Abstract
Cold stimulation of Bailinggu’s mycelia is the main factor that triggers primordia initiation for successful production of fruiting bodies under commercial cultivation. Yet, the molecular-level mechanisms involved in mycelia response to cold stimulation are still unclear. Here, we performed comparative transcriptomic analysis using RNA-Seq technology to better understand the gene expression regulation during different temporal stages of cold stimulation in Bailinggu. A total of 21,558 Bailinggu mycelia unigenes were de novo assembled and annotated from four libraries (control at 25 °C, plus cold stimulation treatments at −3 °C for a duration of 1–2 days, 5–6 days, and 9–10 days). GO and KEGG pathway analysis indicated that functional groups of differentially expressed unigenes associated with cell wall and membrane stabilization, calcium signaling and mitogen-activated protein kinases (MAPK) pathways, and soluble sugars and protein biosynthesis and metabolism pathways play a vital role in Bailinggu’s response to cold stimulation. Six hundred and seven potential EST-based SSRs loci were identified in these unigenes, and 100 EST-SSR primers were randomly selected for validation. The overall polymorphism rate was 92% by using 10 wild strains of Bailinggu. Therefore, these results can serve as a valuable resource for a better understanding of the molecular mechanisms associated with Bailinggu’s response to cold stimulation.
1. Introduction
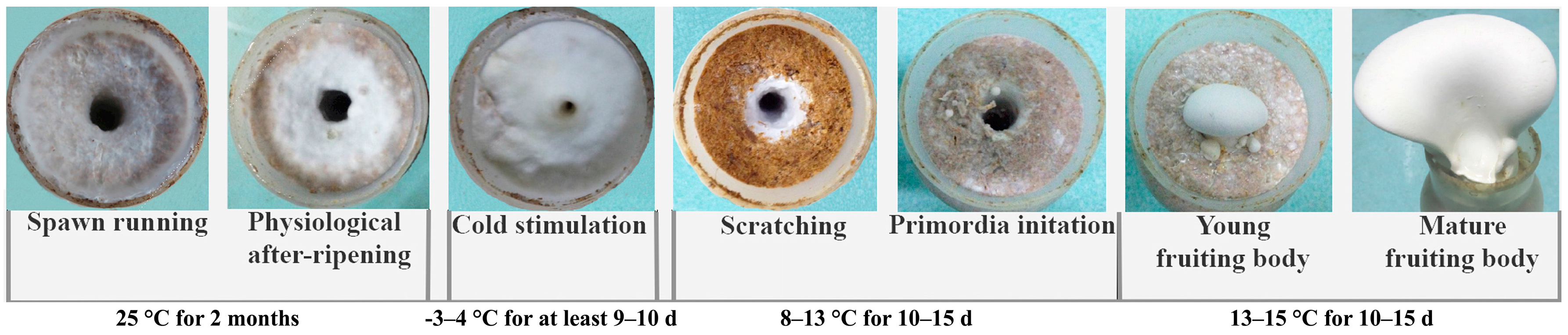
Pleurotus eryngii subsp. tuoliensis [1] is a precious edible mushroom that has been commercially cultivated since 1997 in China [2] and is rapidly growing in popularity throughout Chinese, Japanese, and Korean markets [3,4]. Bailinggu (Figure 1) is the Chinese commercial name for P. eryngii subsp. tuoliensis. Although yields have steadily increased, many factors still limit more substantial increases in Bailinggu production rates under commercial cultivation [5]. In addition to internal physiological factors that influence mycelia ripening, the cultivation cycle of Bailinggu requires special temperature, humidity, and light conditions to initiate primordia and fruiting bodies [6,7]. Among these environmental factors, cold stimulation of mature mycelia is the main trigger for primordia initiation and plays a crucial role in mushroom formation and subsequent yields of Bailinggu [4,6,7]. For example, without cold stimulation at −3 to 4 °C for 5–6 days (d) following physiological after-ripening of mature mycelia, primordia formation is limited or nonexistent. Moreover, cold temperature stimulation of mature mycelia for 9–10 d have the potential to achieve greater consistency in culture bottle budding of primordia and to ensure a high degree of fruiting body uniformity. However, commercial-scale, artificial cold stimulation is expensive and energy intensive. One potential strategy for increasing production while reducing costs and conserving energy is to shorten the duration of cultivation cycles and cold stimulation treatments. To evaluate the feasibility of this strategy, additional studies are needed to better understand the molecular and physiological mechanisms for Bailinggu growth in response to cold stimulation.

