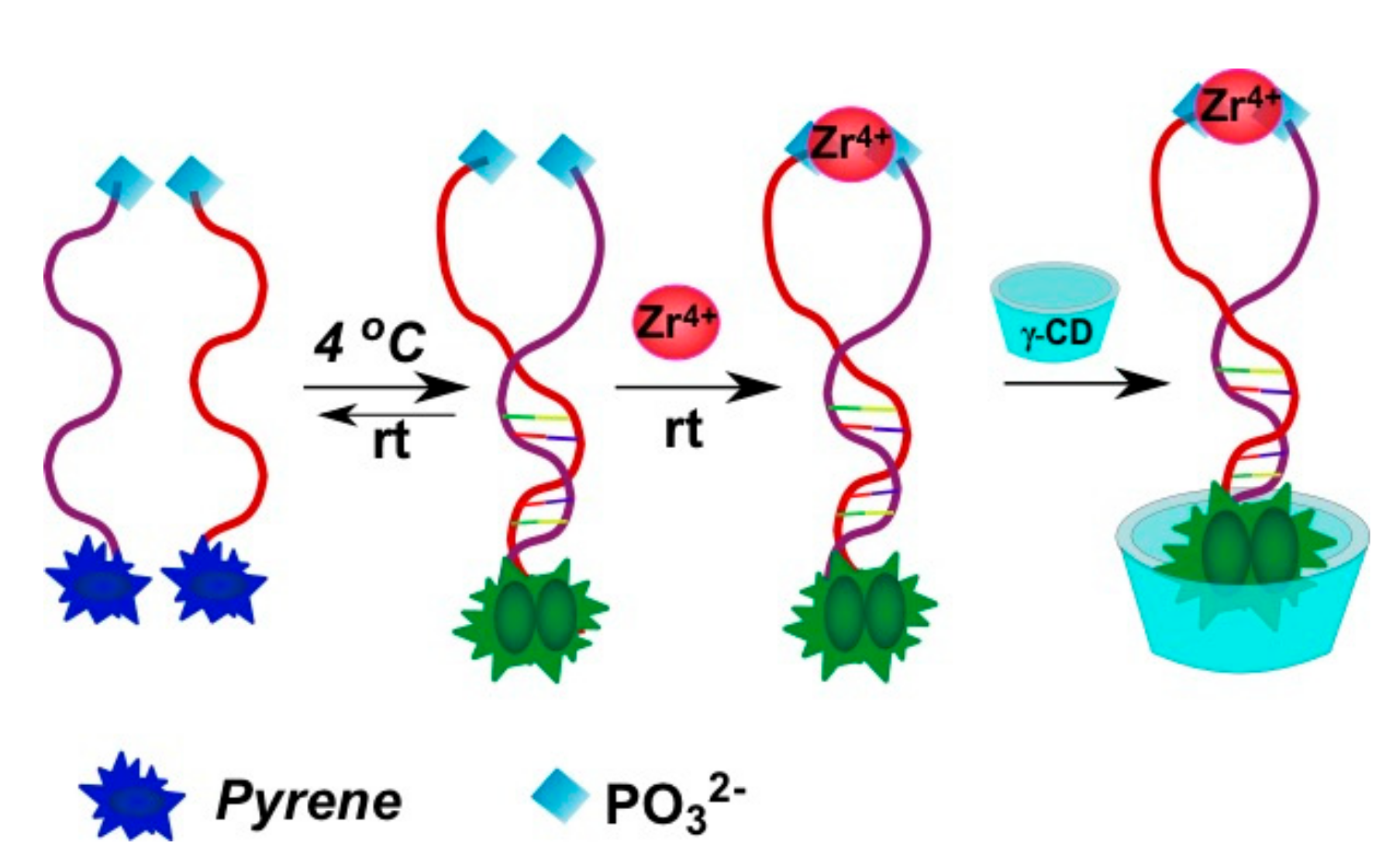
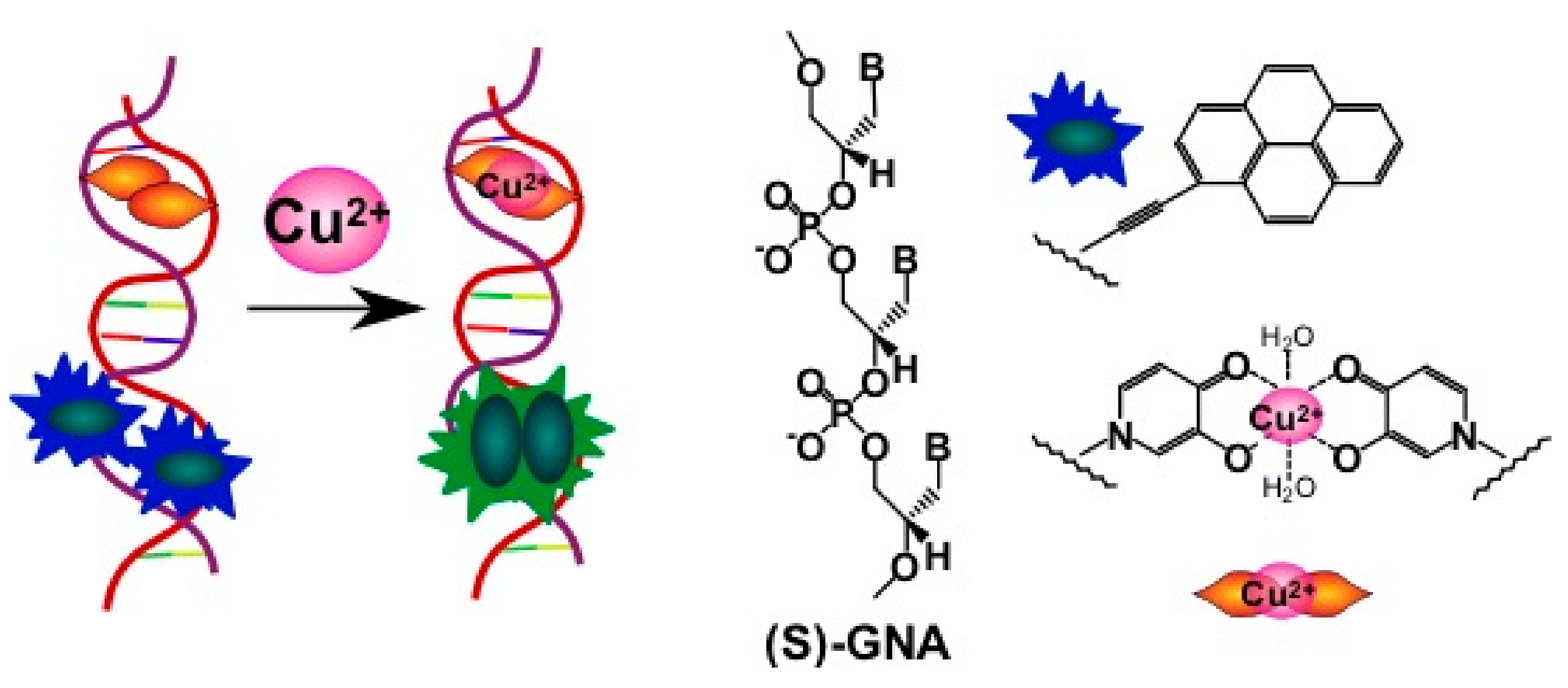
2.3.2. Pyrene-Labeled Oligonucleotide Constructions for the Detection of Metal Ions (K+, Hg2+, Zr4+, Cu2+)
2.3.3. Pyrene-Labeled Oligonucleotide Constructions for Detection of Other Small Molecules
2.3.4. Study of Enzymes and DNAzymes Activity Using Pyrene-Labeled Oligonucleotides
2.3.5. Monitoring Formation of G-Quadruplexes and i-Motifs
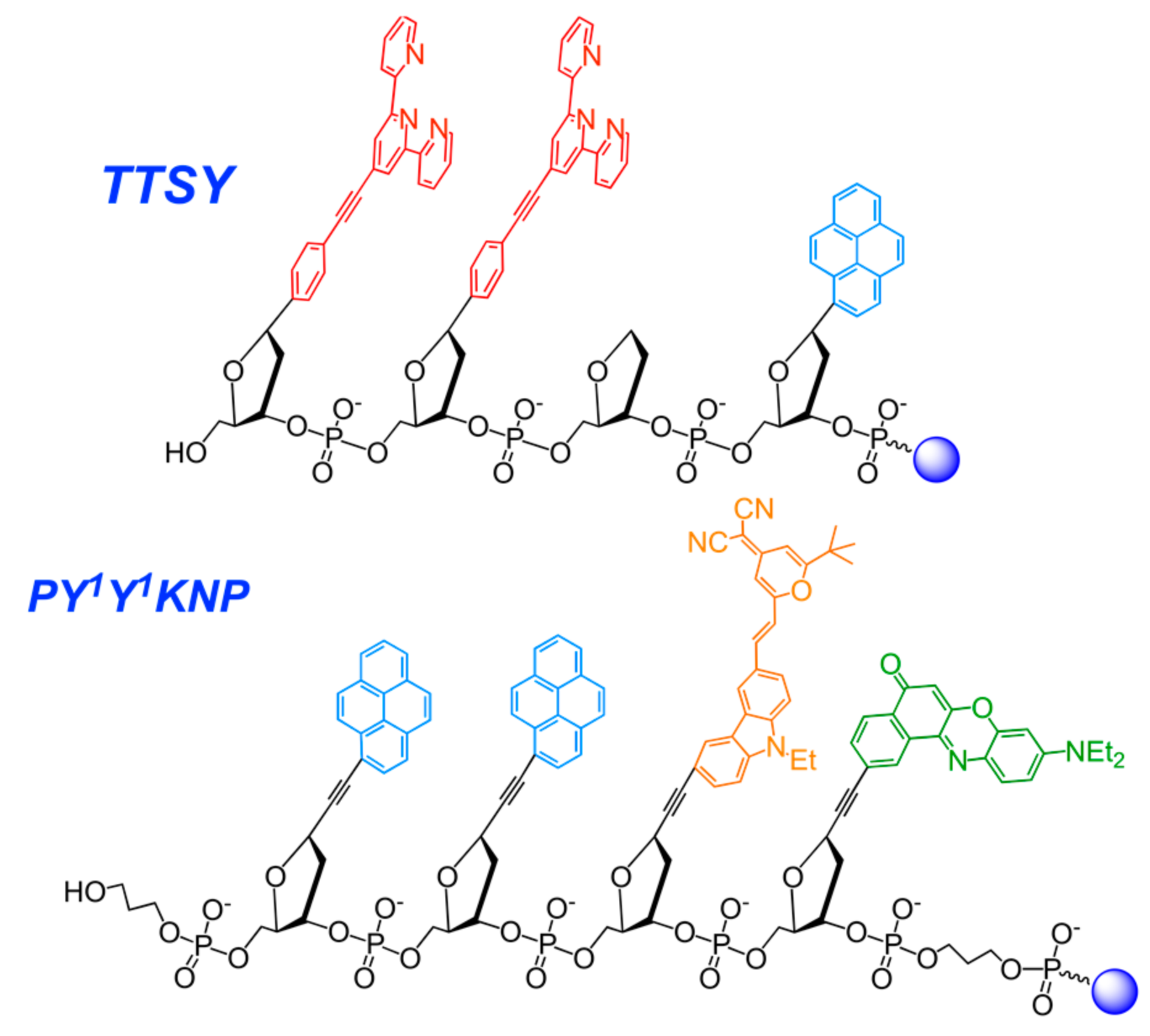
2.3.6. Nucleic Acid-Based Polyfluorophores Comprising Pyrene Derivatives
3. Agents for Targeting of dsDNAs
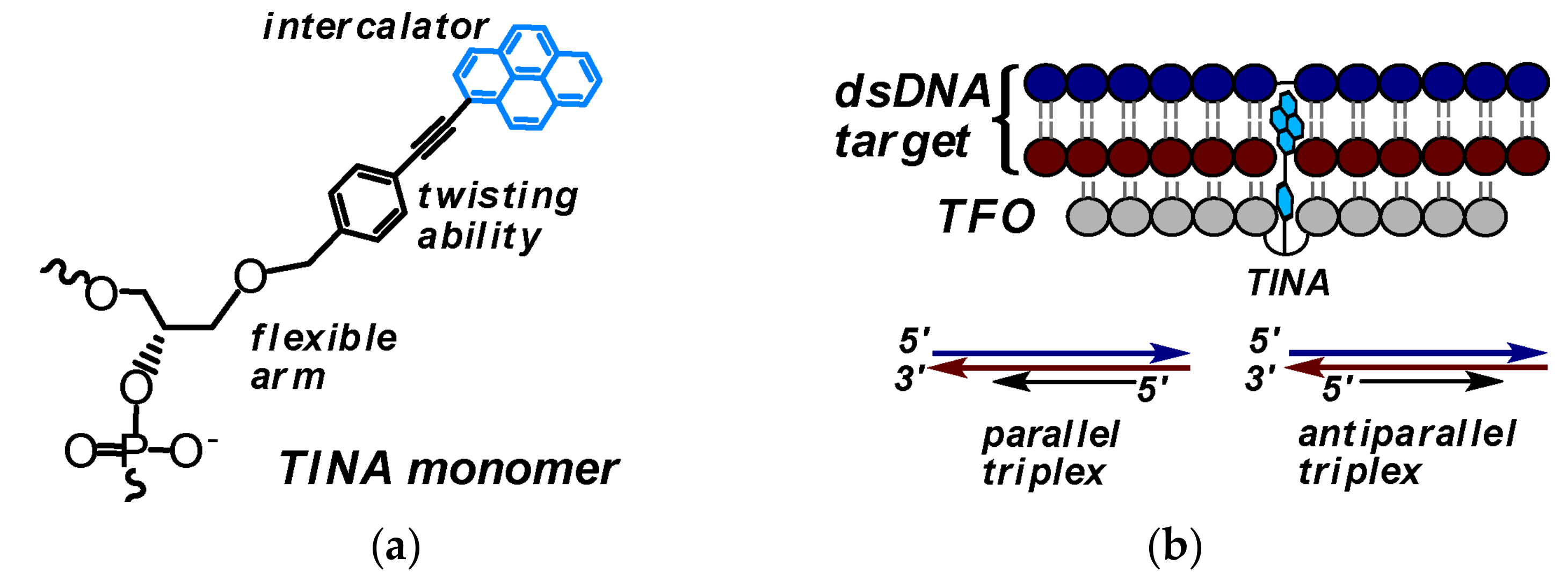
3.1. TINA-Modified Triplex Forming Oligonucleotides
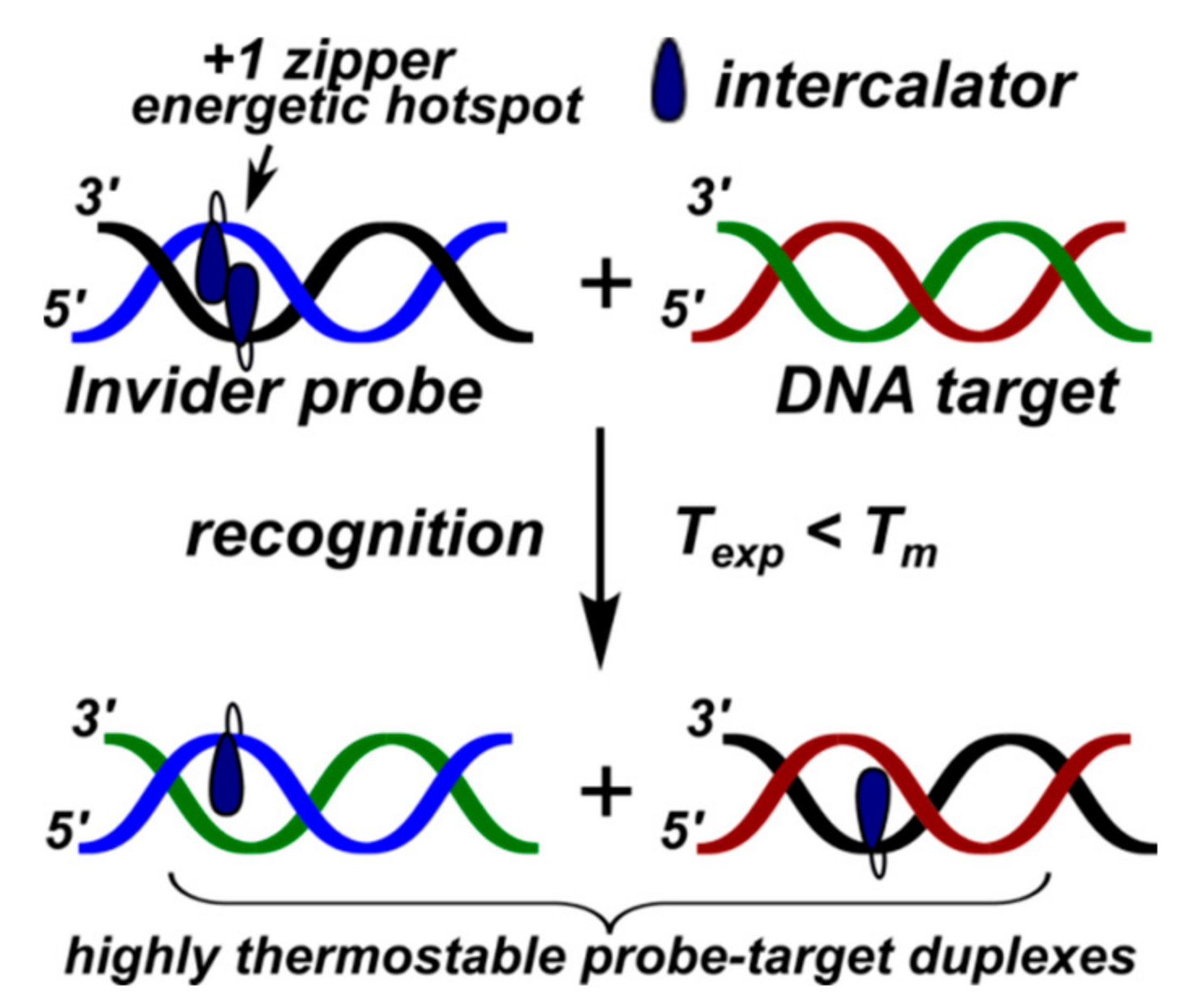
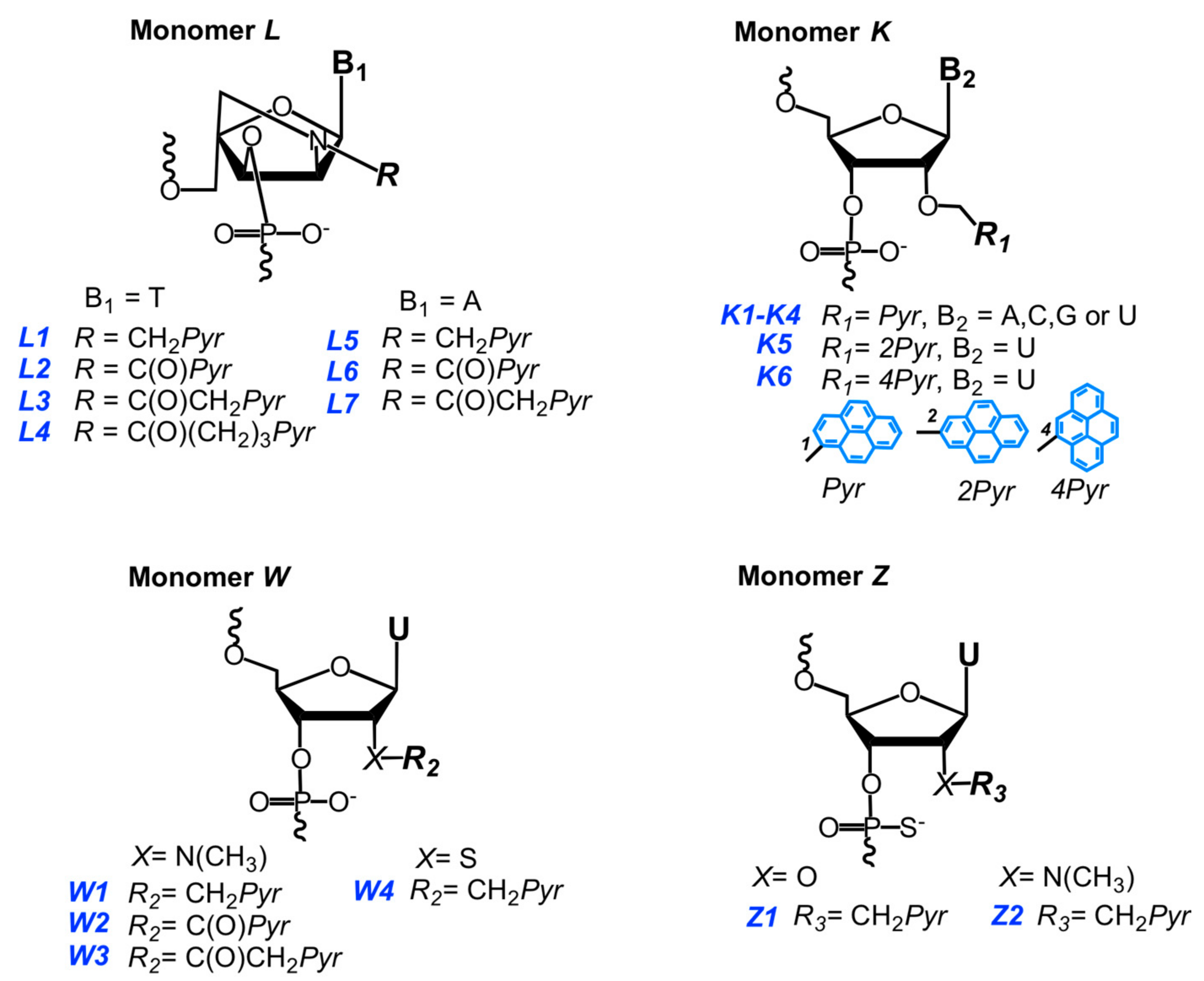
3.2. Invader Probes
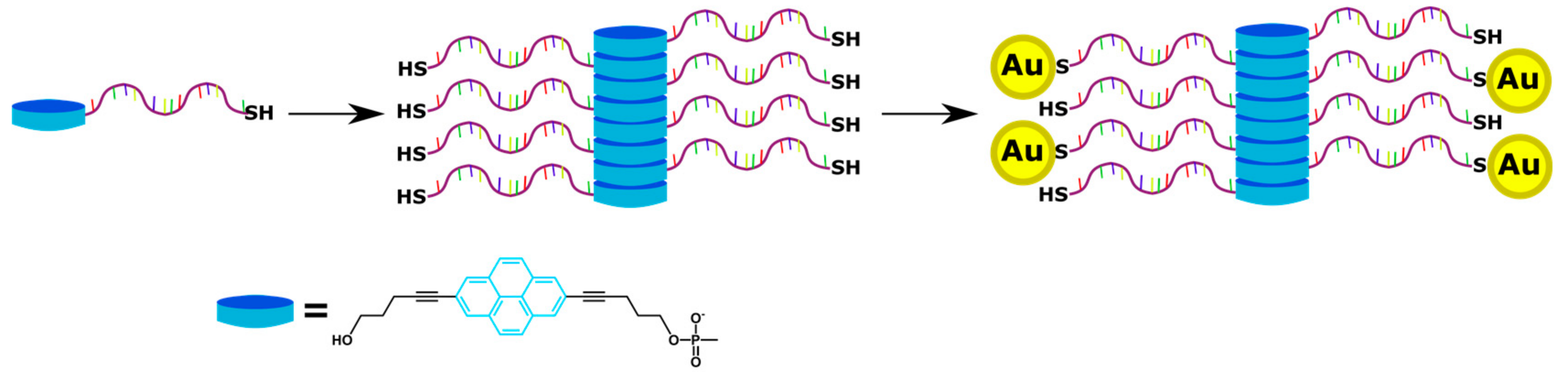
4. Supramolecular Assemblies
5. Miscellaneous Applications
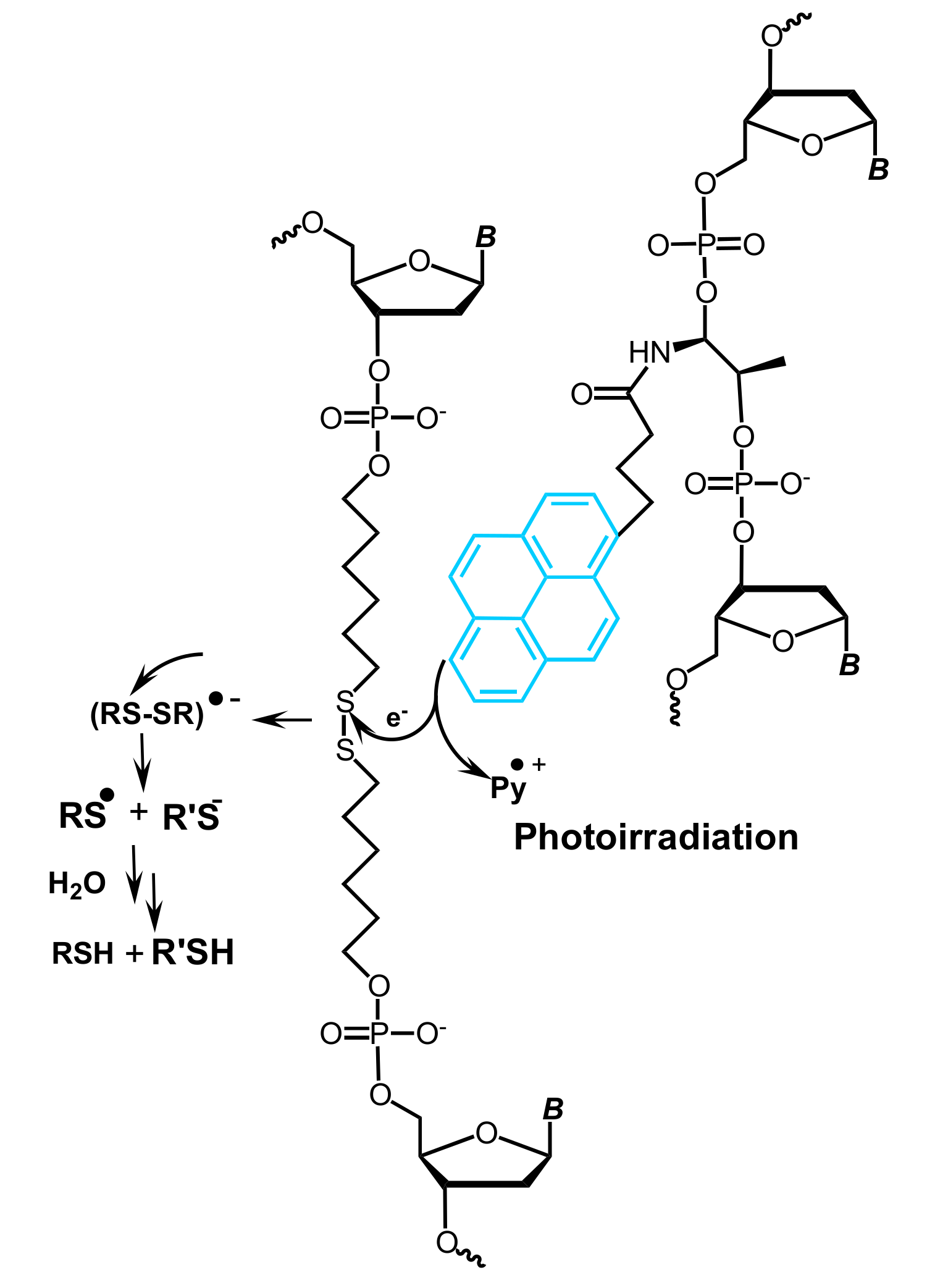
5.1. Pyrene-Mediated Photolysis of Disulfide Bonds in Oligonucleotides
5.2. Pyrene as a Structure Element of RNA Cleaving and Abasic Site Recognizing NA Constructions
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kalyanasundaram, K.; Thomas, J.K. Environmental effects on vibronic band intensities in pyrene monomer fluorescence and their application in studies of micellar systems. J. Am. Chem. Soc. 1977, 99, 2039–2044. [Google Scholar] [CrossRef]
- Manoharan, M.; Tivel, K.L.; Zhao, M.; Nafisi, K.; Netzel, T.L. Base-sequence dependence of emission lifetimes for DNA oligomers and duplexes covalently labeled with pyrene: Relative electron-transfer quenching efficiencies of A, G, C, and T nucleosides toward pyrene. J. Phys. Chem. 1995, 99, 17461–17472. [Google Scholar] [CrossRef]
- Yao, C.; Kraatz, H.-B.; Steer, R.P. Photophysics of pyrene-labelled compounds of biophysical interest. Photochem. Photobiol. Sci. 2005, 4, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, M.E.; Hrdlicka, P.J. Pyrene-functionalized oligonucleotides and locked nucleic acids (LNAs): Tools for fundamental research, diagnostics, and nanotechnology. Chem. Soc. Rev. 2011, 40, 5771–5788. [Google Scholar] [CrossRef] [PubMed]
- Winnik, F.M. Photophysics of preassociated pyrenes in aqueous polymer solutions and in other organized media. Chem. Rev. 1993, 93, 587–614. [Google Scholar] [CrossRef]
- Birks, J.B. Excimers. Rep. Prog. Phys. 1975, 903, 903–974. [Google Scholar] [CrossRef]
- Wu, C.; Wang, C.; Yan, L.; Yang, C.J. Pyrene excimer nucleic acid probes for biomolecule signaling. J. Biomed. Nanotechnol. 2009, 5, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wu, C.; Chen, Y.; Song, Y.; Tan, W.; Yang, C.J. Pyrene excimer for DNA sensors. Curr. Org. Chem. 2011, 15, 465–476. [Google Scholar] [CrossRef]
- Teo, Y.N.; Kool, E.T. DNA-multichromophore systems. Chem. Rev. 2012, 112, 4221–4245. [Google Scholar] [CrossRef] [PubMed]
- Ensslen, P.; Wagenknecht, H.-A. One-dimensional multichromophor arrays based on DNA: From self-assembly to light-harvesting. Acc. Chem. Res. 2015, 48, 2724–2733. [Google Scholar] [CrossRef] [PubMed]
- Bichenkova, E.V.; Savage, H.E.; Sardarian, A.R.; Douglas, K.T. Target-assembled tandem oligonucleotide systems based on exciplexes for detecting DNA mismatches and single nucleotide polymorphisms. Biochem. Biophys. Res. Commun. 2005, 332, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Bichenkova, E.V.; Sardarian, A.R.; Wilton, A.N.; Bonnet, P.; Bryce, R.A.; Douglas, K.T. Exciplex fluorescence emission from simple organic intramolecular constructs in non-polar and highly polar media as model systems for DNA-assembled exciplex detectors. Org. Biomol. Chem. 2006, 4, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Bichenkova, E.V.; Gbaj, A.; Walsh, L.; Savage, H.E.; Rogert, C.; Sardarian, A.R.; Etchells, L.L.; Douglas, K.T. Detection of nucleic acids in situ: Novel oligonucleotide analogues for target-assembled DNA-mounted exciplexes. Org. Biomol. Chem. 2007, 5, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Myznikova, A.; Samokhina, E.; Astakhova, I.K. Rapid genotyping using pyrene−perylene locked nucleic acid complexes. Artif. DNA PNA XNA 2013, 4, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.K.; Pasternak, K.; Campbell, M.A.; Gupta, P.; Wengel, J. A locked nucleic acid-based nanocrawler: Designed and reversible movement detected by multicolor fluorescence. J. Am. Chem. Soc. 2013, 135, 2423–2426. [Google Scholar] [CrossRef] [PubMed]
- Filichev, V.V.; Pedersen, E.B. Stable and selective formation of hoogsteen-type triplexes and duplexes using twisted intercalating nucleic acids (TINA) prepared via postsynthetic Sonogashira solid-phase coupling reactions. J. Am. Chem. Soc. 2005, 127, 14849–14858. [Google Scholar] [CrossRef] [PubMed]
- Hrdlicka, P.J.; Kumar, T.S.; Wengel, J. Targeting of mixed sequence double-stranded DNA using pyrene-functionalized 2′-amino-alpha-l-LNA. Chem. Commun. 2005, 4279–4281. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Fukunaga, Y.; Sasa, K.; Ohtoshi, Y.; Kanaori, K.; Hayashi, H.; Nakano, H.; Yamana, K. Pyrene is highly emissive when attached to the RNA duplex but not to the DNA duplex: The structural basis of this difference. Nucleic Acids Res. 2005, 33, 5887–5895. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Dallmann, A.; Marafini, P.; Gao, R.; Brown, T. Enhanced H-bonding and π-stacking in DNA: A potent duplex-stabilizing and mismatch sensing nucleobase analogue. Chem. Sci. 2014, 5, 3836–3844. [Google Scholar] [CrossRef]
- Lemek, T.; Mazurkiewicz, J.; Stobinski, L.; Lin, H.M.; Tomasik, P. Non-covalent functionalization of multi-walled carbon nanotubes with organic aromatic compounds. J. Nanosci. Nanotechnol. 2007, 7, 3081–3088. [Google Scholar] [CrossRef] [PubMed]
- Di Crescenzo, A.; Ettorre, V.; Fontana, A. Non-covalent and reversible functionalization of carbon nanotubes. Beilstein J. Nanotechnol. 2014, 5, 1675–1690. [Google Scholar] [CrossRef] [PubMed]
- Gaballah, S.T.; Hussein, Y.H.A.; Anderson, N.; Lian, T.T.; Netzel, T.L. Comparison of Py•+/dU•− charge transfer state dynamics in 5-(1-pyrenyl)-2′-deoxyuridine nucleoside conjugates with amido-, ethylenyl-, and ethynyl linkers. J. Phys. Chem. A 2005, 109, 10832–10845. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.V.; Malakhov, A.D.; Stepanova, I.A.; Ustinov, A.V.; Bondarev, S.L.; Paramonov, A.S.; Korshun, V.A. 1-Phenylethynylpyrene (1-PEPy) as refined excimer forming alternative to pyrene: Case of DNA major groove excimer. Bioconjug. Chem. 2007, 18, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.V.; Korshun, V.A.; Wengel, J. Highly fluorescent conjugated pyrenes in nucleic acid probes: (Phenylethynyl)pyrenecarbonyl-functionalized locked nucleic acids. Chem. Eur. J. 2008, 14, 11010–11026. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.K.; Lindegaard, D.; Korshun, V.A.; Wengel, J. Novel interstrand communication systems within DNA duplexes based on 1-, 2- and 4-(phenylethynyl)pyrenes attached to 2′-amino-LNA: High-affinity hybridization and fluorescence sensing. Chem. Commun. 2010, 46, 8362–8364. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Maeda, T.; Mizuno, K.; Fujimoto, K.; Shimizu, H.; Inouye, M. Alkynylpyrenes as improved pyrene-based biomolecular probes with the advantages of high fluorescence quantum yields and long absorption/emission wavelengths. Chem. Eur. J. 2006, 12, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Prokhorenko, I.A.; Malakhov, A.D.; Kozlova, A.A.; Momynaliev, K.; Govorun, V.M.; Korshun, V.A. Phenylethynylpyrene-labeled oligonucleotide probes for excimer fluorescence SNP analysis of 23S rRNA gene in clarithromycin-resistant Helicobacter pylori strains. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2006, 599, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Yamane, A. MagiProbe: A novel fluorescence quenching-based oligonucleotide probe carrying a fluorophore and an intercalator. Nucleic Acids Res. 2002, 30, e97. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.T.; Veedu, R.N.; Seo, Y.J.; Kim, B.H. Quencher-free molecular beacons as probes for oligonucleotides containing CAG repeat sequences. Chem. Commun. 2014, 50, 1561–1563. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.N.; Seo, Y.J. A multiplex fluorophore molecular beacon: detection of the target sequence using large Stokes shift and multiple emission signal properties. Chem. Commun. 2015, 51, 2939–2942. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Takagi, K.; Son, A.; Kurihara, R.; Tanabe, K. Aggregate formation of oligonucleotides that assist molecular imaging for tracking of the oxygen status in tumor tissue. ChemBioChem 2017, 18, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
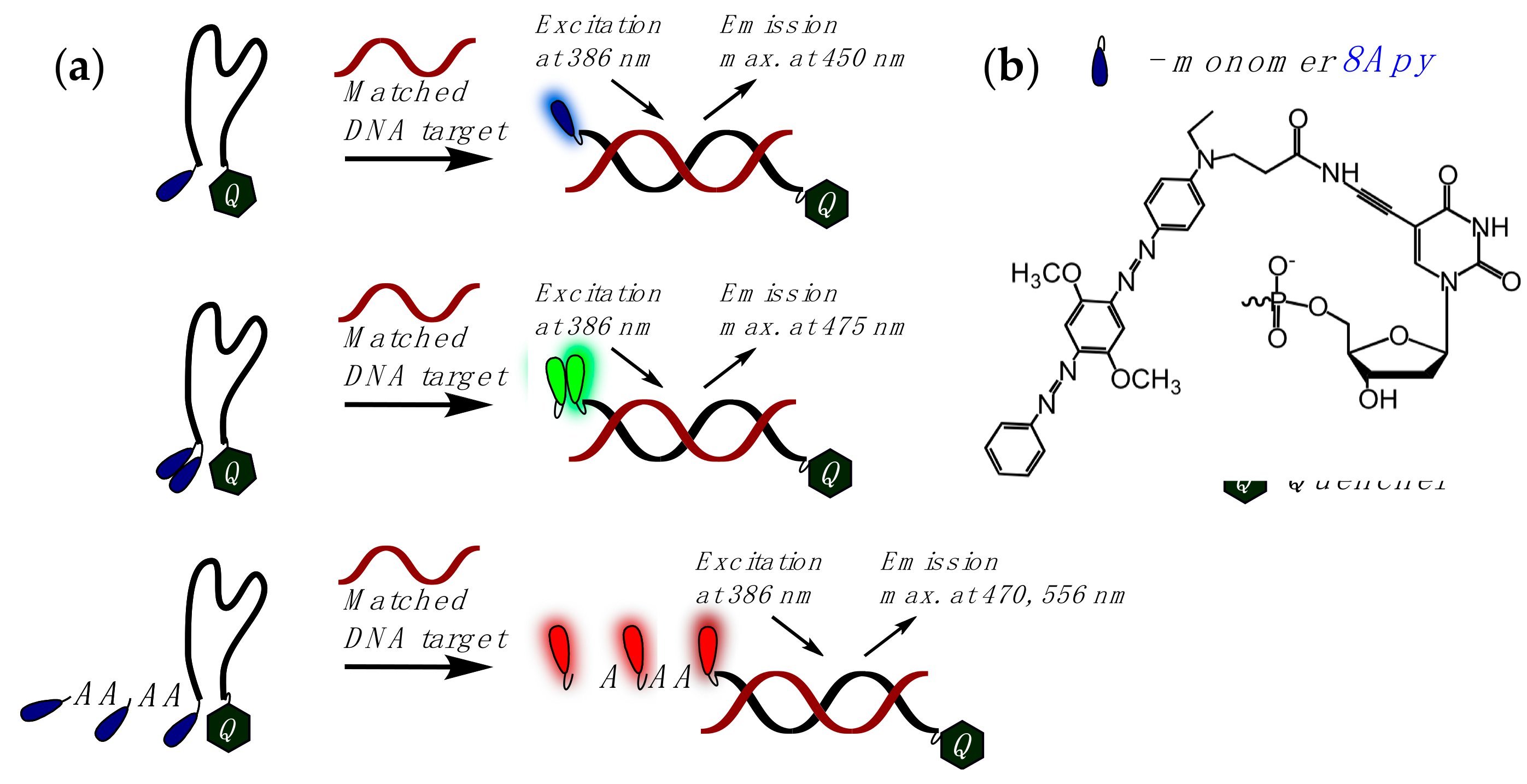
