Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize α-Hairpinin Antimicrobial Peptide
Abstract
:1. Introduction
2. Results
2.1. Synthesis, Folding, Purification and Activity of MBP-1
2.2. Design and Activity of MBP-1 Variants
2.3. Scanning Electron Microscopy
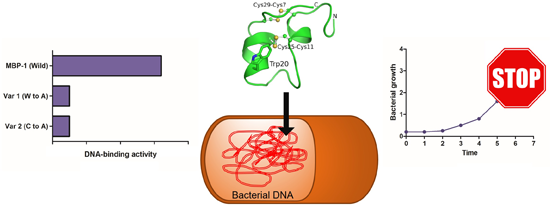
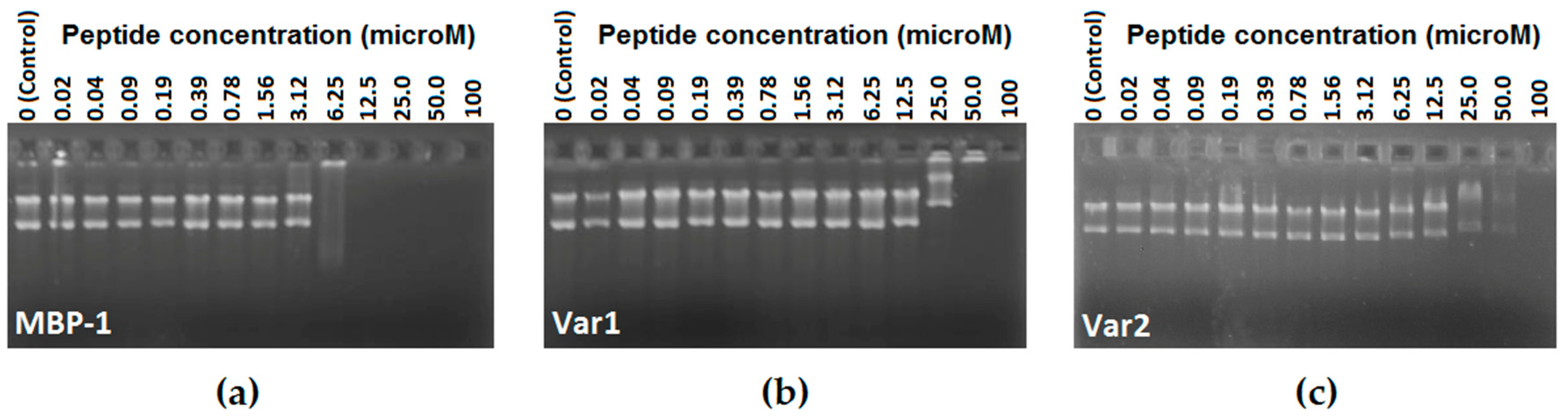
2.4. DNA Binding-Assay
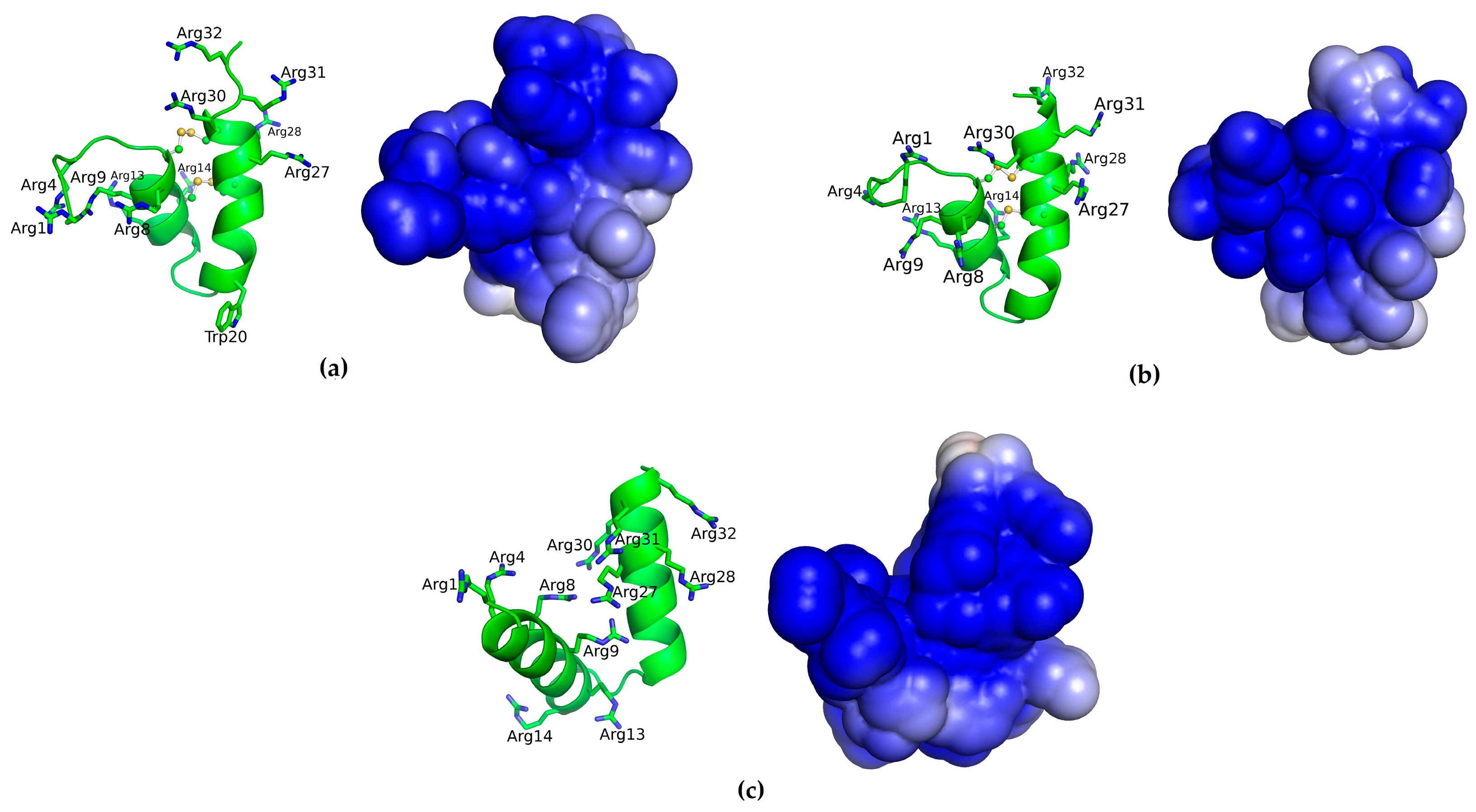
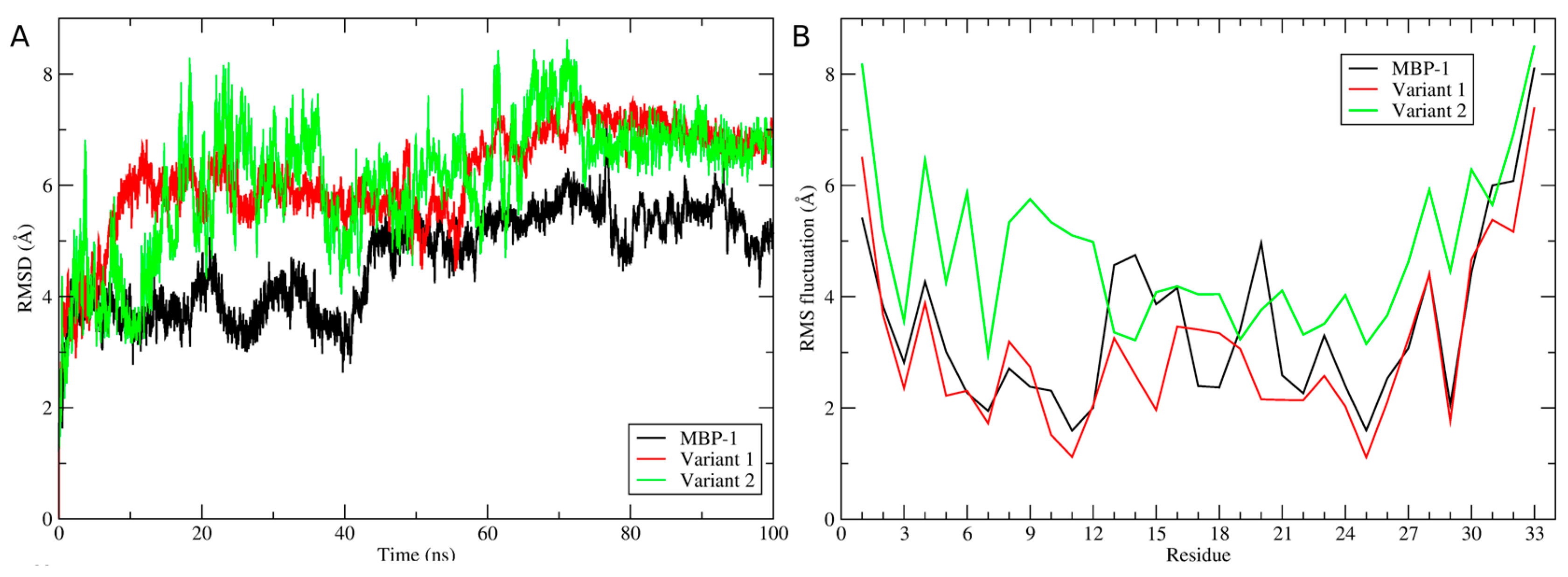
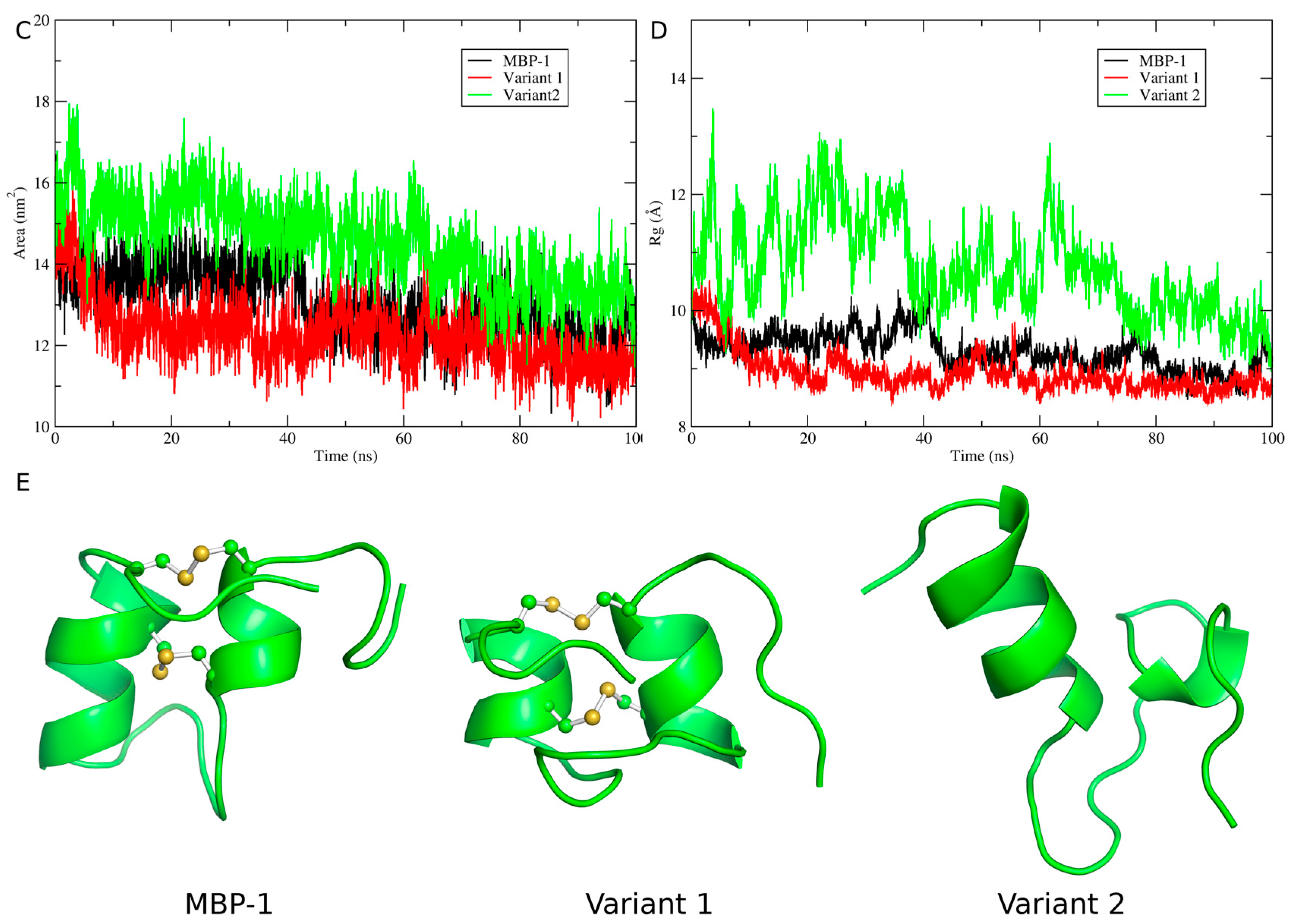
2.5. Structural Analysis
3. Discussion
4. Materials and Methods
4.1. Synthesis, Purification and Folding of MBP-1 and Variants
4.2. Antibacterial Bioassays
4.3. Scanning Electron Microscopy
4.4. DNA Binding-Assay
4.5. Comparative Molecular Modelling
4.6. Molecular Dynamics Simulation and Analyses of Molecular Dynamics Trajectories
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wright, G.D. Q & A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.V.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.Y.; Ng, T.B. Isolation of a new cyclophilin-like protein from chickpeas with mitogenic, antifungal and anti-HIV-1 reverse transcriptase activities. Life Sci. 2002, 70, 1129–1138. [Google Scholar] [CrossRef]
- Lin, P.; Ng, T.B. A novel and exploitable antifungal peptide from kale (Brassica alboglabra) seeds. Peptides 2008, 29, 1664–1671. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Li, Z.; Zhang, R.; Wu, Y.; Li, W.; Cao, Z. StCT2, a new antibacterial peptide characterized from the venom of the scorpion Scorpiops tibetanus. Peptides 2012, 36, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.X.; Liu, Q.H.; Ng, T.B.; Wang, H.X. Isarfelin, a peptide with antifungal and insecticidal activities from Isaria felina. Peptides 2005, 26, 2384–2391. [Google Scholar] [CrossRef] [PubMed]