Figure 1.
Developmental stages of Bailinggu in year-round mushroom factory cultivation cycles. The vegetative (spawning, physiological ripening, cold stimulation) and fructification (primordia, fruiting body) phases are shown.
Cold stimulation of Bailinggu includes chilling (0–4 °C) and freezing (<0 °C) temperatures. Previous research has shown that cold temperature treatment of mature mycelia in Bailinggu following physiological after-ripening triggers a series of physiological and biochemical responses [6,7,8]. For example, cold temperatures activate antioxidative enzymes and other enzymes (e.g., protease, laccase, and amylase) that facilitate rapid conversion of accumulated proteins and polysaccharides to new proteins and soluble sugars [8]. Thus, cold stimulation initiates physiological mechanisms that can create the material conditions for primordia differentiation.
Plants and fungi respond to low temperatures by altering the expression of thousands of genes, thereby changing cellular, physiological, and biochemical processes [9,10,11,12]. Thus, analysis of gene expression would be a valuable tool to understand the transcriptome dynamics and the potential for manipulation of the gene expression pattern in mushrooms. Bailinggu’s genes and their functions associated with energy metabolism, material conversion, and cell growth during cold stimulation have not been studied. Therefore, the molecular-level mechanisms involved in cold temperature gene regulation and signal transduction are still unclear.
With advances in high-throughput sequencing technologies, RNA sequencing (RNA-seq) technology has been successfully used for gene expression profiling and other transcriptome studies in some edible mushrooms, including Agaricus bisporus [13], Lentinula edodes [14], Flammulina velutipes [15], and Laccaria bicolor [16]. In addition, RNA-seq has been applied to quantify RNA levels under chilling stress in edible mushrooms such as Pleurotus ostreatus [9], Tuber melanosporum [10] and under cold stress in other species such as Solanum habrochaites [17], Corylus heterophylla [18], Lilium lancifolium [19], and Vaccinium spp. [20]. Furthermore, the Solexa/Illumina (Illumina) platform has made it possible to perform de novo transcriptome sequencing for species without a sequenced genome such as Bailinggu [21,22,23,24,25]. However, no studies have performed a comparative transcriptomic analysis using next-generation sequencing technologies on Bailinggu during cold stimulation.
Transcriptome sequencing is an efficient, cost-effective way to develop transcript/EST-based simple sequence repeats (EST-SSR) markers, which are important resources for genetic diversity analysis, genetic map construction, and molecular marker-assisted selection in breeding [20,24,26]. Transcriptomic sequencing for SSR mining has been used in a wide range of fungal species and plants, including Auricularia polytricha [22], Agaricus subrufescens [27], Caragana korshinskii [25], and Myrica rubra [28]. However, SSR and EST-SSR molecular markers have yet to be developed for genetic diversity and mapping studies of Bailinggu.
In this study, we used RNA-seq technology to perform an analysis of Bailinggu mycelia transcriptome under the physiological after-ripening stage and several temporal stages of cold stimulation. Our specific objectives were the following: (1) identify relevant functional gene groups and signaling pathways associated with cold stimulation and (2) screen and identify the EST-SSR markers for ongoing genetic diversity and mapping studies of Bailinggu.
2. Results and Discussion
2.1. RNA-Seq and De Novo Assembly
Overall, 78.64 million high-quality, clean 100-bp reads were generated from the four libraries, divided into 18.83, 13.80, 27.14 and 18.86 million from the control (the samples were stored at 25 °C), 1–2 d (the samples were stored at −3 °C), 5–6 d (the samples were stored at −3 °C) and 9–10 d (the samples were stored at −3 °C), respectively. The overall GC content was over 51% in both the control and cold stage samples (Table 1).

Table 1.
Basic statistics of RNA-seq reads obtained from Illumina HiSeq-2000.
De novo assembly with the clean reads resulted in 21,558 unigenes with an average length of 1248 bp and an N50 of 2266 bp (Table 2). Our accuracy check showed that over 88% clean reads mapped to these unigenes, which were then used for further analysis (Table 3).

Table 2.
De novo assembly results using Trinity.

Table 3.