- Kim, K.T.; Heo, W.; Joo, T.; Kim, B.H. Photophysical and structural investigation of a Py A-modified adenine cluster: Its potential use for fluorescent DNA probes exhibiting distinct emission color changes. Org. Biomol. Chem. 2015, 13, 8470–8478. [Google Scholar] [CrossRef] [PubMed]
- Hrdlicka, P.J.; Østergaard, M.E. Fluorophore-functionalised Locked Nucleic Acids (LNAs). In DNA Conjugates and Sensors; Fox, K.R., Brown, T., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2012; pp. 1–33. [Google Scholar] [CrossRef]
- Juskowiak, B. Nucleic acid-based fluorescent probes and their analytical potential. Anal. Bioanal. Chem. 2011, 399, 3157–3176. [Google Scholar] [CrossRef] [PubMed]
- Doluca, O.; Withers, J.M.; Filichev, V.V. Molecular engineering of guanine-rich sequences: Z-DNA, DNA triplexes, and G-quadruplexes. Chem. Rev. 2013, 113, 3044–3083. [Google Scholar] [CrossRef] [PubMed]
- Yamana, K.; Zako, H.; Asazuma, K.; Iwase, R.; Nakano, H.; Murakami, A. Fluorescence detection of specific RNA sequences using 2′-pyrene-modified oligoribonucleotides. Angew. Chem. Int. Ed. 2001, 40, 1104–1106. [Google Scholar] [CrossRef]
- Mahara, A.; Iwase, R.; Sakamoto, T.; Yamaoka, T.; Yamana, K.; Murakami, A. Detection of acceptor sites for antisense oligonucleotides on native folded RNA by fluorescence spectroscopy. Bioorg. Med. Chem. 2003, 11, 2783–2790. [Google Scholar] [CrossRef]
- Waki, R.; Yamayoshi, A.; Kobori, A.; Murakami, A. Development of a system to sensitively and specifically visualize c-fos mRNA in living cells using bispyrene-modified RNA probes. Chem. Commun. 2011, 47, 4204–4206. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Kobori, A.; Yamayoshi, A.; Yoshida, H.; Yamaguchi, M.; Murakami, A. RNA-based diagnosis in a multicellular specimen by whole mount in situ hybridization using an RNA-specific probe. Bioorg. Med. Chem. 2012, 20, 6034–6039. [Google Scholar] [CrossRef] [PubMed]
- Iwase, R.; Mahara, A.; Yamana, K.; Yamaoka, T.; Murakami, A. Study on RNA structure by pyrene-labeled 2′-O-methyloligoribonucleotides. Nucleic Acids Symp. Ser. 1999, 42, 115–116. [Google Scholar] [CrossRef]
- Yamana, K.; Iwase, R.; Furutani, S.; Tsuchida, H.; Zako, H.; Yamaoka, T.; Murakami, A. 2′-Pyrene modified oligonucleotide provides a highly sensitive fluorescent probe of RNA. Nucleic Acids Res. 1999, 27, 2387–2392. [Google Scholar] [CrossRef] [PubMed]
- Mahara, A.; Iwase, R.; Sakamoto, T.; Yamana, K.; Yamaoka, T.; Murakami, A. Bispyrene-conjugated 2′-O-methyloligonucleotide as a highly specific RNA-recognition probe. Angew. Chem. Int. Ed. 2002, 41, 3648–3650. [Google Scholar] [CrossRef]
- Sakamoto, T.; Kobori, A.; Murakami, A. Solid-phase detection of RNA using bispyrene-modified RNA probe. Nucleic Acids Symp. Ser. 2006, 50, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kobori, A.; Shigezawa, M.; Amitani, Y.; Higuchi, M.; Murakami, A. Homogeneous fluorescence assays for RNA diagnosis by pyrene-conjugated 2’-O-methyloligoribonucleotides. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1659–1664. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kobori, A.; Murakami, A. Microarray-based label-free detection of RNA using bispyrene-modified 2’-O-methyl oligoribonucleotide as capture and detection probe. Bioorg. Med. Chem. Lett. 2008, 18, 2590–2593. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Fukuda, M.; Takada, T.; Yamana, K. Highly ordered pyrene π-stacks on an RNA duplex display static excimer fluorescence. Org. Biomol. Chem. 2012, 10, 9620–9626. [Google Scholar] [CrossRef] [PubMed]
- Kobori, A.; Ueda, T.; Sanada, Y.; Yamayoshi, A.; Murakami, A. Dual-fluorescent RNA probes with an extremely large stokes shift. Biosci. Biotechnol. Biochem. 2013, 77, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Hrdlicka, P.J. DNA strands with alternating incorporations of LNA and 2′-O-(pyren-1-yl)methyluridine: SNP-discriminating RNA detection probes. Chem. Sci. 2013, 4, 3447–3454. [Google Scholar] [CrossRef] [PubMed]
- Imincan, G.; Pei, F.; Yu, L.; Jin, H.; Zhang, L.; Yang, X.; Zhang, L.; Tang, X.J. Microenvironmental effect of 2′-O-(1-pyrenylmethyl)uridine modified fluorescent oligonucleotide probes on sensitive and selective detection of target RNA. Anal. Chem. 2016, 88, 4448–4455. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Ohtoshi, Y.; Yamana, K. Helical pyrene-array along the outside of duplex RNA. Chem. Commun. 2005, 5163–5165. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Murakami, Y.; Sasa, K.; Hayashi, H.; Yamana, K. Pyrene-zipper array assembled via RNA duplex formation. J. Am. Chem. Soc. 2008, 130, 6904–6905. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Takada, T.; Yamana, K. Highly ordered pyrene π-stacks on an RNA duplex. In Current Protocols Nucleic Acid Chemistry; Beaucage, S.L., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; Volume 63, pp. 4.66.1–4.66.19. [Google Scholar] [CrossRef]
- Nakamura, M.; Murakami, Y.; Yamana, K. Zipper-like assembly of multi-pyrenes covalently attached to RNA sequences via duplex formation. Nucleic Acids Symp. Ser. 2008, 52, 707–708. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Shimomura, Y.; Ohtoshi, Y.; Sasa, K.; Hayashi, H.; Nakano, H.; Yamana, K. Pyrene aromatic arrays on RNA duplexes as helical templates. Org. Biomol. Chem. 2007, 5, 1945–1951. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Suzuki, J.; Ota, F.; Takada, T.; Akagi, K.; Yamana, K. Helically assembled pyrene arrays on an RNA duplex that exhibit circularly polarized luminescence with excimer formation. Chem. Eur. J. 2016, 22, 9121–9124. [Google Scholar] [CrossRef] [PubMed]
- Didion, B.A.; Karmakar, S.; Guenther, D.C.; Sau, S.P.; Verstegen, J.P.; Hrdlicka, P.J. Invaders: Recognition of double-stranded DNA by using duplexes modified with interstrand zippers of 2′-O-(pyren-1-yl)methyl-ribonucleotides. ChemBioChem 2013, 14, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Denn, B.; Karmakar, S.; Guenther, D.C.; Hrdlicka, P.J. Sandwich assay for mixed-sequence recognition of double-stranded DNA: Invader-based detection of targets specific to foodborne pathogens. Chem. Commun. 2013, 49, 9851–9853. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Madsen, A.S.; Guenther, D.C.; Gibbons, B.C.; Hrdlicka, P.J. Recognition of double-stranded DNA using energetically activated duplexes with interstrand zippers of 1-, 2- or 4-pyrenyl-functionalized O2′-alkylated RNA monomers. Org. Biomol. Chem. 2014, 12, 7758–7773. [Google Scholar] [CrossRef] [PubMed]
- Guenther, D.C.; Karmakar, S.; Hrdlicka, P.J. Bulged Invader probes: Activated duplexes for mixed-sequence dsDNA recognition with improved thermodynamic and kinetic profiles. Chem. Commun. 2015, 51, 15051–15054. [Google Scholar] [CrossRef] [PubMed]
- Sau, S.P.; Madsen, A.S.; Podbevsek, P.; Andersen, N.K.; Kumar, T.S.; Andersen, S.; Rathje, R.L.; Anderson, B.A.; Guenther, D.C.; Karmakar, S.; et al. Identification and characterization of second-generation invader locked nucleic acids (LNAs) for mixed-sequence recognition of double-stranded DNA. J. Org. Chem. 2013, 78, 9560–9570. [Google Scholar] [CrossRef] [PubMed]
- Guenther, D.C.; Anderson, G.H.; Karmakar, S.; Anderson, B.; Didion, B.; Guo, W.; Verstegen, J.P.; Hrdlicka, P.J. Invader probes: Harnessing the energy of intercalation to facilitate recognition of chromosomal DNA for diagnostic applications. Chem. Sci. 2015, 6, 5006–5015. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.A.; Hrdlicka, P.J. Synthesis and characterization of oligodeoxyribonucleotides modified with 2′-thio-2′-deoxy-2′-S-(pyren-1-yl)methyluridine. Bioorg. Med. Chem. Lett. 2015, 25, 3999–4004. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Guenther, D.C.; Hrdlicka, P.J. Recognition of mixed-sequence DNA duplexes: Design guidelines for invaders based on 2′-O-(pyren-1-yl)methyl-RNA monomers. J. Org. Chem. 2013, 78, 12040–12048. [Google Scholar] [CrossRef] [PubMed]
- Hrdlicka, P.J.; Karmakar, S. 25 years and still going strong: 2′-O-(pyren-1-yl)methylribonucleotides—Versatile building blocks for applications in molecular biology, diagnostics and materials science. Org. Biomol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, K.K.; Pasternak, A.; Jensen, T.B.; Wengel, J. Pyrene-modified unlocked nucleic acids: Synthesis, thermodynamic studies, and fluorescent properties. ChemBioChem 2012, 13, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, K.K.; Okholm, A.; Kjems, J.; Wengel, J. A quencher-free molecular beacon design based on pyrene excimer fluorescence using pyrene-labeled UNA (unlocked nucleic acid). Bioorg. Med. Chem. 2013, 21, 6186–6190. [Google Scholar] [CrossRef] [PubMed]
- Perlíková, P.; Ejlersen, M.; Langkjaer, N.; Wengel, J. Bis-pyrene-modified unlocked nucleic acids: Synthesis, hybridization studies, and fluorescent properties. ChemMedChem 2014, 9, 2120–2127. [Google Scholar] [CrossRef] [PubMed]