- Molhoek, E.M.; van Dijk, A.; Veldhuizen, E.J.; Dijk-Knijnenburg, H.; Mars-Groenendijk, R.H.; Boele, L.C.; Kaman-van Zanten, W.E.; Haagsman, H.P.; Bikker, F.J. Chicken cathelicidin-2-derived peptides with enhanced immunomodulatory and antibacterial activities against biological warfare agents. Int. J. Antimicrob. Agents 2010, 36, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Kamysz, W.; Okroj, M.; Lukasiak, J. Novel properties of antimicrobial peptides. Acta Biochem. Pol. 2003, 50, 461–469. [Google Scholar]
- Hwang, P.M.; Vogel, H.J. Structure-function relationships of antimicrobial peptides. Biochem. Cell Biol. 1998, 76, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Nolde, S.B.; Vassilevski, A.A.; Rogozhin, E.A.; Barinov, N.A.; Balashova, T.A.; Samsonova, O.V.; Baranov, Y.V.; Feofanov, A.V.; Egorov, T.A.; Arseniev, A.S.; et al. Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli). J. Biol. Chem. 2011, 286, 25145–25153. [Google Scholar] [CrossRef] [PubMed]
- Oparin, P.B.; Mineev, K.S.; Dunaevsky, Y.E.; Arseniev, A.S.; Belozersky, M.A.; Grishin, E.V.; Egorov, T.A.; Vassilevski, A.A. Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defense peptides. Biochem. J. 2012, 446, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Berkut, A.A.; Usmanova, D.R.; Peigneur, S.; Oparin, P.B.; Mineev, K.S.; Odintsova, T.I.; Tytgat, J.; Arseniev, A.S.; Grishin, E.V.; Vassilevski, A.A. Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design. J. Biol. Chem. 2014, 289, 14331–14340. [Google Scholar] [CrossRef] [PubMed]
- Duvick, J.P.; Rood, T.; Rao, A.G.; Marshak, D.R. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J. Biol. Chem. 1992, 267, 18814–18820. [Google Scholar] [PubMed]