Clean reads mapping to unigenes.
Recent studies indicated that the overlapping RNA-seq method is a useful tool for identifying differential gene expression (DEG) in libraries that represent different points in time [29]. We performed the overlapping RNA-seq on four libraries (Control, 1–2 d, 5–6 d, and 9–10 d) to better elucidate year-round cultivation at different temporal stages of cold stimulation. Therefore, the four library datasets generated represent a comprehensive view of Bailinggu response to temperature changes and the cold stimulation process.
2.2. Transcriptome Annotation and Functional Classification
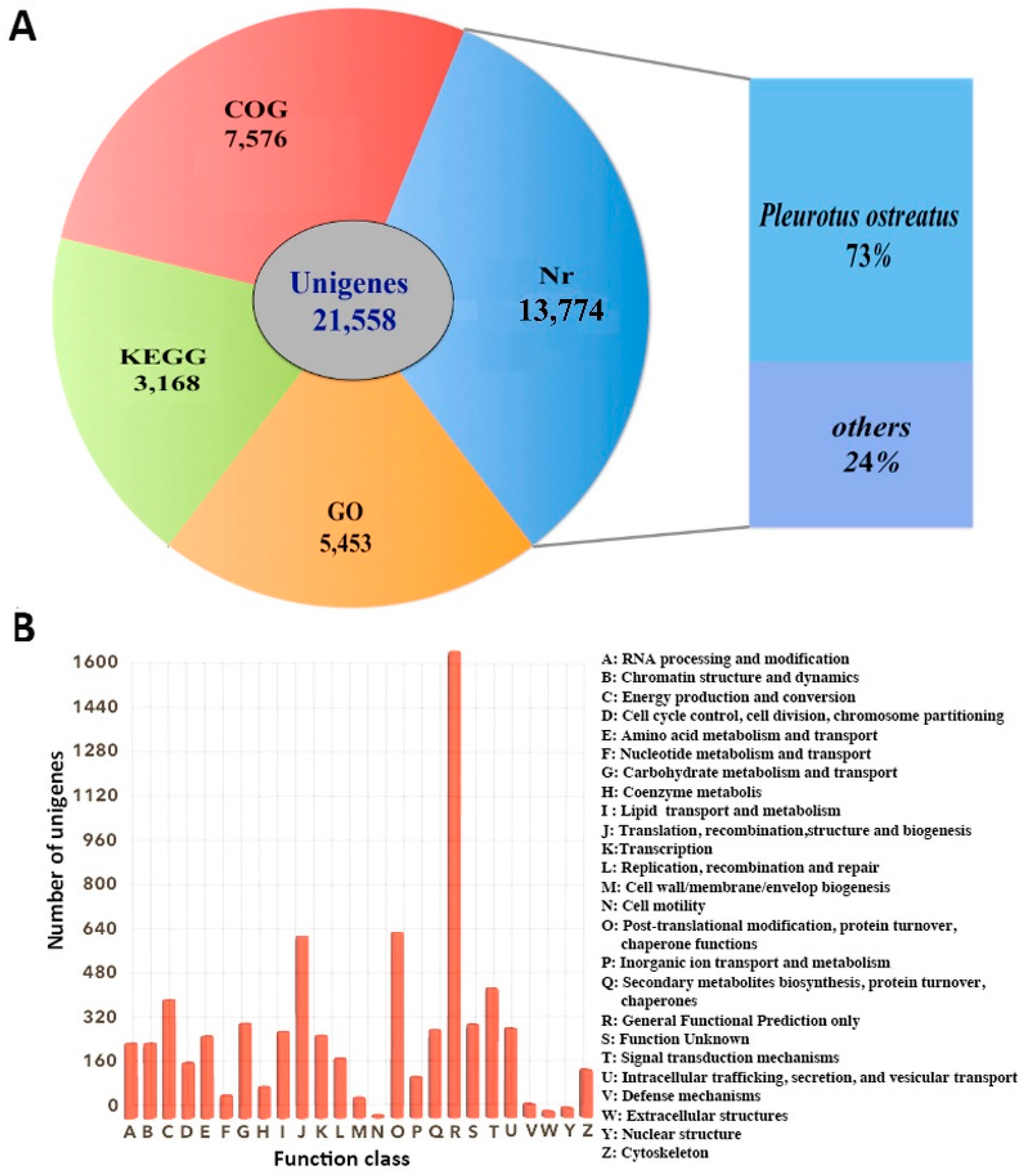
Annotation and functional classification results of 21,558 unigenes were obtained using the five datasets (Figure 2A). A total of 13,774 (63.89%) unigenes showed hitting with known proteins in the NR database, followed by Interproscan database (11,334, 52.57%), COG database (7576, 35.14%), GO database (5453, 25.29%), and KEGG database (3168, 14.70%). Distribution analysis based on BLASTx searches showed that 73% of the annotated unigenes have homologs in Pleurotus ostreatus, which is the species with a sequenced genome most closely related to P. eryngii subsp. tuoliensis (Bailinggu). COG results showed that a total of 7576 unigenes were assigned to 25 classifications (Figure 2B). The top five of COG categories were associated with (C) Energy production and conversion, (G) Carbohydrate metabolism and transport, (J) Translation, recombination, structure and biogenesis, (O) Post-translational modification, protein turnover, chaperone functions and (T) Signal transduction mechanisms.