- Ejlersen, M.; Langkjær, N.; Wengel, J. 3′-Pyrene-modified unlocked nucleic acids: Synthesis, fluorescence properties and a surprising stabilization effect on duplexes and triplexes. Org. Biomol. Chem. 2017, 2073–2085. [Google Scholar] [CrossRef] [PubMed]
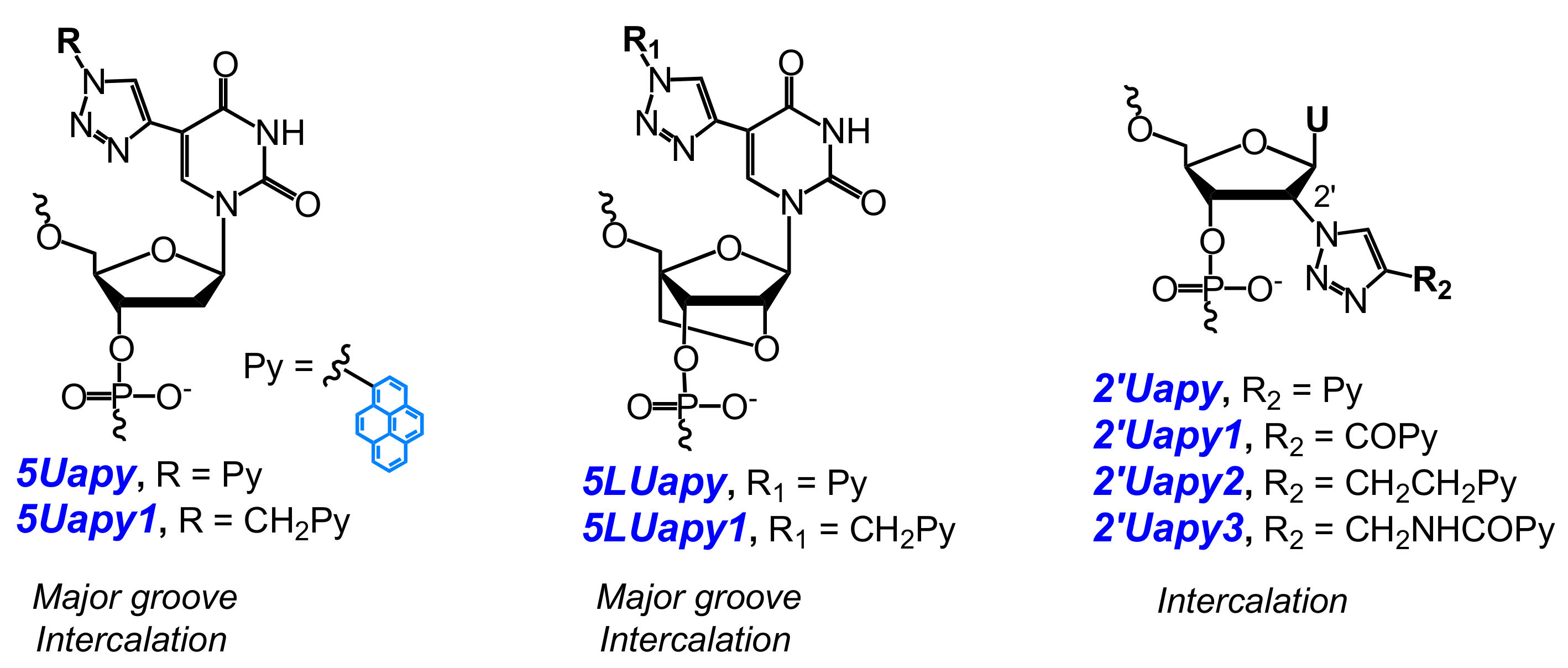
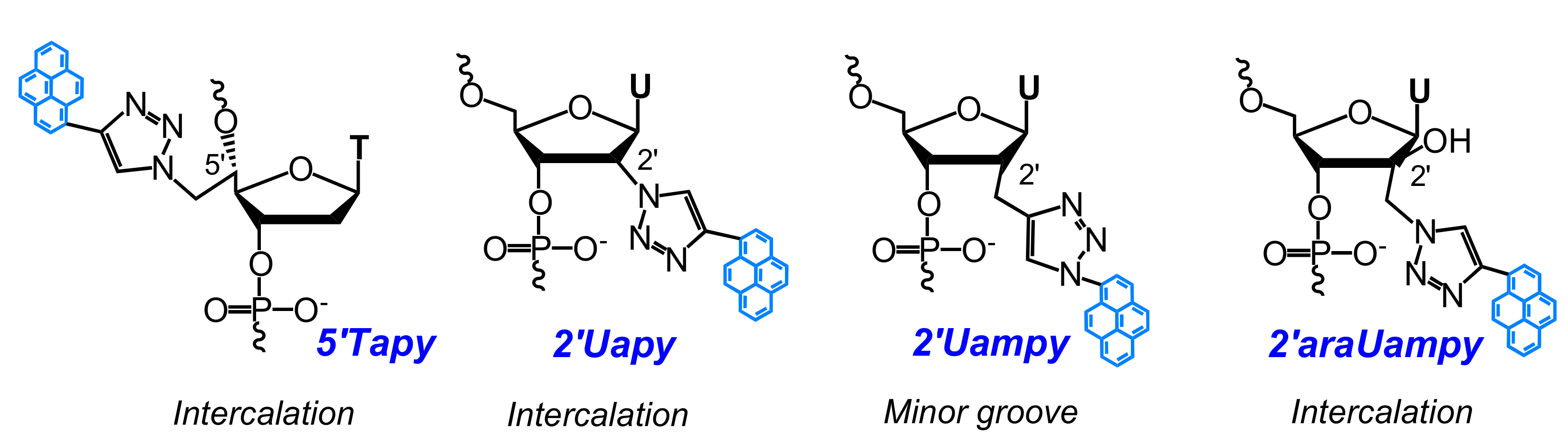
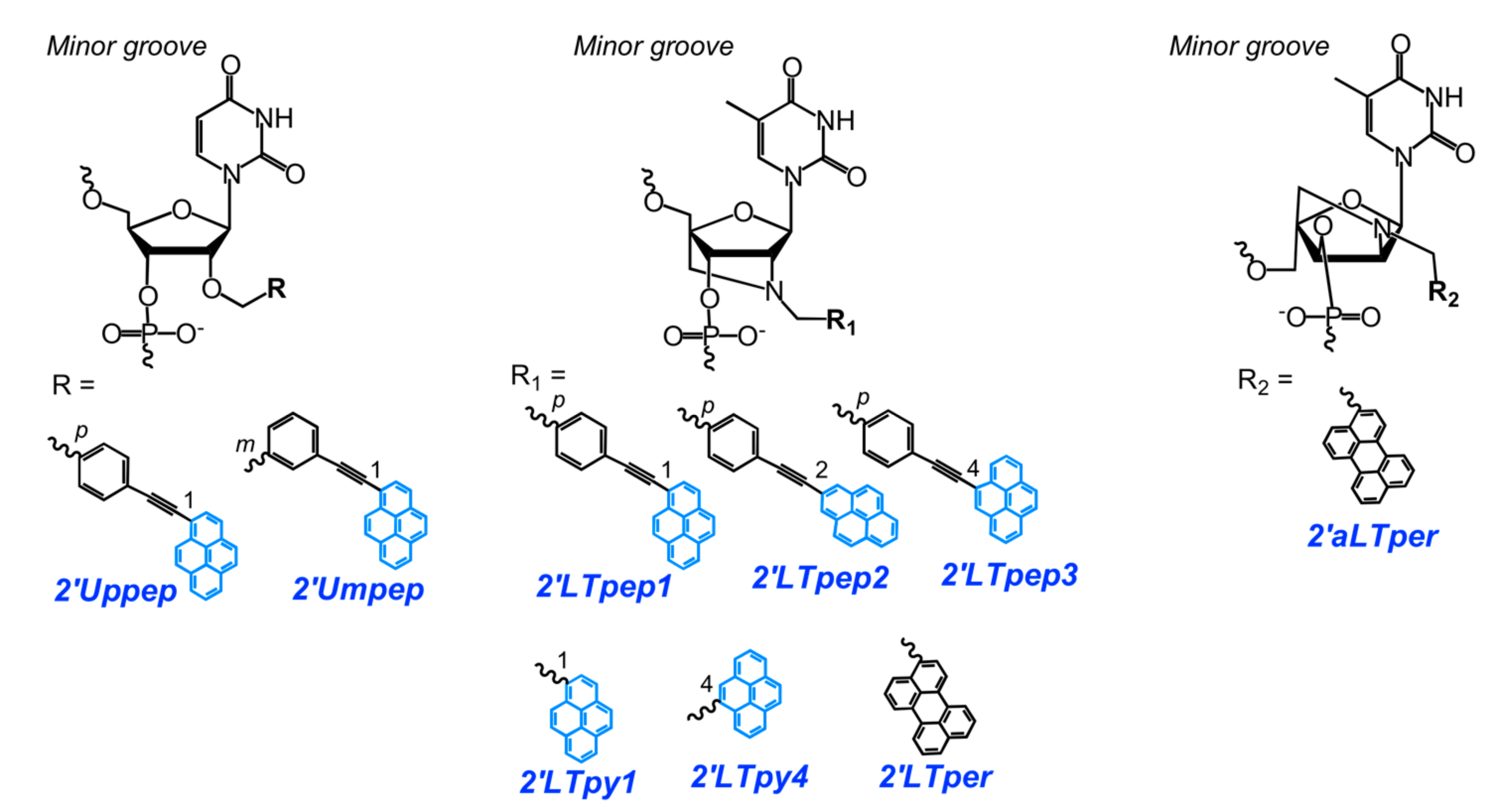
- Ostergaard, M.E.; Guenther, D.C.; Kumar, P.; Baral, B.; Deobald, L.; Paszczynski, A.J.; Sharma, P.K.; Hrdlicka, P.J. Pyrene-functionalized triazole-linked 2′-deoxyuridines-probes for discrimination of single nucleotide polymorphisms (SNPs). Chem. Commun. 2010, 46, 4929–4931. [Google Scholar] [CrossRef] [PubMed]
- Sau, S.P.; Hrdlicka, P.J. C2′-pyrene-functionalized triazole-linked DNA: Universal DNA/RNA hybridization probes. J. Org. Chem. 2011, 77, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Kaura, M.; Kumar, P.; Hrdlicka, P.J.; Guenther, D.C.; Anderson, B.A.; Ytreberg, F.M.; Deobald, L.; Paszczynski, A.J.; Sharma, P.K.; Hrdlicka, P.J. Synthesis and hybridization properties of oligonucleotides modified with 5-(1-aryl-1,2,3-triazol-4-yl)-2′-deoxyuridines. Org. Biomol. Chem. 2012, 10, 8575–8578. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Shaikh, K.I.; Jørgensen, A.S.; Kumar, S.; Nielsen, P. Three pyrene-modified nucleotides: Synthesis and effects in secondary nucleic acid structures. J. Org. Chem. 2012, 77, 9562–9573. [Google Scholar] [CrossRef] [PubMed]
- Ingale, S.A.; Seela, F. Nucleoside and oligonucleotide pyrene conjugates with 1,2,3-triazolyl or ethynyl linkers: Synthesis, duplex stability, and fluorescence changes generated by the DNA-dye connector. Tetrahedron 2014, 70, 380–391. [Google Scholar] [CrossRef]
- Kumar, P.; Ostergaard, M.E.; Baral, B.; Anderson, B.A.; Guenther, D.C.; Kaura, M.; Raible, D.J.; Sharma, P.K.; Hrdlicka, P.J. Synthesis and biophysical properties of C5-functionalized LNA (locked nucleic acid). J. Org. Chem. 2014, 79, 5047–5061. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sharma, P.K.; Nielsen, P. Synthesis, hybridization and fluorescence properties of a 2′-C-pyrene-triazole modified arabino-uridine nucleotide. Bioorg. Med. Chem. 2017, 25, 2084–2090. [Google Scholar] [CrossRef] [PubMed]
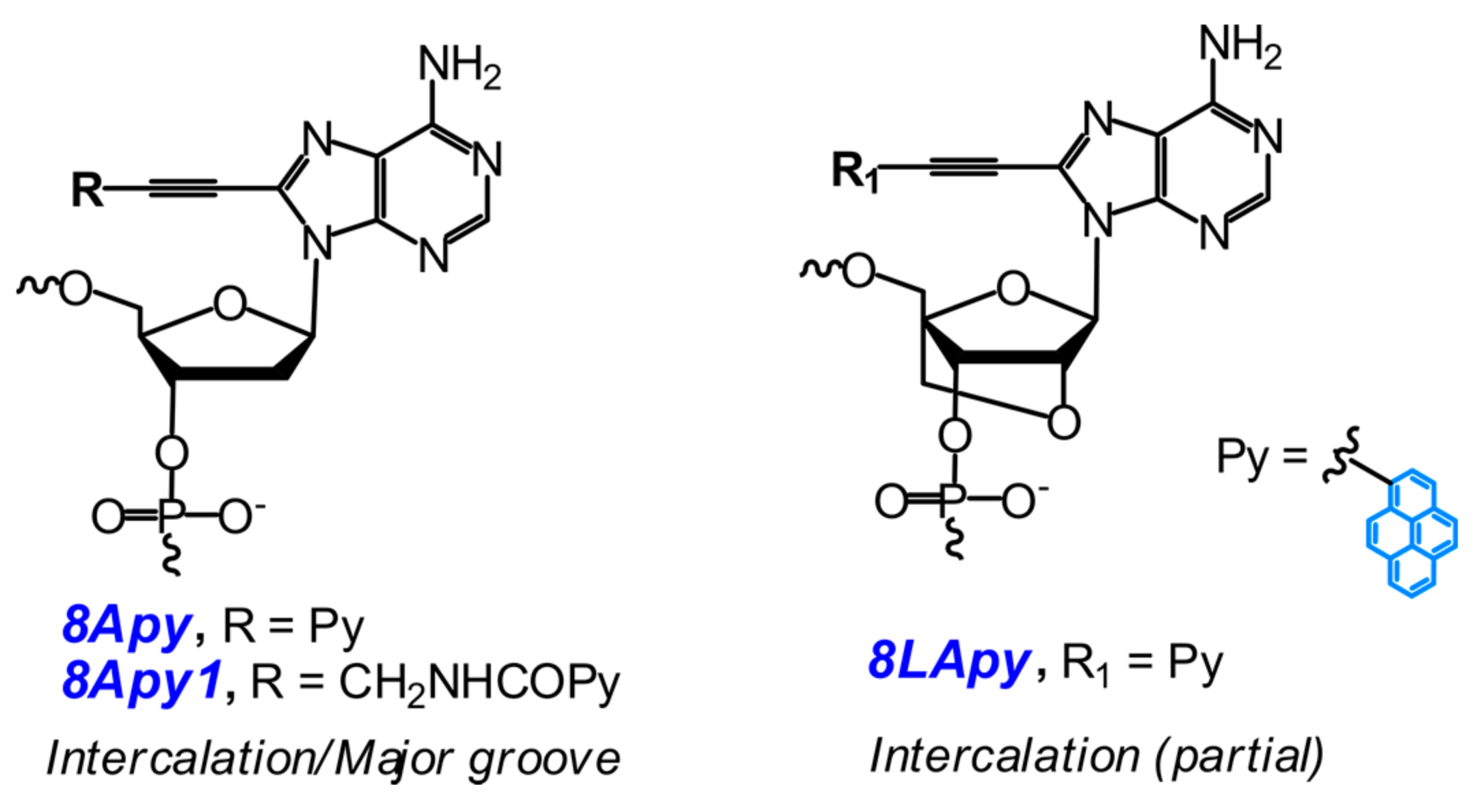
- Ingale, S.A.; Pujari, S.S.; Sirivolu, V.R.; Ding, P.; Xiong, H.; Mei, H.; Seela, F. 7-Deazapurine and 8-aza-7-deazapurine nucleoside and oligonucleotide pyrene “click” conjugates: Synthesis, nucleobase controlled fluorescence quenching, and duplex stability. J. Org. Chem. 2012, 77, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Ingale, S.A.; Seela, F. Stepwise click functionalization of DNA through a bifunctional azide with a chelating and a nonchelating azido group. J. Org. Chem. 2013, 78, 3394–3399. [Google Scholar] [CrossRef] [PubMed]
- Ingale, S.A.; Leonard, P.; Yang, H.; Seela, F. 5-Nitroindole oligonucleotides with alkynyl side chains: Universal base pairing, triple bond hydration and properties of pyrene “click” adducts. Org. Biomol. Chem. 2014, 12, 8519–8532. [Google Scholar] [CrossRef] [PubMed]
- Aparin, I.O.; Farzan, V.M.; Veselova, O.A.; Chistov, A.A.; Podkolzin, A.T.; Ustinov, A.V.; Shipulin, G.A.; Formanovsky, A.A.; Korshun, V.A.; Zatsepin, T.S. 1-Phenylethynylpyrene (PEPy) as a novel blue-emitting dye for qPCR assay. Analyst 2016, 141, 1331–1338. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Seela, F. “Bis-Click” ligation of DNA: Template-controlled assembly, circularisation and functionalisation with bifunctional and trifunctional azides. Chem. Eur. J. 2017, 23, 3375–3385. [Google Scholar] [CrossRef] [PubMed]
- Farzan, V.M.; Ulashchik, E.A.; Martynenko-Makaev, Y.V.; Kvach, M.V.; Aparin, I.O.; Brylev, V.A.; Prikazchikova, T.A.; Maklakova, S.Y.; Majouga, A.G.; Ustinov, A.V.; et al. Automated solid-phase click synthesis of oligonucleotide conjugates: From small molecules to diverse N-acetylgalactosamine clusters. Bioconjug. Chem. 2017, 28, 2599–2607. [Google Scholar] [CrossRef] [PubMed]
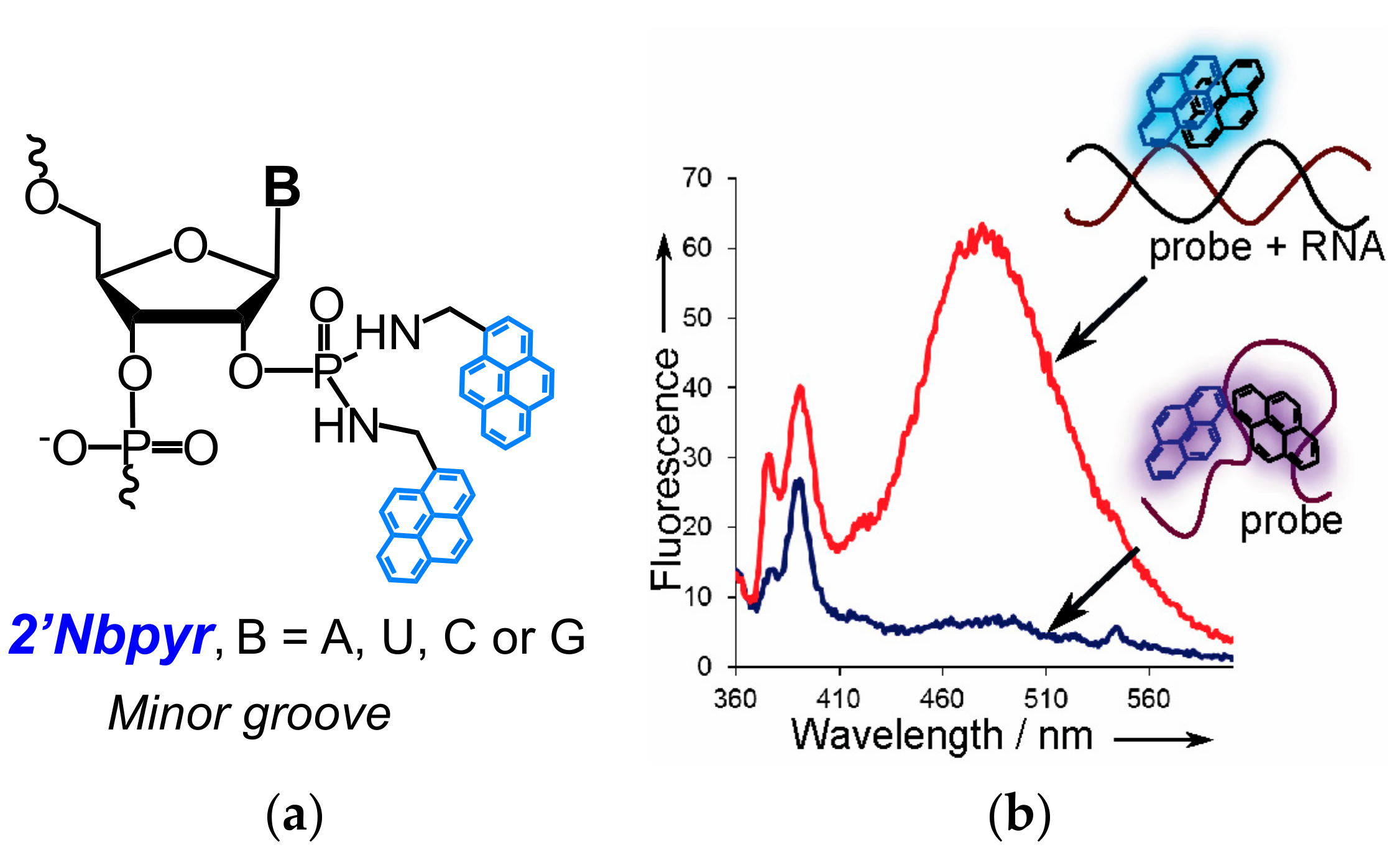
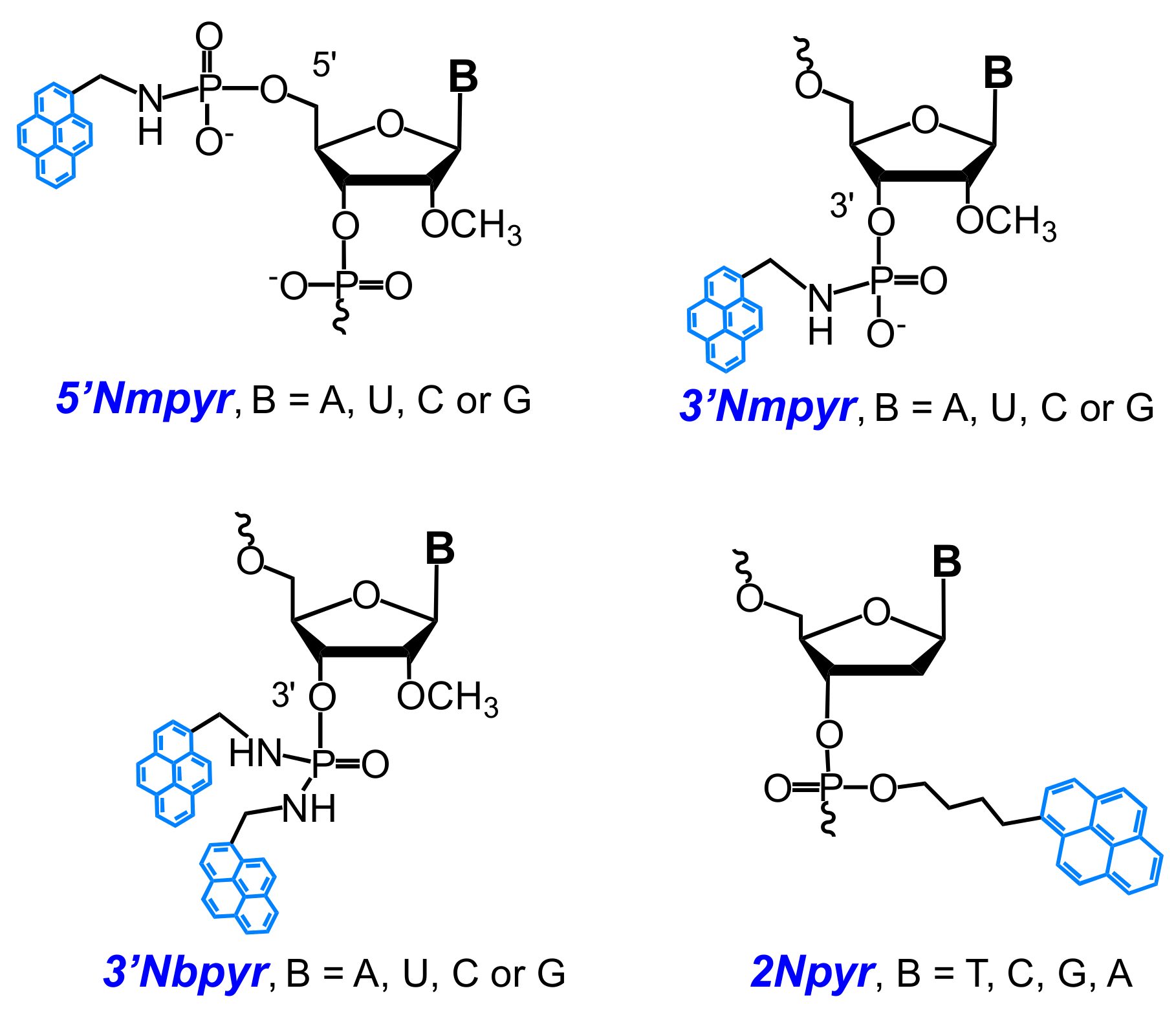
- Krasheninina, O.A.; Novopashina, D.S.; Venyaminova, A.G. Oligo(2′-O-methylribonucleotides) containing insertions of 2′-bispyrenylmethylphosphorodiamidate nucleoside derivatives as prospective fluorescent probes for RNA detection. Russ. J. Bioorg. Chem. 2011, 37, 244–248. [Google Scholar] [CrossRef]
- Krasheninina, O.A.; Novopashina, D.S.; Lomzov, A.A.; Venyaminova, A.G. 2′-Bispyrene-modified 2′-O-methyl RNA probes as useful tools for the detection of RNA: Synthesis, fluorescent properties, and duplex stability. ChemBioChem 2014, 15, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
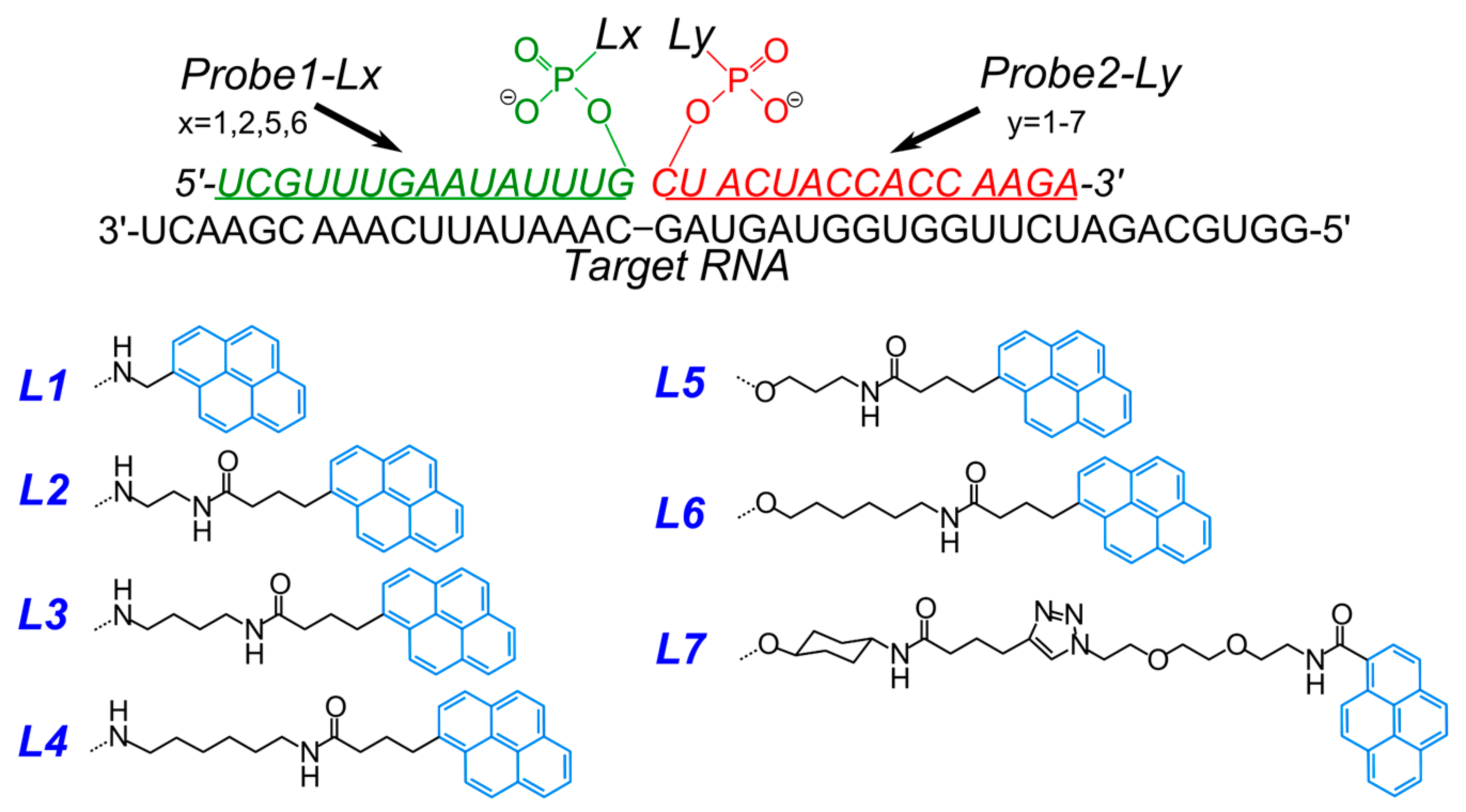
- Krasheninina, O.A.; Lomzov, A.A.; Fishman, V.S.; Novopashina, D.S.; Venyaminova, A.G. Rational design and studies of excimer forming novel dual probes to target RNA. Bioorg. Med. Chem. 2017, 25, 2244–2250. [Google Scholar] [CrossRef] [PubMed]
- Krasheninina, O.A.; Fishman, V.S.; Novopashina, D.S.; Venyaminova, A.G. 5′-Bispyrene molecular beacons for RNA detection. Russ. J. Bioorg. Chem. 2017, 43. [Google Scholar] [CrossRef]
- Kostenko, E.; Dobrikov, M.; Pyshnyi, D.; Petyuk, V.; Komarova, N.; Vlassov, V.; Zenkova, M. 5′-Bis-pyrenylated oligonucleotides displaying excimer fluorescence provide sensitive probes of RNA sequence and structure. Nucleic Acids Res. 2001, 29, 3611–3620. [Google Scholar] [CrossRef] [PubMed]
- Novopashina, D.S.; Totskaya, O.S.; Kholodar, S.A.; Meshchaninova, M.I.; Ven’yaminova, A.G. Oligo(2′-O-methylribonucleotides) and their derivatives: III. 5’-Mono- and 5’-bispyrenyl derivatives of oligo(2′-O-methylribonucleotides) and their 3’-modified analogues: Synthesis and properties. Russ. J. Bioorg. Chem. 2008, 34, 602–612. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M. Binary probes for nucleic acid analysis. Chem. Rev. 2010, 110, 4709–4723. [Google Scholar] [CrossRef] [PubMed]
- Marti, A.A.; Li, X.; Jockusch, S.; Li, Z.; Raveendra, B.; Kalachikov, S.; Russo, J.J.; Morozova, I.; Puthanveettil, S.V.; Ju, J.; et al. Pyrene binary probes for unambiguous detection of mRNA using time-resolved fluorescence spectroscopy. Nucleic Acids Res. 2006, 34, 3161–3168. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.K.; Samokhina, E.; Babu, B.R.; Wengel, J. Novel (phenylethynyl)pyrene-LNA constructs for fluorescence SNP sensing in polymorphic nucleic acid targets. ChemBioChem 2012, 13, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Kholodar, S.A.; Novopashina, D.S.; Meschaninova, M.I.; Venyaminova, A.G. Multipyrene tandem probes for point mutations detection in DNA. J. Nucleic Acids 2013, 2013, 860457. [Google Scholar] [CrossRef] [PubMed]
- Dembska, A.; Juskowiak, B. Pyrene functionalized molecular beacon with pH-sensitive i-motif in a loop. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 150, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.-M.; Fu, T.; Zhang, X.-B.; Wang, N.-N.; Tan, W.; Shen, G.-L.; Yu, R.-Q. Efficient fluorescence turn-on probe for zirconium via a target-triggered DNA molecular beacon strategy. Anal. Chem. 2012, 84, 2124–2128. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Ichimura, M.; Kato, M.; Suzuki, K.; Takahashi, Y.; Shinozuka, K. Development of the excimer probe responsible for DNA target bearing the silylated pyrenes at base moiety. Bioorg. Med. Chem. Lett. 2014, 24, 4372–4375. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Qing, Z.; He, X.; Wang, K.; He, D.; Shi, H.; Yang, X.; Qing, T.; Yang, X. Ligation-rolling circle amplification combined with γ-cyclodextrin mediated stemless molecular beacon for sensitive and specific genotyping of single-nucleotide polymorphism. Talanta 2014, 125, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hudson, R.H.E. PNA molecular beacons assembled by post-synthetic click chemistry functionalization. ChemBioChem 2015, 16, 2156–2161. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Park, Y.; Kim, B.H. Quencher-free molecular aptamer beacons (QF-MABs) for detection of ATP. Bioorg. Med. Chem. Lett. 2015, 25, 4597–4600. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yang, R.; Shi, M.; Wu, C.; Fang, X.; Li, Y.; Li, J.; Tan, W. Rationally designed molecular beacons for bioanalytical and biomedical applications. Chem. Soc. Rev. 2015, 44, 3036–3055. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, M.E.; Cheguru, P.; Papasani, M.R.; Hill, R.A.; Hrdlicka, P.J. Glowing locked nucleic acids: Brightly fluorescent probes for detection of nucleic acids in cells. J. Am. Chem. Soc. 2010, 132, 14221–14228. [Google Scholar] [CrossRef] [PubMed]