- Marcus, J.P.; Green, J.L.; Goulter, K.C.; Manners, J.M. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 1999, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Shimada, T.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. J. Biol. Chem. 1999, 274, 2563–2570. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, X.X.; Xia, H.C.; Zeng, R.; Hu, W.G.; Li, Z.; Zhang, Z.C. Purification and characterization of Luffin P1, a ribosome-inactivating peptide from the seeds of Luffa cylindrica. Peptides 2003, 24, 799–805. [Google Scholar] [CrossRef]
- Conners, R.; Konarev, A.V.; Forsyth, J.; Lovegrove, A.; Marsh, J.; Joseph-Horne, T.; Shewry, P.; Brady, R.L. An unusual helix-turn-helix protease inhibitory motif in a novel trypsin inhibitor from seeds of Veronica (Veronica hederifolia L.). J. Biol. Chem. 2007, 282, 27760–27768. [Google Scholar] [CrossRef] [PubMed]
- Yeung, H.; Squire, C.J.; Yosaatmadja, Y.; Panjikar, S.; Lopez, G.; Molina, A.; Baker, E.N.; Harris, P.W.; Brimble, M.A. Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily. Angew. Chem. Int. Ed. 2016, 55, 7930–7933. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Franco, O.L. Theoretical structural insights into the snakin/GASA family. Peptides 2013, 44, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Franco, O.L. Peptide promiscuity: An evolutionary concept for plant defense. FEBS Lett. 2011, 585, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wu, C.; Liu, W.; Zhang, J. Disulfide Bond Formation in Peptides by Dimethyl Sulfoxide. Scope and Applications. J. Am. Chem. Soc. 1991, 113, 6657–6662. [Google Scholar] [CrossRef]
- Hale, J.D.; Hancock, R.E. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Franco, O.L.; Alencar, S.A. Computational analyses and prediction of guanylin deleterious SNPs. Peptides 2015, 69, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Purohit, R. Use of long term molecular dynamics simulation in predicting cancer associated SNPs. PLoS Comput. Biol. 2014, 10, e1003318. [Google Scholar] [CrossRef] [PubMed]