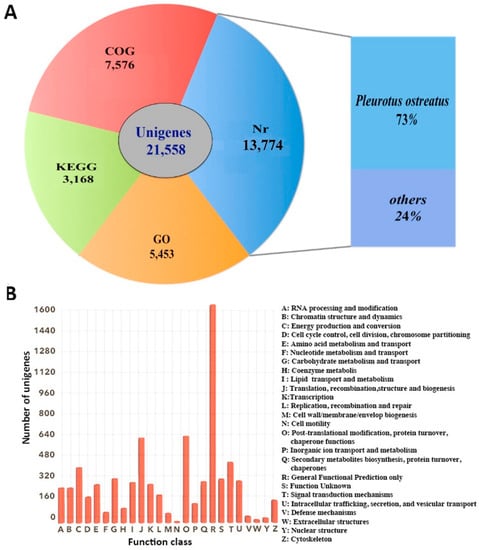
Figure 2.
Unigene annotation. (A) Number of unigenes blasted to Non-Redundant (NR), Clusters of Orthologous Groups (COG), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (e < 0.00001); (B) COG classification. A total of 7576 unigenes were assigned to 25 classifications. The capital letters on the x-axis indicate the COG categories as listed on the right of the histogram.
2.3. Identification of DEGs Involved in Different Stages of Cold Stimulation
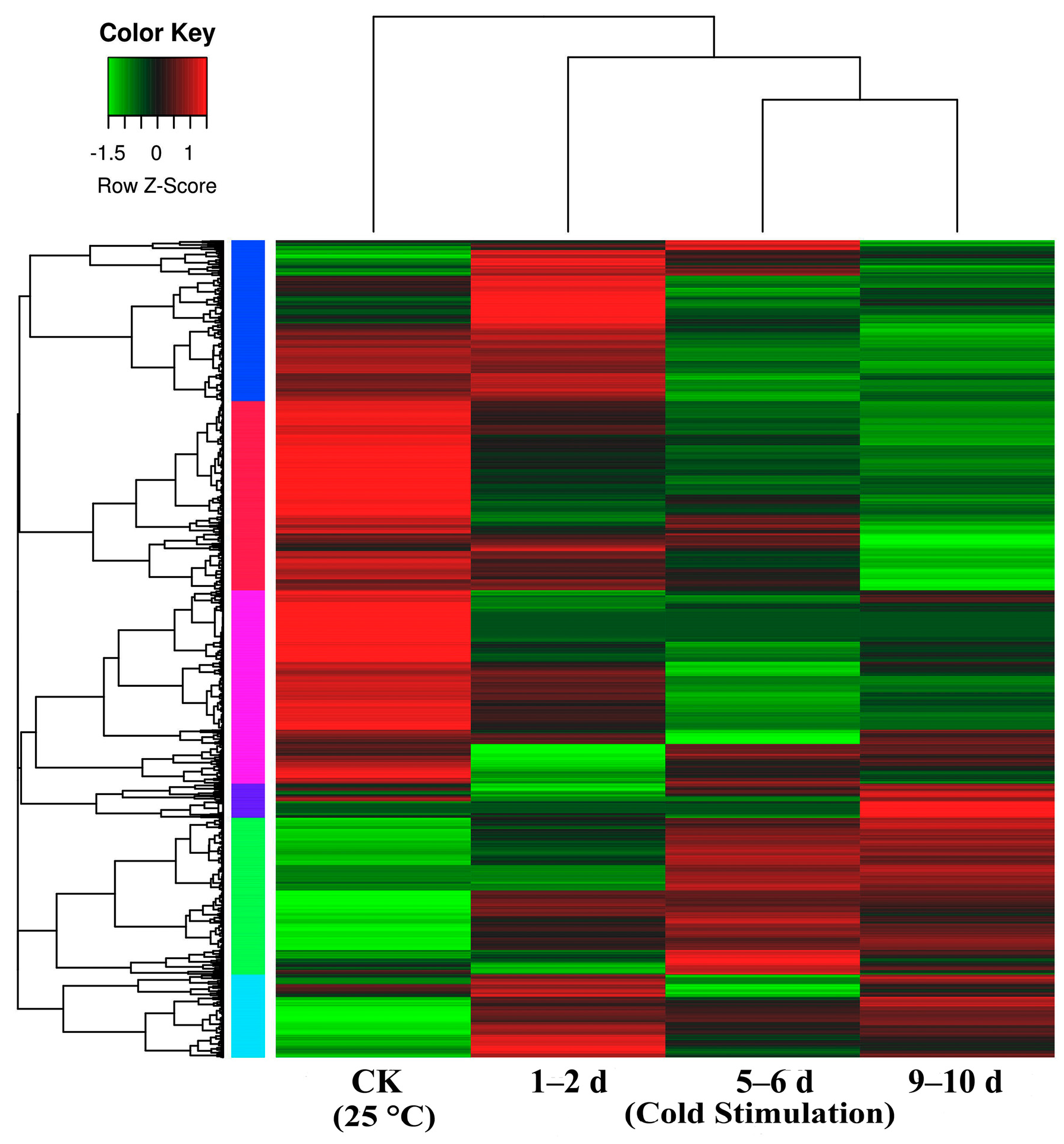
We compared libraries for gene expression changes that were ≥2-fold (Figure 3, Table S1). Overall, we detected 2799, 3312, and 2945 up-regulated DEGs and 2504, 3063, and 3091 down-regulated DEGs between the control and each of the cold stage libraries (1–2 d, 5–6 d, and 9–10 d) (Table S1), respectively. Additionally, we detected 2493, and 1997 up-regulated DEGs and 2392 and 2446 down-regulated DEGs between the 1–2 d and 5–6 d libraries and the 5–6 d and 9–10 d libraries, respectively.