- Conlon, P.; Yang, C.J.; Wu, Y.; Chen, Y.; Martinez, K.; Kim, Y.; Stevens, N.; Marti, A.A.; Jockusch, S.; Turro, N.J.; et al. Pyrene excimer signaling molecular beacons for probing nucleic acids. J. Am. Chem. Soc. 2008, 130, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Hwang, G.T.; Kim, B.H. Quencher-free molecular beacon systems with two pyrene units in the stem region. Tetrahedron Lett. 2006, 47, 4037–4039. [Google Scholar] [CrossRef]
- Seo, Y.J.; Ryu, J.H.; Kim, B.H. Quencher-free, end-stacking oligonucleotides for probing single-base mismatches in DNA. Org. Lett. 2005, 7, 4931–4933. [Google Scholar] [CrossRef] [PubMed]
- Varghese, R.; Wagenknecht, H.-A.; Fujiwara, Y.; Mårtensson, J.; Brown, T.; Wilhelmsson, L.M.; Albinsson, B.; Akinsd, D.L.; Puthanveettil, S.V.; Ju, J.; et al. Red–white–blue emission switching molecular beacons: Ratiometric multicolour DNA hybridization probes. Org. Biomol. Chem. 2010, 8, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, Y.; Chen, Y.; Zhu, Z.; Yang, X.; Yang, C.J.; Wang, K.; Tan, W. Pyrene-excimer probes based on the hybridization chain reaction for the detection of nucleic acids in complex biological fluids. Angew. Chem. Int. Ed. 2011, 50, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Aparin, I.O.; Proskurin, G.V.; Golovin, A.V.; Ustinov, A.V.; Formanovsky, A.A.; Zatsepin, T.S.; Korshun, V.A. Fine tuning of pyrene excimer fluorescence in molecular beacons by alteration of the monomer structure. J. Org. Chem. 2017, 82, 10015–10024. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Shimizu, H.; Inouye, M. Unambiguous detection of target DNAs by excimer−monomer switching molecular beacons. J. Org. Chem. 2004, 69, 3271–3275. [Google Scholar] [CrossRef] [PubMed]
- Yamana, K.; Ohshita, Y.; Fukunaga, Y.; Nakamura, M.; Maruyama, A. Bis-pyrene-labeled molecular beacon: A monomer-excimer switching probe for the detection of DNA base alteration. Bioorg. Med. Chem. 2008, 16, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, C.J.; Wu, Y.; Conlon, P.; Kim, Y.; Lin, H.; Tan, W. Light-switching excimer beacon assays for ribonuclease H kinetic study. ChemBioChem 2008, 9, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Häner, R.; Biner, S.M.; Langenegger, S.M.; Meng, T.; Malinovskii, V.L. A highly sensitive, excimer-controlled molecular beacon. Angew. Chem. Int. Ed. 2010, 49, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Dembska, A.; Kierzek, E.; Juskowiak, B. Studying the influence of stem composition in pH-sensitive molecular beacons onto their sensing properties. Anal. Chim. Acta 2017, 990, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, X.; Wang, K.; Wang, Q.; Guo, Q.; Huang, J.; Liu, J.; Guo, X.; Li, W.; He, L. Amplified fluorescence detection of DNA based on catalyzed dynamic assembly and host–guest interaction between β-cyclodextrin polymer and pyrene. Talanta 2015, 144, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Misra, A. SNP genotyping: Technologies and biomedical applications. Annu. Rev. Biomed. Eng. 2007, 9, 289–320. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, M.E.; Kumar, P.; Baral, B.; Guenther, D.C.; Anderson, B.A.; Ytreberg, F.M.; Deobald, L.; Paszczynski, A.J.; Sharma, P.K.; Hrdlicka, P.J. C5-Functionalized DNA, LNA, and α-L-LNA: Positional control of polarity-sensitive fluorophores leads to improved SNP-typing. Chem. Eur. J. 2011, 17, 3157–3165. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Baral, B.; Anderson, B.A.; Guenther, D.C.; Østergaard, M.E.; Sharma, P.K.; Hrdlicka, P.J. C5-alkynyl-functionalized α-L-LNA: Synthesis, thermal denaturation experiments and enzymatic stability. J. Org. Chem. 2014, 79, 5062–5073. [Google Scholar] [CrossRef] [PubMed]
- Kaura, M.; Hrdlicka, P.J. Locked nucleic acid (LNA) induced effect on the hybridization and fluorescence properties of oligodeoxyribonucleotides modified with nucleobase-functionalized DNA monomers. Org. Biomol. Chem. 2015, 13, 7236–7247. [Google Scholar] [CrossRef] [PubMed]
- Kaura, M.; Hrdlicka, P.J. Efficient discrimination of single nucleotide polymorphisms (SNPs) using oligonucleotides modified with C5-pyrene-functionalized DNA and flanking locked nucleic acid (LNA) monomers. Chem. Asian J. 2016, 11, 1366–1369. [Google Scholar] [CrossRef] [PubMed]
- Kaura, M.; Kumar, P.; Hrdlicka, P.J. Synthesis, hybridization characteristics, and fluorescence properties of oligonucleotides modified with nucleobase-functionalized locked nucleic acid adenosine and cytidine monomers. J. Org. Chem. 2014, 79, 6256–6268. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, I.V.; Ustinov, A.V.; Korshun, V.A.; Wengel, J. LNA for optimization of fluorescent oligonucleotide probes: Improved spectral properties and target binding. Bioconjug. Chem. 2011, 22, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Astakhova, K.; Golovin, A.V.; Prokhorenko, I.A.; Ustinov, A.V.; Stepanova, I.A.; Zatsepin, T.S.; Korshun, V.A. Design of 2′-phenylethynylpyrene excimer forming DNA/RNA probes for homogeneous SNP detection: The attachment manner matters. Tetrahedron 2017, 73, 3220–3230. [Google Scholar] [CrossRef]
- Novopashina, D.S.; Meschaninova, M.I.; Kholodar, S.A.; Lomzov, A.A.; Venyaminova, A.G. New eximer-based tandem systems for SNP detection. Nucleic Acids Symp. Ser. 2008, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Kholodar, S.A.; Novopashina, D.S.; Meschaninova, M.I.; Lomzov, A.A.; Venyaminova, A.G. Multipyrene tandem probes for detection of C677T polymorphism in MTHFR gene. Nucleic Acids Symp. Ser. 2009, 53, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, H.; Wang, Z.; Feng, M.; Jin, H.; Wu, Y.; Zhang, L.; Zhang, L.; Tang, X. Sensitive detection of single-nucleotide mutation in the BRAF mutation site (V600E) of human melanoma using phosphate–pyrene-labeled DNA probes. Anal. Chem. 2016, 88, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.N.; Seo, Y.J. Single excitation three color folded DNA probe for SNP typing. Bioorg. Med. Chem. Lett. 2015, 25, 5286–5290. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, P.-J.J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.-H.; Zhang, T.; Luo, H.; Yen, T.; Chen, P.-W.; Han, Y.; Lo, Y.-H. Nucleic acid aptamers: An emerging tool for biotechnology and biomedical sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, M.; Timoshenko, V.; Vorobjev, P.; Venyaminova, A. Aptamers against immunologic targets: Diagnostic and therapeutic prospects. Nucleic Acid Ther. 2016, 26, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent aptamers: Versatile tools for diagnostic and therapeutic applications. Molecules 2016, 21, 1613. [Google Scholar] [CrossRef] [PubMed]
- Davydova, A.; Vorobjeva, M.; Pyshnyi, D.; Altman, S.; Vlassov, V.; Venyaminova, A. Aptamers against pathogenic microorganisms. Crit. Rev. Microbiol. 2016, 42, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Ilgu, M.; Nilsen-Hamilton, M. Aptamers in analytics. Analyst 2016, 141, 1551–1568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, H.; Aldalbahi, A.; Zuo, X.; Fan, C.; Mi, X. Fluorescent biosensors enabled by graphene and graphene oxide. Biosens. Bioelectron. 2017, 89, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Kamekawa, N.; Shimomura, Y.; Nakamura, M.; Yamana, K. Pyrene-modified DNA aptamer as a fluorescent biosensor with high affinity and specificity for ATP sensing. Chem. Lett. 2006, 35, 660–661. [Google Scholar] [CrossRef]
- Musumeci, D.; Oliviero, G.; Roviello, G.N.; Bucci, E.M.; Piccialli, G. G-Quadruplex-forming oligonucleotide conjugated to magnetic nanoparticles: Synthesis, characterization, and enzymatic stability assays. Bioconjug. Chem. 2012, 23, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Fukuda, M.; Tamura, T.; Sakaguchi, R.; Nakata, E.; Morii, T. Simultaneous detection of ATP and GTP by covalently linked fluorescent ribonucleopeptide sensors. J. Am. Chem. Soc. 2013, 135, 3465–3473. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhao, Q. Rapid fluorescence detection of immunoglobulin E using an aptamer switch based on a binding-induced pyrene excimer. Anal. Methods 2017, 9, 3962–3967. [Google Scholar] [CrossRef]
- Yang, C.J.; Jockusch, S.; Vicens, M.; Turro, N.J.; Tan, W. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc. Natl. Acad. Sci. USA 2005, 102, 17278–17283. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Cheng, L. Detection of thrombin using an excimer aptamer switch labeled with dual pyrene molecules. Anal. Bioanal. Chem. 2013, 405, 8233–8239. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Yan, L.; Wang, C.; Lin, H.; Wang, C.; Chen, X.; Yang, C.J. A general excimer signaling approach for aptamer sensors. Biosens. Bioelectron. 2010, 25, 2232–2237. [Google Scholar] [CrossRef] [PubMed]