- Chitrala, K.N.; Yeguvapalli, S. Computational screening and molecular dynamic simulation of breast cancer associated deleterious non-synonymous single nucleotide polymorphisms in TP53 gene. PLoS ONE 2014, 9, e104242. [Google Scholar] [CrossRef] [PubMed]
- Rogozhin, E.A.; Slezina, M.P.; Slavokhotova, A.A.; Istomina, E.A.; Korostyleva, T.V.; Smirnov, A.N.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. A novel antifungal peptide from leaves of the weed Stellaria media L. Biochimie 2015, 116, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Alfred, R.L.; Palombo, E.A.; Panozzo, J.F.; Bhave, M. The antimicrobial domains of wheat puroindolines are cell-penetrating peptides with possible intracellular mechanisms of action. PLoS ONE 2013, 8, e75488. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.H.; Park, K.H.; Park, Y.; Jeon, Y.J.; Kim, Y.; Park, I.S.; Hahm, K.S.; Shin, S.Y. Investigating the effects of positive charge and hydrophobicity on the cell selectivity, mechanism of action and anti-inflammatory activity of a Trp-rich antimicrobial peptide indolicidin. FEMS Microbiol. Lett. 2009, 292, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G.; Agerberth, B.; Boman, A. Mechanisms of action on Escherichia coli of cecropin P1 and PR-39, two antibacterial peptides from pig intestine. Infect. Immun. 1993, 61, 2978–2984. [Google Scholar] [PubMed]
- Haney, E.F.; Petersen, A.P.; Lau, C.K.; Jing, W.; Storey, D.G.; Vogel, H.J. Mechanism of action of puroindoline derived tryptophan-rich antimicrobial peptides. Biochim. Biophys. Acta 2013, 1828, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, A.Y.; Tarnovskaya, S.I.; Chernova, I.A.; Shataeva, L.K.; Skorik, Y.A. The interaction of amino acids, peptides, and proteins with DNA. Int. J. Biol. Macromol. 