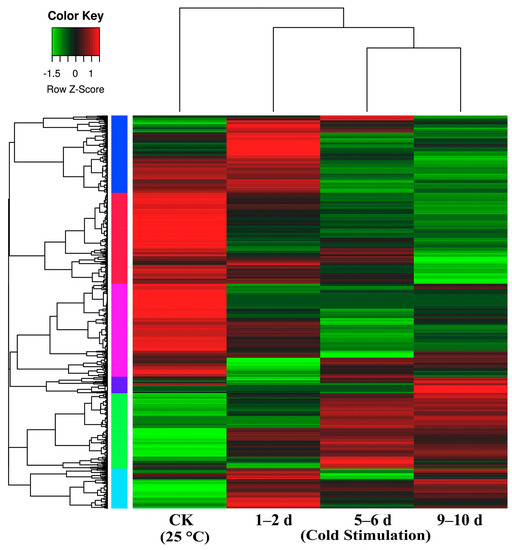
Figure 3.
Heat map representation of cluster analysis for the expression patterns of over 7000 differential gene expressions (DEGs) (p value: < 0.05, fold change: ≥ 2 fold (log2 |FC > 1|)) between control (CK = 25 °C) and different stages of cold stimulation (−3 °C for a duration of 1–2 d, 5–6 d, and 9–10 d) using the FPKM. The samples of the three stages of cold stimulation cluster together; notably, the 5–6 d and 9–10 d samples cluster particularly close.
Zampieri et al. (2011) [10] demonstrated that DEGs listed as environmental stress-responsive genes might help Tuber melanosporum adapt to cold temperatures that regulate fructification. The WEGO and KEGG results for our four libraries showed that DEGs listed as responsive to cold stimulation were involved in cell wall and membrane system stabilization, calcium signaling, osmotic regulation, antioxidant enzymatic defense system, and soluble sugars and protein biosynthesis and metabolism.