- Freeman, R.; Li, Y.; Tel-Vered, R.; Sharon, E.; Elbaz, J.; Willner, I. Self-assembly of supramolecular aptamer structures for optical or electrochemical sensing. Analyst 2009, 134, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Yamana, K.; Ohtani, Y.; Nakano, H.; Saito, I. Bis-pyrene labeled DNA aptamer as an intelligent fluorescent biosensor. Bioorg. Med. Chem. Lett. 2003, 13, 3429–3431. [Google Scholar] [CrossRef]
- Ohtani, Y.; Yamana, K.; Nakano, H. Molecular sensing by pyrene excimer signal of fluorescent DNA aptamer. Nucleic Acids Symp. Ser. 2002, 2, 169–170. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, Z.; Bamrungsap, S.; Zhu, G.; You, M.; He, X.; Wang, K.; Tan, W. Competition-mediated pyrene-switching aptasensor: Probing lysozyme in human serum with a monomer-excimer fluorescence switch. Anal. Chem. 2010, 82, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, J.; Jiang, Y.; Jin, J.; Wang, K.; Yang, R.; Tan, W. Design of aptamer-based sensing platform using triple-helix molecular switch. Anal. Chem. 2011, 83, 6586–6592. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.-S.; Hu, P.; Zhou, H.; Shen, G.; Yu, R. Fluorescent oligonucleotide probe based on G-quadruplex scaffold for signal-on ultrasensitive protein assay. Biomaterials 2010, 31, 1918–1924. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Yang, X.; Wang, K.; Wang, Q.; Guo, Q.; Huang, J.; Liu, J.; Song, C. Amplified fluorescence detection of adenosine via catalyzed hairpin assembly and host–guest interactions between β-cyclodextrin polymer and pyrene. Analyst 2016, 141, 2502–2507. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Lian, Y.; Li, J.; Zheng, J.; Hu, Y.; Liu, J.; Huang, J.; Yang, R. Molecule-binding dependent assembly of split aptamer and γ-cyclodextrin: A sensitive excimer signaling approach for aptamer biosensors. Anal. Chim. Acta 2013, 799, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Sun, S.; Guo, X.; Yang, X.; Huang, J.; Wang, K.; Wang, Q.; Liu, J.; He, L. Competitive host–guest interaction between β-cyclodextrin polymer and pyrene-labeled probes for fluorescence analyses. Anal. Chem. 2015, 87, 2665–2671. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, J.; Gao, X.; Jin, J.; Wang, K.; Tan, W.; Yang, R. Modulating molecular level space proximity: A simple and efficient strategy to design structured DNA probes. Anal. Chem. 2010, 82, 3914–3921. [Google Scholar] [CrossRef] [PubMed]
- Qing, Z.; He, X.; Huang, J.; Wang, K.; Zou, Z.; Qing, T.; Mao, Z.; Shi, H.; He, D. Target-catalyzed dynamic assembly-based pyrene excimer switching for enzyme-free nucleic acid amplified detection. Anal. Chem. 2014, 86, 4934–4939. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yang, X.; Liu, P.; Wang, K.; Wang, Q.; Guo, Q.; Huang, J.; Li, W.; Xu, F.; Song, C. Multiple amplification detection of microRNA based on the host–guest interaction between β-cyclodextrin polymer and pyrene. Analyst 2015, 140, 4291–4297. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Yang, X.; Wang, K.; Wang, Q.; Liu, J.; Huang, J.; Zhou, M.; Guo, X. Steric hindrance regulated supramolecular assembly between β-cyclodextrin polymer and pyrene for alkaline phosphatase fluorescent sensing. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 156, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, P.; Yang, X.; Wang, K.; Wang, Q.; Guo, Q.; Huang, J.; Liu, J.; Song, C.; Li, W. A multiple amplification strategy for nucleic acid detection based on host–guest interaction between the β-cyclodextrin polymer and pyrene. Analyst 2015, 140, 2016–2022. [Google Scholar] [CrossRef] [PubMed]
- Nagatoishi, S.; Nojima, T.; Juskowiak, B.; Takenaka, S. A Pyrene-labeled G-quadruplex oligonucleotide as a fluorescent probe for potassium ion detection in biological applications. Angew. Chem. Int. Ed. 2005, 44, 5067–5070. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Gu, H.; Ma, C. An aptamer-based fluorescent biosensor for potassium ion detection using a pyrene-labeled molecular beacon. Anal. Biochem. 2010, 400, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Huang, H.; Zhao, C. An aptamer-based and pyrene-labeled fluorescent biosensor for homogeneous detection of potassium ions. Anal. Sci. 2010, 26, 1261–1264. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Muto, Y.; Inouye, M. A general and versatile molecular design for host molecules working in water: A duplex-based potassium sensor consisting of three functional regions. Chem. Commun. 2005, 4780–4782. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, Y.; Wang, L.; Zhang, J.; Song, S.; Fan, C.; Wang, S. Optical detection of mercury(II) in aqueous solutions by using conjugated polymers and label-free oligonucleotides. Adv. Mater. 2007, 19, 1471–1474. [Google Scholar] [CrossRef]
- Gao, X.; Deng, T.; Li, J.; Yang, R.; Shen, G.; Yu, R. New probe design strategy by cooperation of metal/DNA-ligation and supermolecule inclusion interaction: Application to detection of mercury ions(II). Analyst 2013, 138, 2755–2760. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Nie, Y.; Hu, Y.; Li, J.; Li, Y.; Jiang, Y.; Yang, R. Time-resolved fluorescent detection of Hg2+ in a complex environment by conjugating magnetic nanoparticles with a triple-helix molecular switch. Chem. Commun. 2013, 49, 6915–6917. [Google Scholar] [CrossRef] [PubMed]
- Wegner, S.V.; Okesli, A.; Chen, P.; He, C. Design of an emission ratiometric biosensor from MerR family proteins: A sensitive and selective sensor for Hg2+. J. Am. Chem. Soc. 2007, 129, 3474–3475. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ma, X.; Wang, J.; Zhang, L. Pyrene acetylide nucleotides in GNA: Probing duplex formation and sensing of copper(II) ions. Org. Biomol. Chem. 2009, 7, 2297–2302. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Muto, Y.; Inouye, M. A DNA duplex-based, tailor-made fluorescent sensor for porphyrin derivatives. Bioconjug. Chem. 2008, 19, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Deng, T.; Li, J.; Xu, W.; Shen, G.; Yu, R. Cyclodextrin supramolecular inclusion-enhanced pyrene excimer switching for time-resolved fluorescence detection of biothiols in serum. Biosens. Bioelectron. 2015, 68, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, K.; Yamada, S.; Inouye, M. Synthesis of versatile fluorescent sensors based on Click chemistry: Detection of unsaturated fatty acids by their pyrene-emission switching. Chem. Commun. 2009, 7164–7166. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Gao, Y.; Li, B.; Jin, Y. Cyclic up-regulation fluorescence of pyrene excimer for studying polynucleotide kinase activity based on dual amplification. Biosens. Bioelectron. 2016, 80, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Yang, X.; Wang, K.; Wang, Q.; Liu, J.; Huang, J.; He, L.; Liu, P.; Qing, Z.; Liu, W. A sensitive detection of T4 polynucleotide kinase activity based on β-cyclodextrin polymer enhanced fluorescence combined with an exonuclease reaction. Chem. Commun. 2015, 51, 1815–1818. [Google Scholar] [CrossRef] [PubMed]
- Beharry, A.A.; Lacoste, S.; O’Connor, T.R.; Kool, E.T. Fluorescence monitoring of the oxidative repair of DNA alkylation damage by ALKBH3, a prostate cancer marker. J. Am. Chem. Soc. 2016, 138, 3647–3650. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.K.; Ono, T.; Wang, S.; Jiang, W.; Franzini, R.M.; Jung, J.W.; Chan, K.M.; Kool, E.T. In vitro fluorogenic real-time assay of the repair of oxidative DNA damage. ChemBioChem 2015, 16, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-W.; Edwards, S.K.; Kool, E.T. Selective fluorogenic chemosensors for distinct classes of nucleases. ChemBioChem 2013, 14, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Wang, S.; Koo, C.-K.; Engstrom, L.; David, S.S.; Kool, E.T. Direct fluorescence monitoring of DNA base excision repair. Angew. Chem. Int. Ed. 2012, 51, 1689–1692. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Edwards, S.K.; Wang, S.; Jiang, W.; Kool, E.T. Monitoring eukaryotic and bacterial UDG repair activity with DNA-multifluorophore sensors. Nucleic Acids Res. 2013, 41, e127. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhu, J.; He, J. Conformational studies of 10–23 DNAzyme in solution through pyrenyl-labeled 2′-deoxyadenosine derivatives. Org. Biomol. Chem. 2016, 14, 9846–9858. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Lee, I.J.; Yi, J.W.; Kim, B.H. Probing the stable G-quadruplex transition using quencher-free end-stacking ethynyl pyrene–adenosine. Chem. Commun. 2007, 117, 2817–2819. [Google Scholar] [CrossRef] [PubMed]
- Dembska, A.; Juskowiak, B. The fluorescence properties and lifetime study of G-quadruplexes single- and double-labeled with pyrene. J. Fluoresc. 2010, 20, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
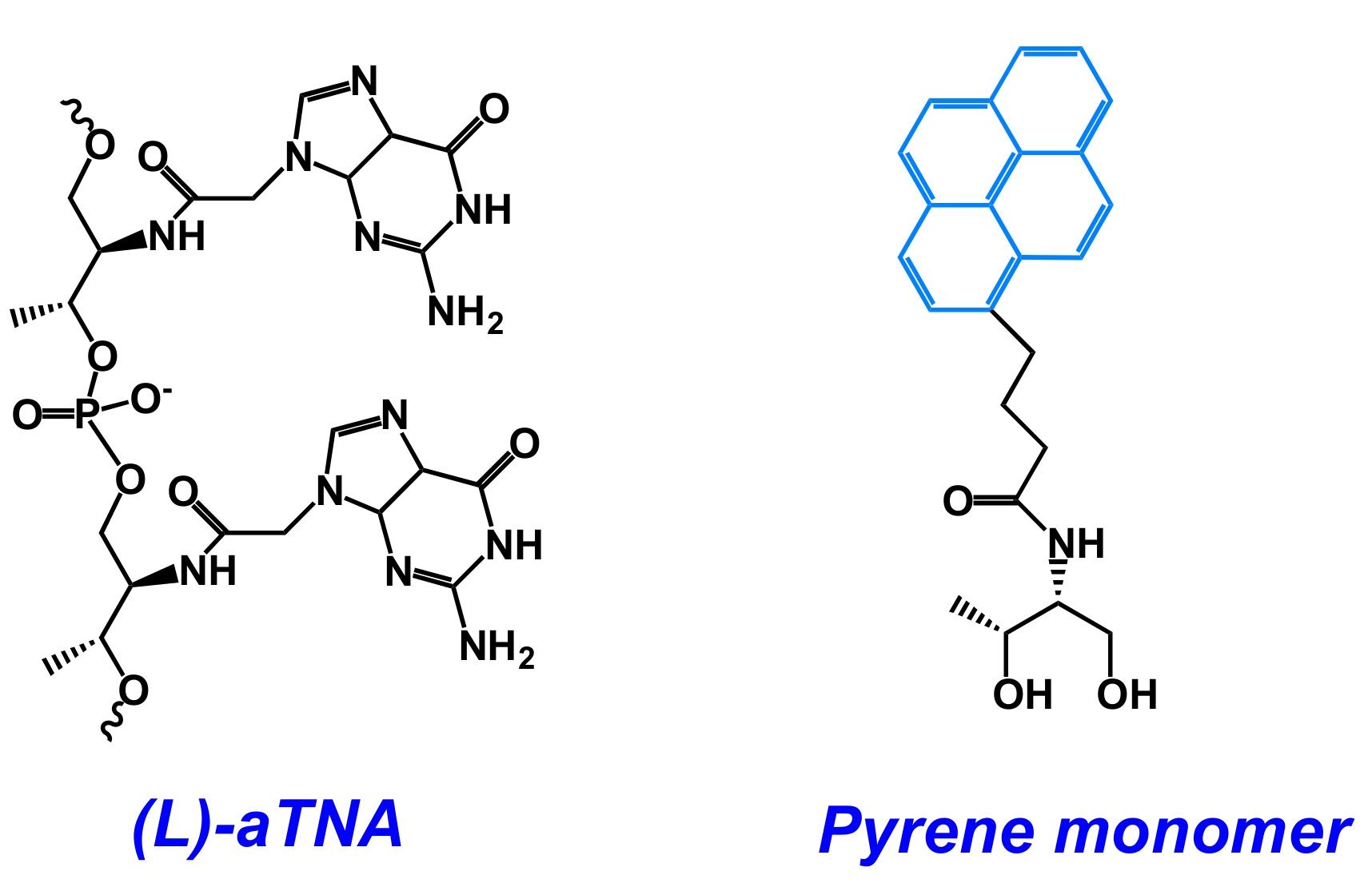