2015, 78, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Hocquellet, A.; le Senechal, C.; Garbay, B. Importance of the disulfide bridges in the antibacterial activity of human hepcidin. Peptides 2012, 36, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Campopiano, D.J.; Clarke, D.J.; Polfer, N.C.; Barran, P.E.; Langley, R.J.; Govan, J.R.; Maxwell, A.; Dorin, J.R. Structure-activity relationships in defensin dimers: A novel link between beta-defensin tertiary structure and antimicrobial activity. J. Biol. Chem. 2004, 279, 48671–48679. [Google Scholar] [CrossRef] [PubMed]
- Varkey, J.; Nagaraj, R. Antibacterial activity of human neutrophil defensin HNP-1 analogs without cysteines. Antimicrob. Agents Chemother. 2005, 49, 4561–4566. [Google Scholar] [CrossRef] [PubMed]
- Hoover, D.M.; Rajashankar, K.R.; Blumenthal, R.; Puri, A.; Oppenheim, J.J.; Chertov, O.; Lubkowski, J. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 2000, 275, 32911–32918. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Kerscher, B.; Dall’Angelo, S.; Sani, M.; Longhi, R.; Baloban, M.; Wilson, H.M.; Mergaert, P.; Zanda, M.; Ferguson, G.P. Role of cysteine residues and disulfide bonds in the activity of a legume root nodule-specific, cysteine-rich peptide. J. Biol. Chem. 2012, 287, 10791–10798. [Google Scholar] [CrossRef] [PubMed]
- Tamamura, H.; Ikoma, R.; Niwa, M.; Funakoshi, S.; Murakami, T.; Fujii, N. Antimicrobial activity and conformation of tachyplesin I and its analogs. Chem. Pharm. Bull. 1993, 41, 978–980. [Google Scholar] [CrossRef] [PubMed]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 2012, 80, 1715–1735. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., 3rd; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces; Pullman, B., Ed.; Springer: Houten, The Netherlands, 1981; Volume 14, pp. 331–342. [Google Scholar]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Kollman, P.A. SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds MBP-1, Var 1 e Var 2 are available from the authors.

| Peptide | Sequence | Mass before Oxidation | Mass after Oxidation | MIC against E. coli |
|---|---|---|---|---|
| MBP-1 | RSGRGECRRQCLRRHEGQPWETQECMRRCRRRG | 4125.69 m/z | 4121.04 m/z | 50 μM |
| Var 1 | RSGRGECRRQCLRRHEGQPAETQECMRRCRRRG | 4008.05 m/z | 4004.01 m/z | >400 μM |
| Var 2 | RSGRGEARRQALRRHEGQPWETQEAMRRARRRG | 4001.40 m/z | 4001.38 m/z | >400 μM |
| Peptide | Solvation Potential Energy (KJ/mol) |
|---|---|
| MBP-1 | 3399.486 |
| Var 1 | 3373.256 |
| Var 2 | 3487.306 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, D.A.; Porto, W.F.; Silva, M.Z.; Da Silva, T.R.; Franco, O.L. Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize α-Hairpinin Antimicrobial Peptide. Molecules 2016, 21, 1062. https://doi.org/10.3390/molecules21081062
Sousa DA, Porto WF, Silva MZ, Da Silva TR, Franco OL. Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize α-Hairpinin Antimicrobial Peptide. Molecules. 2016; 21(8):1062. https://doi.org/10.3390/molecules21081062
Chicago/Turabian StyleSousa, Daniel A., William F. Porto, Maria Z. Silva, Tatiane R. Da Silva, and Octávio L. Franco. 2016. "Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize α-Hairpinin Antimicrobial Peptide" Molecules 21, no. 8: 1062. https://doi.org/10.3390/molecules21081062
APA StyleSousa, D. A., Porto, W. F., Silva, M. Z., Da Silva, T. R., & Franco, O. L. (2016). Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize α-Hairpinin Antimicrobial Peptide. Molecules, 21(8), 1062. https://doi.org/10.3390/molecules21081062