2.4. Genes Involved in Cell Wall and Membrane System Stabilization
A wide range of studies indicate that the cell wall and membrane systems are the primary site for the action of cold-responsive proteins in fungi and plants [23,30,31]. We found unigenes with up- or down-regulated expression during the cold stimulation process that associated with WEGO terms related to “cell wall” and “membrane”. These changes in cell wall and membrane proteins during cold stimulation could be important to maintain cell wall integrity. These associations included unigenes encoding for hydrophobin (TR9892|c2_g1), cell wall-associated hydrolase (TR7761|c1_g2), chitin synthase (TR4601|c1_g4) and plasma membrane proteolipid 3 (TR3136|c0_g1 and TR8280|c0_g1). Hydrophobins are proteins unique to fungi and are a type of cell wall protein [32,33,34] involved in fungal growth and development [35,36,37]; response to environmental factors such as light, nutrient availability, and high temperatures [38]; and resistance to pathogens [39]. We found that unigene (TR9892|c2_g1) encoding for the PoH2-type hydrophobin [40] exhibited up-regulated expression during the entire cold stimulation process. We suggest that PoH2-type hydrophobin in Bailinggu might play important roles associated with cold stimulation and the vegetative mycelium development.
Cold tolerance in fungi and plants requires lipid remodeling at membrane system and gene encoding phospholipase and fatty acid desaturases have the potential to alter phospholipid and lipids composition [41,42,43,44]. We found up-regulated unigenes of delta 9-fatty acid desaturase (TR7433|c4_g1) and phospholipase D (TR7300|c4_g1) between the control and the cold stage libraries. Previous studies have demonstrated that the delta 9-fatty acid desaturase gene (in Saccharomyces cerevisiae and Thetrahymena thermophila) and the phospholipase gene contribute to chilling or cold tolerance by altering phospholipid and lipid composition [45,46,47,48]. Similarly, our results indicate that changes in the composition of membrane phospholipid and lipids during the cold stimulation process in Bailinggu may function to stabilize membrane structure.
2.5. Genes Involved in Calcium Signaling and Osmotic Regulation
As a secondary messenger, Ca2+ is required for cold-induced gene expression in fungi and plants and may activate a variety of signaling pathways during cold stress [49,50,51,52]. We found that unigenes encoding calcium sensor and receptor (TR6457|c2_g2, TR7548|c1_g1, TR7021|c7_g1, TR6437|c1_g1 and TR7554|c2_g3) such as calcium-dependent protein kinase and two-component histidine kinase were expressed at higher levels during the cold stimulation stages. These genes have also been associated with cold temperature response in previous studies [21,23,53,54] and, therefore, are likely part of the cold stimulation signal transduction pathways in Bailinggu.
Mitogen-activated protein kinases (MAPK) pathways may be triggered by receptors/sensors such as histidine kinases under various abiotic stress stimuli, including cold stress in plants and fungi [55,56,57,58,59]. We found that unigenes encoding MAPK kinase (TR7227|c5_g1) and MAPK kinase kinase (TR4680|c1_g1) in the MAPK signaling pathways (ko04011) respond to cold stimulation. Moreover, we found down-regulated unigene expression of tyrosine phosphatase (TR5178|c3_g1) between control and the cold stimulation stages. Lee and Esselman (2002) [60] suggested that the reduced tyrosine phosphatase activity leads to an increase in the output of MAPK pathways. Our results suggest a similar calcium/calmodulin-triggering mechanism in Bailinggu in response to cold stimulation can regulate the expression and activity of kinases in the MAPK pathway. We indicate that Hog1-type MAPK in Bailinggu associated with osmotic regulation during the cold stimulation.
2.6. Genes Involved in Antioxidant Enzymatic Defense System
The antioxidant enzymatic defense system and the organic osmolytes are closely related to plant and fungi cold tolerance [9,19]. We found that unigenes encoding enzymes associated with antioxidative enzymes were expressed at higher levels in the cold stimulation stages of Bailinggu. For example, unigenes encoding catalase (TR3796|c0_g1) and peroxidase (TR7183|c0_g1) exhibited higher expression levels in the 5–6 d cold stimulation library. In addition, our data analysis revealed down-regulated gene expression of delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDH, TR7530|c1_g1), which is involved in proline catabolism [61,62] during the cold stimulation process. When P5CDH activity is limited, the P5C-proline cycle can transfer more electrons to the mitochondrial electron transport chain and generate reactive oxygen species (ROS) [63]. Under cold stress, the accumulation of ROS leads to activation of the antioxidant enzyme defense system [20,63]. These results suggest that the antioxidant enzymatic defense system and proline metabolism are positively correlated with detoxification of ROS caused by cold stimulation of Bailinggu.