- Kumar, V.; Gothelf, K.V. Synthesis and biophysical properties of L-aTNA based G-quadruplexes. Org. Biomol. Chem. 2016, 14, 1540–1544. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, S.K.; Hariharan, M. Non-natural G-quadruplex in a non-natural environment. Photochem. Photobiol. Sci. 2014, 13, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Kim, K.T.; Kim, B.H. G-Quadruplex formation using fluorescent oligonucleotides as a detection method for discriminating AGG trinucleotide repeats. Chem. Commun. 2016, 52, 12757–12760. [Google Scholar] [CrossRef] [PubMed]
- Seio, K.; Tokugawa, M.; Tsunoda, H.; Ohkubo, A.; Arisaka, F.; Sekine, M. Assembly of pyrene-modified DNA/RNA duplexes incorporating a G-rich single strand region. Bioorg. Med. Chem. Lett. 2013, 23, 6822–6824. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.S.; Seo, Y.J. Synthesis and characterization of a fluorescent adenosine derivative for detection of intermolecular RNA G-quadruplexes. Tetrahedron Lett. 2014, 55, 1461–1463. [Google Scholar] [CrossRef]
- Switalska, A.; Kierzek, R.; Dembska, A.; Juskowiak, B. Spectroscopic study of fluorescent probes based on G-quadruplex oligonucleotides labeled with ethynylpyrenyldeoxyuridine. Int. J. Biol. Macromol. 2017, 105, 862–872. [Google Scholar] [CrossRef] [PubMed]
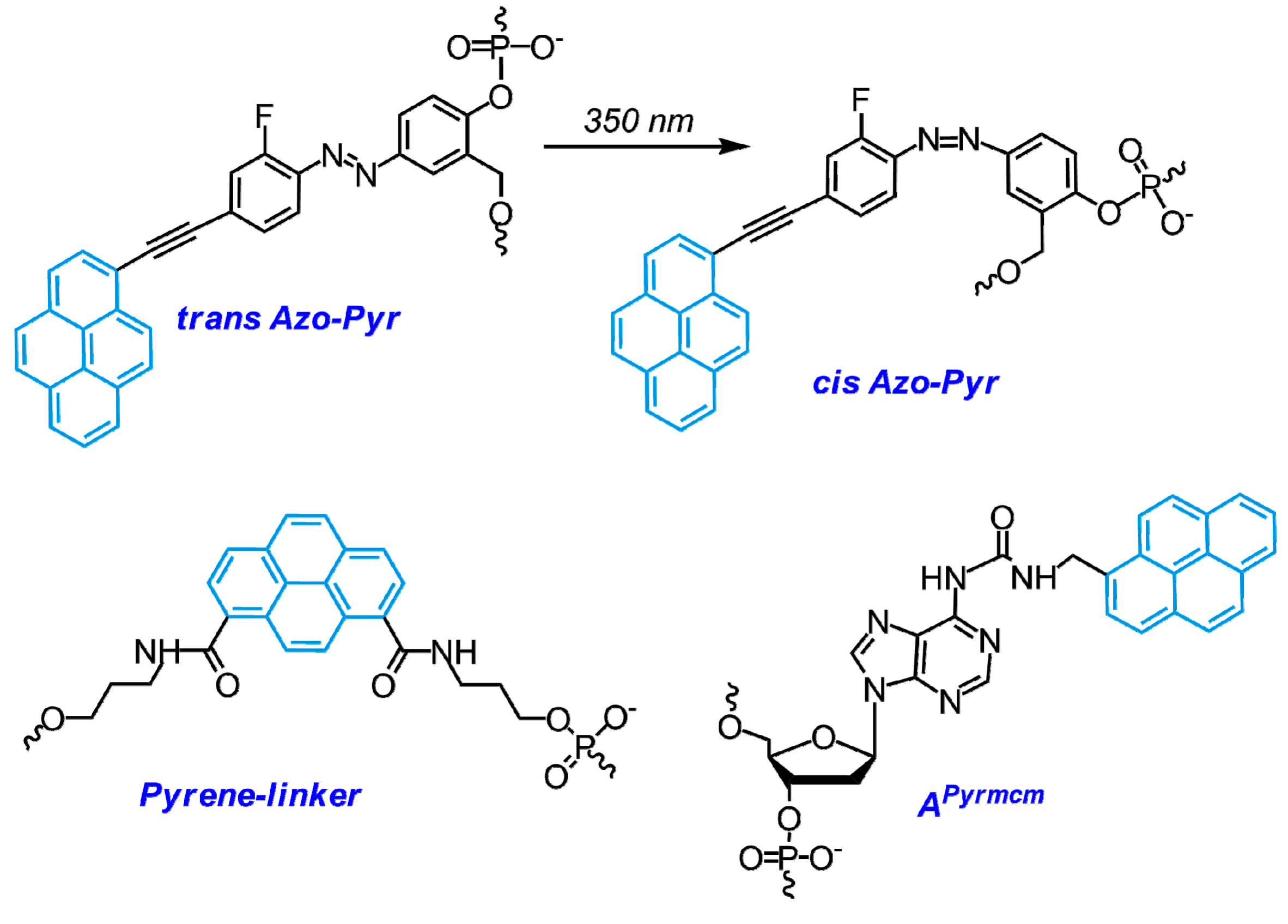
- Joo, H.N.; Van Thi Nguyen, T.; Chae, H.K.; Seo, Y.J. pH-Dependant fluorescence switching of an i-motif structure incorporating an isomeric azobenzene/pyrene fluorophore. Bioorg. Med. Chem. Lett. 2017, 27, 2415–2419. [Google Scholar] [CrossRef] [PubMed]
- Dembska, A. The analytical and biomedical potential of cytosine-rich oligonucleotides: A review. Anal. Chim. Acta 2016, 930, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Perlíková, P.; Karlsen, K.K.; Pedersen, E.B.; Wengel, J. Unlocked nucleic acids with a pyrene-modified uracil: Synthesis, hybridization studies, fluorescent properties and i-motif stability. ChemBioChem 2014, 15, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.J.; Yi, J.W.; Kim, B.H. Probe for i-motif structure and G-rich strands using end-stacking ability. Chem. Commun. 2009, 5383. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.J.; Kim, B.H. Monitoring i-motif transitions through the exciplex emission of a fluorescent probe incorporating two PyA units. Chem. Commun. 2012, 48, 2074–2076. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Seo, Y.J.; Kim, B.H. Fluorescence modification of the AAAA (4A) loop: Toward a probe of the structural dynamics of the i-motif of the retinoblastoma gene. Chem. Commun. 2014, 50, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Kim, I.S.; Park, J.W.; Seo, Y.J. Multiplex fluorophore systems on DNA with new diverse fluorescence properties and ability to sense the hybridization dynamics. Chem. Commun. 2014, 50, 7273–7276. [Google Scholar] [CrossRef] [PubMed]
- Dembska, A.; Rzepecka, P.; Juskowiak, B. Spectroscopic characterization of i-motif forming c-myc derived sequences double-labeled with pyrene. J. Fluoresc. 2013, 23, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tanaka, A.; Cho, D.W.; Fujitsuka, M.; Majima, T. Efficient electron transfer in i-motif DNA with a tetraplex structure. Angew. Chem. Int. Ed. 2013, 52, 12937–12941. [Google Scholar] [CrossRef] [PubMed]
- Dembska, A.; Rzepecka, P.; Juskowiak, B. Steady-state fluorescence and lifetime emission study of pH-sensitive probes based on i-motif forming oligonucleotides single and double labeled with pyrene. Chemosensors 2015, 3, 211–223. [Google Scholar] [CrossRef]
- Seo, Y.J.; Kim, B.H. Probing the B-to-Z-DNA duplex transition using terminally stacking ethynyl pyrene-modified adenosine and uridine bases. Chem. Commun. 2006, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.N.; Gao, J.; Kool, E.T. Oligodeoxyfluorosides: Strong sequence dependence of fluorescence emission. Tetrahedron 2007, 63, 3427–3433. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.N.; Kool, E.T. Fluorescent DNA base replacements: Reporters and sensors for biological systems. Org. Biomol. Chem. 2006, 4, 4265–4274. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Jiang, W.; Kool, E.T. Pattern-based detection of anion pollutants in water with DNA polyfluorophores. Chem. Sci. 2015, 6, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.; Chan, K.M.; Kool, E.T. DNA as an environmental sensor: Detection and identification of pesticide contaminants in water with fluorescent nucleobases. Org. Biomol. Chem. 2017, 15, 1801–1809. [Google Scholar] [CrossRef] [PubMed]
- Teo, Y.N.; Wilson, J.N.; Kool, E.T. Polyfluorophores on a DNA backbone: A multicolor set of labels excited at one wavelength. J. Am. Chem. Soc. 2009, 131, 3923–3933. [Google Scholar] [CrossRef] [PubMed]
- Kölmel, D.K.; Barandun, L.J.; Kool, E.T. Efficient synthesis of fluorescent alkynyl C-nucleosides via Sonogashira coupling for the preparation of DNA-based polyfluorophores. Org. Biomol. Chem. 2016, 14, 6407–6412. [Google Scholar] [CrossRef] [PubMed]
- Ensslen, P.; Gärtner, S.; Glaser, K.; Colsmann, A.; Wagenknecht, H.-A. A DNA-fullerene conjugate as a template for supramolecular chromophore assemblies: Towards DNA-based solar cells. Angew. Chem. Int. Ed. 2016, 55, 1904–1908. [Google Scholar] [CrossRef] [PubMed]
- Ensslen, P.; Brandl, F.; Sezi, S.; Varghese, R.; Kutta, R.-J.; Dick, B.; Wagenknecht, H.-A. DNA-based oligochromophores as light-harvesting systems. Chem. Eur. J. 2015, 21, 9349–9354. [Google Scholar] [CrossRef] [PubMed]
- Simon, P.; Cannata, F.; Concordet, J.-P.; Giovannangeli, C. Targeting DNA with triplex-forming oligonucleotides to modify gene sequence. Biochimie 2008, 90, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Tomizaki, A.; Matsueda, N.; Okamura, H.; Sasaki, S. Enhancement of TFO triplex formation by conjugation with pyrene via click chemistry. Chem. Pharm. Bull. (Tokyo) 2015, 63, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Pabon-Martinez, Y.V.; Xu, Y.; Villa, A.; Lundin, K.E.; Geny, S.; Nguyen, C.-H.; Pedersen, E.B.; Jørgensen, P.T.; Wengel, J.; Nilsson, L.; et al. LNA effects on DNA binding and conformation: From single strand to duplex and triplex structures. Sci. Rep. 2017, 7, 11043. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, H.; Dong, B.; Nie, K.; Liu, Z.; He, N. Recognition mechanisms and applications of peptide nucleic acids targeting double-stranded DNA. Curr. Med. Chem. 2016, 23, 4681–4705. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, T.; Yoshida, J.; Yamamoto, Y.; Sumaoka, J.; Tedeschi, T.; Corradini, R.; Sforza, S.; Komiyama, M. Chiral introduction of positive charges to PNA for double-duplex invasion to versatile sequences. Nucleic Acids Res. 2008, 36, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Boutorine, A.S.; Novopashina, D.S.; Krasheninina, O.A.; Nozeret, K.; Venyaminova, A.G. Fluorescent probes for nucleic acid visualization in fixed and live cells. Molecules 2013, 18, 15357–15397. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.-W.; Bao, G.; Leong, K.W. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef] [PubMed]
- Bomholt, N.; Osman, A.M.A.; Pedersen, E.B. High physiological thermal triplex stability optimization of twisted intercalating nucleic acids (TINA). Org. Biomol. Chem. 2008, 6, 3714–3722. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, M.; Cogoi, S.; Filichev, V.V.; Bomholt, N.; Pedersen, E.B.; Xodo, L.E. Purine twisted-intercalating nucleic acids: A new class of anti-gene molecules resistant to potassium-induced aggregation. Nucleic Acids Res. 2008, 36, 3494–3507. [Google Scholar] [CrossRef] [PubMed]
- Filichev, V.V.; Gaber, H.; Olsen, T.R.; Jørgensen, P.T.; Jessen, C.H.; Pedersen, E.B. Twisted intercalating nucleic acids—Intercalator influence on parallel triplex stabilities. Eur. J. Org. Chem. 2006, 2006, 3960–3968. [Google Scholar] [CrossRef]
- Filichev, V.V.; Nielsen, M.C.; Bomholt, N.; Jessen, C.H.; Pedersen, E.B. High thermal stability of 5′-5′-linked alternate hoogsteen triplexes at physiological pH. Angew. Chem. Int. Ed. 2006, 45, 5311–5315. [Google Scholar] [CrossRef] [PubMed]
- Schneider, U.V.; Mikkelsen, N.D.; Jøhnk, N.; Okkels, L.M.; Westh, H.; Lisby, G. Optimal design of parallel triplex forming oligonucleotides containing twisted intercalating nucleic acids—TINA. Nucleic Acids Res. 2010, 38, 4394–4403. [Google Scholar] [CrossRef] [PubMed]
- Doluca, O.; Boutorine, A.S.; Filichev, V.V. Triplex-forming twisted intercalating nucleic acids (TINAs): Design rules, stabilization of antiparallel DNA triplexes and inhibition of G-quartet-dependent self-association. ChemBioChem 2011, 12, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Vasilyeva, S.V.; Filichev, V.V.; Boutorine, A.S. Application of Cu(I)-catalyzed azide–alkyne cycloaddition for the design and synthesis of sequence specific probes targeting double-stranded DNA. Beilstein J. Org. Chem. 2016, 12, 1348–1360. [Google Scholar] [CrossRef] [PubMed]
- Filichev, V.V.; Astakhova, I.V.; Malakhov, A.D.; Korshun, V.A.; Pedersen, E.B. 1-, 2-, and 4-Ethynylpyrenes in the structure of twisted intercalating nucleic acids: Structure, thermal stability, and fluorescence relationship. Chem. Eur. J. 2008, 14, 9968–9980. [Google Scholar] [CrossRef] [PubMed]
- Géci, I.; Filichev, V.V.; Pedersen, E.B. Stabilization of parallel triplexes by twisted intercalating nucleic acids (TINAs) incorporating 1,2,3-triazole units and prepared by microwave-accelerated click chemistry. Chem. Eur. J. 2007, 13, 6379–6386. [Google Scholar] [CrossRef] [PubMed]
- Fatthalla, M.I.; Pedersen, E.B. Unexpected hydration of a triple bond during DNA synthesis: Conjugating 3-(pyren-1-ylethynyl)indole to DNA for triplex studies. Eur. J. Org. Chem. 2016, 2016, 3528–3535. [Google Scholar] [CrossRef]
- Van Daele, I.; Bomholt, N.; Filichev, V.V.; Van Calenbergh, S.; Pedersen, E.B. Triplex formation by pyrene-labelled probes for nucleic acid detection fluorescence assays. ChemBioChem 2008, 9, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Doluca, O.; Hale, T.K.; Edwards, P.J.B.; González, C.; Filichev, V.V. Assembly dependent fluorescence enhancing nucleic acids in sequence-specific detection of double-stranded DNA. ChemPlusChem 2014, 79, 58–66. [Google Scholar] [CrossRef]
- Schneider, U.V.; Géci, I.; Jøhnk, N.; Mikkelsen, N.D.; Pedersen, E.B.; Lisby, G. Increasing the analytical sensitivity by oligonucleotides modified with para- and ortho- Twisted Intercalating Nucleic Acids—TINA. PLoS ONE 2011, 6, e20565. [Google Scholar] [CrossRef] [PubMed]
- Schneider, U.V.; Mikkelsen, N.D.; Lindqvist, A.; Okkels, L.M.; Jøhnk, N.; Lisby, G. Improved efficiency and robustness in qPCR and multiplex end-point PCR by twisted intercalating nucleic acid modified primers. PLoS ONE 2012, 7, e38451. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Paramasivam, M.; Filichev, V.V.; Géci, I.; Pedersen, E.B.; Xodo, L.E. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009, 52, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, E.B.; Nielsen, J.T.; Nielsen, C.; Filichev, V.V. Enhanced anti-HIV-1 activity of G-quadruplexes comprising locked nucleic acids and intercalating nucleic acids. Nucleic Acids Res. 2011, 39, 2470–2481. [Google Scholar] [CrossRef] [PubMed]
- Membrino, A.; Cogoi, S.; Pedersen, E.B.; Xodo, L.E. G4-DNA Formation in the HRAS Promoter and Rational Design of Decoy Oligonucleotides for Cancer Therapy. PLoS ONE 2011, 6, e24421. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Zorzet, S.; Rapozzi, V.; Gé Ci, I.; Pedersen, E.B.; Xodo, L.E. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013, 41, 4049–4064. [Google Scholar] [CrossRef] [PubMed]
- Doluca, O.; Withers, J.M.; Loo, T.S.; Edwards, P.J.B.; González, C.; Filichev, V.V. Interdependence of pyrene interactions and tetramolecular G4-DNA assembly. Org. Biomol. Chem. 2015, 13, 3742–3748. [Google Scholar] [CrossRef] [PubMed]
- Mutsamwira, S.; Ainscough, E.W.; Partridge, A.C.; Derrick, P.J.; Filichev, V.V. G-Quadruplex supramolecular assemblies in photochemical upconversion. Chem. Eur. J. 2016, 22, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.A.; Pedersen, E.B.; Khaireldin, N.A. Studying the influence of the pyrene intercalator TINA on the stability of DNA i-motifs. Nucleosides Nucleotides Nucleic Acids 2012, 31, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Rohrbach, F.; Fatthalla, M.I.; Kupper, T.; Pötzsch, B.; Müller, J.; Petersen, M.; Pedersen, E.B.; Mayer, G. Chemical maturation of a bivalent aptamer by single domain variation. ChemBioChem 2012, 13, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Geny, S.; Moreno, P.M.D.; Krzywkowski, T.; Gissberg, O.; Andersen, N.K.; Isse, A.J.; El-Madani, A.M.; Lou, C.; Pabon, Y.V.; Anderson, B.A.; et al. Next-generation bis-locked nucleic acids with stacking linker and 2′-glycylamino-LNA show enhanced DNA invasion into supercoiled duplexes. Nucleic Acids Res. 2016, 44, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Le, B.T.; Filichev, V.V.; Veedu, R.N. Investigation of twisted intercalating nucleic acid (TINA)-modified antisense oligonucleotides for splice modulation by induced exon-skipping in vitro. RSC Adv. 2016, 6, 95169–95172. [Google Scholar] [CrossRef]
- Güixens-Gallardo, P.; Hocek, M.; Perlíková, P. Inhibition of non-templated nucleotide addition by DNA polymerases in primer extension using twisted intercalating nucleic acid modified templates. Bioorg. Med. Chem. Lett. 2016, 26, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Mutsamwira, S.; Ainscough, E.W.; Partridge, A.C.; Derrick, P.J.; Filichev, V.V. DNA duplex as a scaffold for a ground state complex formation between a zinc cationic porphyrin and phenylethynylpyren-1-yl. J. Photochem. Photobiol. A Chem. 2014, 288, 76–81. [Google Scholar] [CrossRef]
- Mutsamwira, S.; Ainscough, E.W.; Partridge, A.C.; Derrick, P.J.; Filichev, V.V. DNA-based assemblies for photochemical upconversion. J. Phys. Chem. B 2015, 119, 14045–14052. [Google Scholar] [CrossRef] [PubMed]
- Filichev, V.V.; Vester, B.; Hansen, L.H.; Pedersen, E.B. Easily denaturing nucleic acids derived from intercalating nucleic acids: Thermal stability studies, dual duplex invasion and inhibition of transcription start. Nucleic Acids Res. 2005, 33, 7129–7137. [Google Scholar] [CrossRef] [PubMed]
- Sau, S.P.; Kumar, T.S.; Hrdlicka, P.J. Invader LNA: Efficient targeting of short double stranded DNA. Org. Biomol. Chem. 2010, 8, 2028–2036. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.A.; Onley, J.J.; Hrdlicka, P.J. Recognition of double-stranded DNA using energetically activated duplexes modified with N2′-pyrene-, perylene-, or coronene-functionalized 2′-N-methyl-2′-amino-DNA monomers. J. Org. Chem. 2015, 80, 5395–5406. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.A.; Karmakar, S.; Hrdlicka, P.J. Mixed-sequence recognition of double-stranded DNA using enzymatically stable phosphorothioate invader probes. Molecules 2015, 20, 13780–13793. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.A.; Hrdlicka, P.J. Merging two strategies for mixed-sequence recognition of double-stranded DNA: Pseudocomplementary Invader probes. J. Org. Chem. 2016, 81, 3335–3346. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Madsen, A.S.; Wengel, J.; Hrdlicka, P.J. Synthesis and hybridization studies of 2′-amino-alpha-L-LNA and tetracyclic “locked LNA. ” J. Org. Chem. 2006, 71, 4188–4201. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Madsen, A.S.; Ostergaard, M.E.; Sau, S.P.; Wengel, J.; Hrdlicka, P.J. Functionalized 2′-amino-alpha-L-LNA: Directed positioning of intercalators for DNA targeting. J. Org. Chem. 2009, 74, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Andersen, N.K.; Anderson, B.A.; Wengel, J.; Hrdlicka, P.J. Synthesis and characterization of oligodeoxyribonucleotides modified with 2′-amino-α-l-LNA adenine monomers: High-affinity targeting of single-stranded DNA. J. Org. Chem. 2013, 78, 12690–12702. [Google Scholar] [CrossRef] [PubMed]
- Malinovskii, V.L.; Samain, F.; Haner, R. Helical arrangement of interstrand stacked pyrenes in a DNA framework. Angew. Chem. Int. Ed. 2007, 46, 4464–4467. [Google Scholar] [CrossRef] [PubMed]
- Bittermann, H.; Siegemund, D.; Malinovskii, V.L.; Häner, R. Dialkynylpyrenes: Strongly fluorescent, environment-sensitive DNA building blocks. J. Am. Chem. Soc. 2008, 130, 15285–15287. [Google Scholar] [CrossRef] [PubMed]
- Garo, F.; Häner, R. A DNA-based light-harvesting antenna. Angew. Chem. Int. Ed. 2012, 51, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Trkulja, I.; Häner, R. Triple-helix mediated excimer and exciplex formation. Bioconjug. Chem. 2007, 18, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Trkulja, I.; Haner, R. Monomeric and heterodimer triple helical DNA mimic. J. Am. Chem. Soc. 2007, 129, 7982–7989. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Vybornyi, M.; Häner, R. From ribbons to networks: Hierarchical organization of DNA-grafted supramolecular polymers. J. Am. Chem. Soc. 2015, 137, 14051–14054. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Vybornyi, M.; Haner, R. Pathway diversity in the self-assembly of DNA-derived bioconjugates. Bioconjug. Chem. 2016, 27, 2755–2761. [Google Scholar] [CrossRef] [PubMed]
- Winiger, C.B.; Langenegger, S.M.; Khorev, O.; Häner, R. Influence of perylenediimide–pyrene supramolecular interactions on the stability of DNA-based hybrids: Importance of electrostatic complementarity. Beilstein J. Org. Chem. 2014, 10, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Winiger, C.B.; Langenegger, S.M.; Calzaferri, G.; Häner, R. Formation of two homo-chromophoric H-aggregates in DNA-assembled alternating dye stacks. Angew. Chem. Int. Ed. 2015, 54, 3643–3647. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Vybornyi, M.; Häner, R. Functional DNA-grafted supramolecular polymers—Chirality, cargo binding and hierarchical organization. Chem. Commun. 2017, 53, 5179–5181. [Google Scholar] [CrossRef] [PubMed]