2.7. Genes Involved in Soluble Sugars and Protein Biosynthesis and Metabolism
Using physiological, biochemical, and growth tests on mature mycelia during cold stimulation, we identified abundant conversion of biomacromolecules and biosynthesis of new compounds such as glycogen breakdown and protein synthesis and metabolism [7]. Our KEGG analysis found that the most abundant unigenes were up- or down-regulated expressions involved in the “starch and sucrose metabolism” (ko00500), “glycolysis/gluconeogenesis” (ko00010), “amino sugar and nucleotide sugar metabolism” (ko00520), and “protein processing in endoplasmic reticulum” (ko04141) pathways during cold stimulation.
The accumulation of soluble sugars has been associated with adaption to cold stress in fungi and plants [21,64,65]. In “starch and sucrose metabolism”, “glycolysis” and “amino sugar and nucleotide sugar metabolism” pathways, we observed unigenes relating to trehalose, glucose, and fructose biosynthesis. The DEGs include unigenes encoding starch 1,4-α-glucan branching enzyme (glycoside hydrolase family 13 protein, TR6876|c2_g1), trehalose 6-phosphate synthase (glycosyltransferase family 20 protein, TR6743|c2_g1), glucan 1,3-beta-glucosidase (glycoside hydrolase family 5 protein, TR6979|c4_g1), glycoside hydrolase family 16 protein (TR1573|c0_g1), glycoside hydrolase family 47 protein (TR3403|c1_g1), and other different glycosyl hydrolase families. Ramírez et al. (2011) [9] revealed that the above genes encoding for carbohydrate active enzymes of Pleurotus ostreatus were involved in cold stress. These data suggest that the transformation and combination of soluble sugars might be necessary for cold tolerance and energy preservation in Bailinggu during cold stimulation.
Moreover, the cold-responsive genes encoding molecular chaperones, including heat shock proteins Hsp20 (TR2339|c0_g1), Hsp70 (TR2377|c0_g1), and Hsp90 (TR5261|c1_g1) were identified in the “protein processing in endoplasmic reticulum” pathway between the control and the cold stage libraries. These genes were also induced in black truffle, S. cerevisiae, and other plants in response to cold stress [10,31,65,66]. This result suggests that the induction of various heat shock proteins might be necessary for fungi and plants to adapt to 4 °C and cold temperatures by stabilizing proteins against cold-induced denaturation.
Overall, the above results provide evidence to support that carbohydrate metabolism plays the vital role during the cold stimulation process of Bailinggu.
2.8. Computational Identification and Prediction of Transcription Factor
We identified 37 TF families from the FTFD and Plant TFDB pipeline (Table S2). TFs comprised the major TF families in most fungi and plants such as, Myb, bHLH, zinc fingers, WRKY, bZIP, and homebox, and the special families in most fungi such as Zn2Cys6 and HMG TFs.
The TFs controlling the expression of cold-induced genes that increase fungi and plant cold tolerance have been identified [67,68]. In this study, many TFs that play a role in cold stress response were identified, including C3H zinc finger, C2H2 zinc finger, GATA zinc finger, Myb, bZIP, bHLH, WRKY, AP2, NAC, ERF, and RAV TFs. Previous studies also demonstrated that these TF families play diverse roles in fungi and plant developmental processes and environmental responses such as chilling and cold stress resistance [14,22,62,69,70,71]. For example, Myb, bHLH, C2H2 zinc finger and WRKY motifs affect seed germination and growth and induce the enhancement of cold tolerance in higher plants [72,73,74,75,76]. Together, these results might indicate that Myb, bHLH, C2H2 zinc finger and WRKY are also important players in Bailinggu response to cold stimulation.