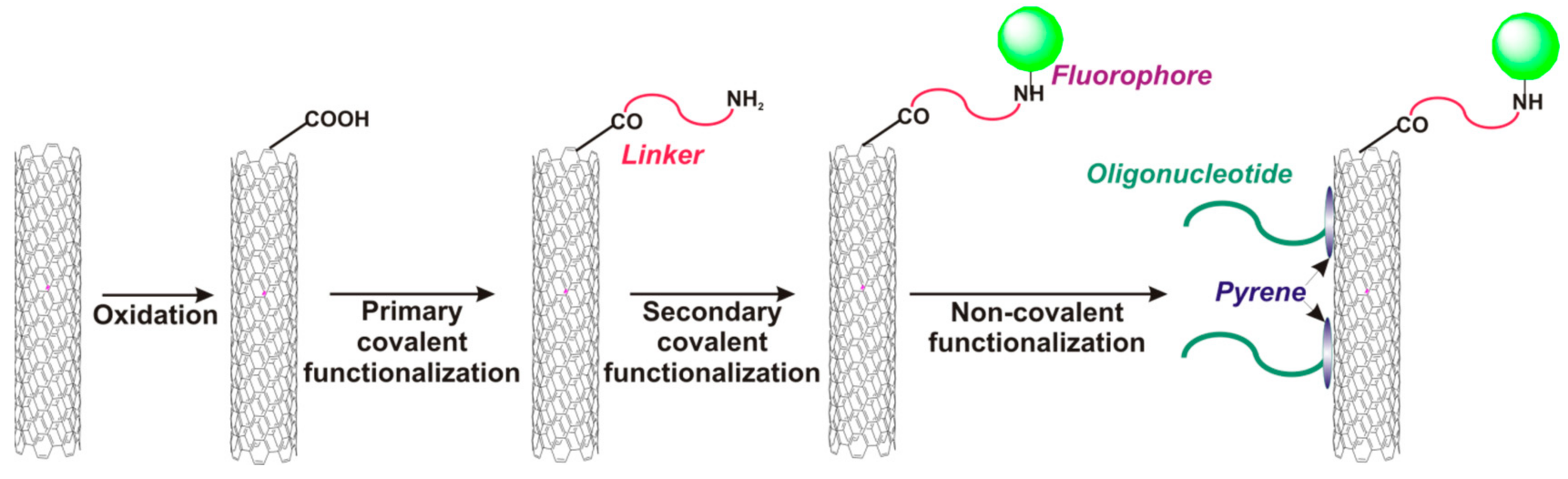
- Taft, B.J.; Lazareck, A.D.; Withey, G.D.; Yin, A.; Xu, J.M.; Kelley, S.O. Site-specific assembly of DNA and appended cargo on arrayed carbon nanotubes. J. Am. Chem. Soc. 2004, 126, 12750–12751. [Google Scholar] [CrossRef] [PubMed]
- Loukanov, A.; Filipov, C.; Lecheva, M.; Emin, S. Immobilization and stretching of 5′-pyrene-terminated DNA on carbon film deposited on electron microscope grid. Microsc. Res. Tech. 2015, 78, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- So, H.-M.; Park, D.-W.; Chang, H.; Lee, J.-O. Carbon nanotube biosensors with aptamers as molecular recognition elements. In Carbon Nanotubes Methods in Molecular Biology (Methods and Protocols); Balasubramanian, K., Burghard, M., Eds.; Humana Press: Mumbai, India, 2010. [Google Scholar]
- Joiner, C.S.; Gruner, G.; Star, A. Nanotube Sensor for DNA Detection. U.S. Patent 2007/0178477 A1, 2 August 2007. [Google Scholar]
- Baek, Y.-K.; Jung, D.-H.; Yoo, S.M.; Shin, S.; Kim, J.-H.; Jeon, H.-J.; Choi, Y.-K.; Lee, S.Y.; Jung, H.-T. Label-free detection of DNA hybridization using pyrene-functionalized single-walled carbon nanotubes: Effect of chemical structures of pyrene molecules on DNA sensing performance. J. Nanosci. Nanotechnol. 2011, 11, 4210–4216. [Google Scholar] [CrossRef] [PubMed]
- Fedorovskaya, E.O.; Apartsin, E.K.; Novopashina, D.S.; Venyaminova, A.G.; Kurenya, A.G.; Bulusheva, L.G.; Okotrub, A.V. RNA-modified carbon nanotube arrays recognizing RNA via electrochemical capacitance response. Mater. Des. 2016, 100, 67–72. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, F.; Cai, X.; Zheng, M.; Gao, F.; Jiang, S.; Wang, Q. Application of graphene–pyrenebutyric acid nanocomposite as probe oligonucleotide immobilization platform in a DNA biosensor. Mater. Sci. Eng. C 2013, 33, 3851–3857. [Google Scholar] [CrossRef] [PubMed]
- Maehashi, K.; Katsura, T.; Kerman, K.; Takamura, Y.; Matsumoto, K.; Tamiya, E. Label-free protein biosensor based on aptamer-modified carbon nanotube field-effect transistors. Anal. Chem. 2007, 79, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Maehashi, K.; Matsumoto, K. Label-free electrical detection using carbon nanotube-based biosensors. Sensors 2009, 9, 5368–5378. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.P.; Hong, S.; Sim, S.J. Apta-biosensors for nonlabeled real time detection of human IgE based on carbon nanotube field effect transistors. J. Nanosci. Nanotechnol. 2011, 11, 4182–4187. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, F.; Loeian, S.; Panchapakesan, B. Ultrasensitive label-free sensing of IL-6 based on PASE functionalized carbon nanotube micro-arrays with RNA-aptamers as molecular recognition elements. Biosensors 2017, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Apartsin, E.K.; Buyanova, M.Y.; Novopashina, D.S.; Venyaminova, A.G. Hybrids of siRNA with carbon nanotubes as RNA interference instruments. In Nanobiophysics: Fundamentals and Applications; Karachevtsev, V., Ed.; Pan Stanford Publishing: Singapore, 2015; pp. 33–57. ISBN 9789814613965. [Google Scholar] [CrossRef]
- Apartsin, E.K.; Buyanova, M.Y.; Novopashina, D.S.; Ryabchikova, E.I.; Venyaminova, A.G. Non-covalent immobilization of oligonucleotides on single-walled carbon nanotubes. In Nanomaterials Imaging Techniques, Surface Studies, and Applications; Springer Proceedings in Physics volume, 146, Fesenko, O., Yatsenko, L., Brodin, M., Eds.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Apartsin, E.K.; Buyanova, M.Y.; Novopashina, D.S.; Ryabchikova, E.I.; Filatov, A.V.; Zenkova, M.A.; Venyaminova, A.G. Novel multifunctional hybrids of single-walled carbon nanotubes with nucleic acids: synthesis and interactions with living cells. ACS Appl. Mater. Interfaces 2014, 6, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Permyakova, E.S.; Novopashina, D.S.; Venyaminova, A.G.; Apartsin, E.K. Non-covalent anchoring of oligonucleotides on single-walled carbon nanotubes via short bioreducible linker. RSC Adv. 2017, 7, 29212–29217. [Google Scholar] [CrossRef]
- Apartsin, E.K.; Novopashina, D.S.; Nastaushev, Y.V.; Ven’yaminova, A.G. Fluorescently labeled single-walled carbon nanotubes and their hybrids with oligonucleotides. Nanotechnol. Russ. 2012, 7, 99–109. [Google Scholar] [CrossRef]
- Novopashina, D.S.; Apartsin, E.K.; Venyaminova, A.G. Fluorecently labeled bionanotransporters of nucleic acid based on carbon nanotubes. Ukr. J. Phys. 2012, 57, 718–722. [Google Scholar]
- You, M.; Zhu, Z.; Liu, H.; Gulbakan, B.; Han, D.; Wang, R.; Williams, K.R.; Tan, W. Pyrene-assisted efficient photolysis of disulfide bonds in DNA-based molecular engineering. ACS Appl. Mater. Interfaces 2010, 2, 3601–3605. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.-C.; You, M.; Tan, W.; Ye, B.-C. Mercury(II) ion detection via pyrene-mediated photolysis of disulfide bonds. Chem. Eur. J. 2012, 18, 1286–1289. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Chen, Y.; Zhang, X.; Liu, H.; Wang, R.; Wang, K.; Williams, K.R.; Tan, W. An autonomous and controllable light-driven DNA walking device. Angew. Chem. Int. Ed. 2012, 51, 2457–2460. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Goetzfried, M.A.; Hidaka, K.; You, M.; Tan, W.; Sugiyama, H.; Endo, M. Direct visualization of walking motions of photocontrolled nanomachine on the DNA nanostructure. Nano Lett. 2015, 15, 6672–6676. [Google Scholar] [CrossRef] [PubMed]
- Mastroyiannopoulos, N.P.; Uney, J.B.; Phylactou, L.A. The application of ribozymes and DNAzymes in muscle and brain. Molecules 2010, 15, 5460–5472. [Google Scholar] [CrossRef] [PubMed]
- Fokina, A.A.; Stetsenko, D.A.; François, J.-C. DNA enzymes as potential therapeutics: Towards clinical application of 10–23 DNAzymes. Expert Opin. Biol. Ther. 2015, 15, 689–711. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. Catalytic DNA: Scope, applications, and biochemistry of deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. DNA catalysis: The chemical repertoire of DNAzymes. Molecules 2015, 20, 20777–20804. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, H.; Hayashi, H.; Zhao, J.; Liang, X.; Yamazawa, A.; Kuramochi, T.; Matsunaga, D.; Aiba, Y.; Kashida, H.; Komiyama, M. Enhancement of RNA cleavage activity of 10–23 DNAzyme by covalently introduced intercalator. Chem. Commun. 2006, 5, 5062–5064. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Liang, X.; Zhao, J.; Komiyama, M.; Asanuma, H. Activation of DNA enzyme 10–23 by tethering an intercalator to its backbone. Nucleic Acids Symp. Ser. 2006, 50, 167–168. [Google Scholar] [CrossRef] [PubMed]
- Looser, V.; Langenegger, S.M.; Häner, R.; Hartig, J.S. Pyrene modification leads to increased catalytic activity in minimal hammerhead ribozymes. Chem. Commun. 2007, 4357–4359. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Rana, J.; Roy, J.; Huang, H. Cleavage of pyrene-stabilized RNA bulge loops by trans-(±)-cyclohexane-1,2-diamine. Chem. Cent. J. 2012, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Roday, S.; Sturm, M.B.; Blakaj, D.M.; Schramm, V.L. Detection of an abasic site in RNA with stem-loop DNA beacons: Application to an activity assay for Ricin Toxin A-Chain. J. Biochem. Biophys. Methods 2008, 70, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhang, R.; Wang, Z.; Tian, J.; Chen, X. Recent advances in high-performance fluorescent and bioluminescent RNA imaging probes. Chem. Soc. Rev. 2017, 46, 2824–2843. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Yue, S.; Zhang, S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef] [PubMed]







© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krasheninina, O.A.; Novopashina, D.S.; Apartsin, E.K.; Venyaminova, A.G. Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides. Molecules 2017, 22, 2108. https://doi.org/10.3390/molecules22122108
Krasheninina OA, Novopashina DS, Apartsin EK, Venyaminova AG. Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides. Molecules. 2017; 22(12):2108. https://doi.org/10.3390/molecules22122108
Chicago/Turabian StyleKrasheninina, Olga A., Darya S. Novopashina, Evgeny K. Apartsin, and Alya G. Venyaminova. 2017. "Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides" Molecules 22, no. 12: 2108. https://doi.org/10.3390/molecules22122108
APA StyleKrasheninina, O. A., Novopashina, D. S., Apartsin, E. K., & Venyaminova, A. G. (2017). Recent Advances in Nucleic Acid Targeting Probes and Supramolecular Constructs Based on Pyrene-Modified Oligonucleotides. Molecules, 22(12), 2108. https://doi.org/10.3390/molecules22122108