2.9. Validation of Transcriptome Data by qRT-PCR
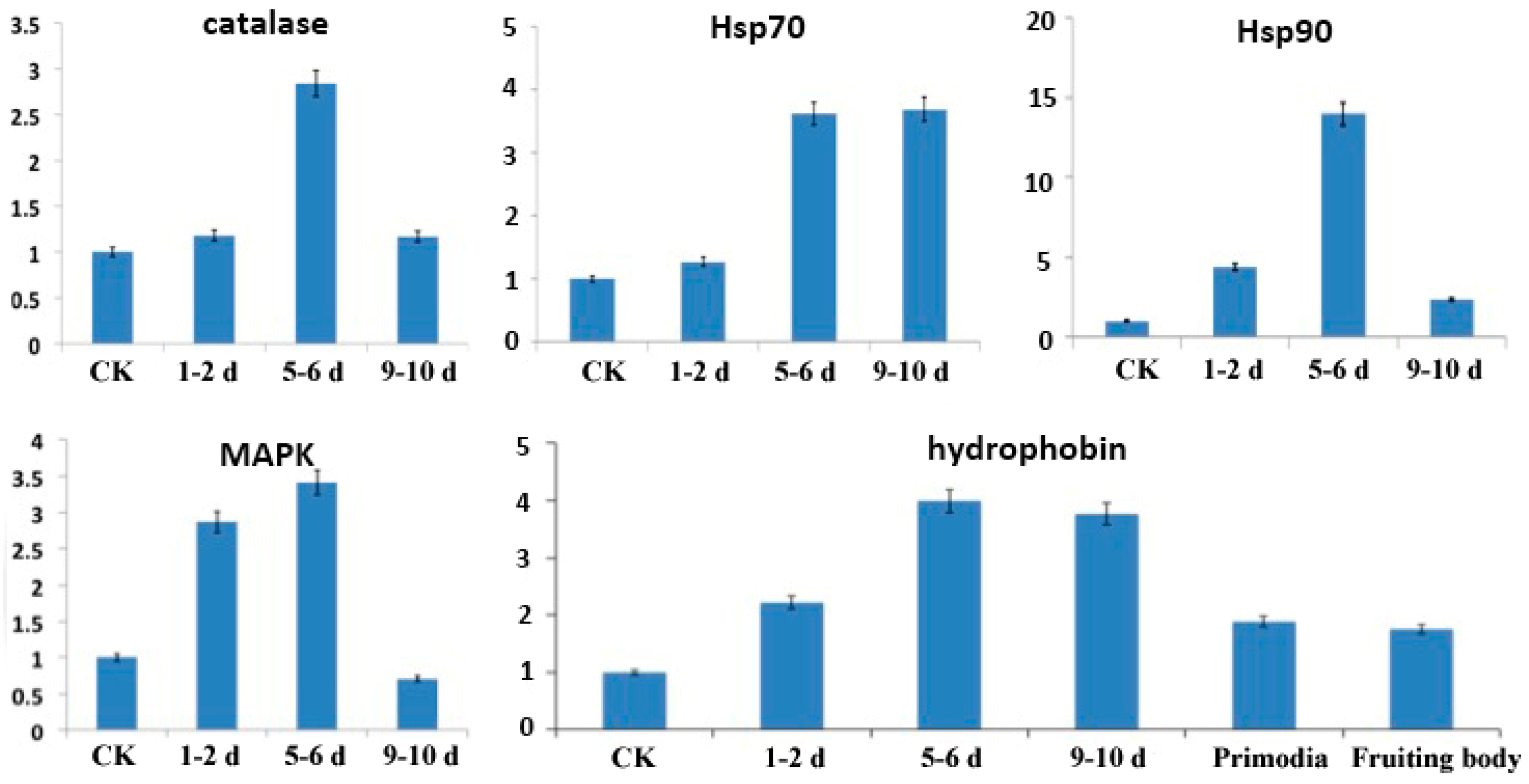
To validate the results of the RNA-seq analysis, five genes were selected to confirm differential expression by quantitative real-time PCR (qRT-PCR). These five genes—PoH2-type hydrophobin, MAPK (mitogen-activated protein kinase hog1), catalase, Hsp70 (heat shock protein 70) and Hsp90 (heat shock protein 90)—are known to relate to cold stress. These genes showed expression patterns similar to the differential analysis results from the RNA-seq output of the bioinformation analysis (Figure 4). That is, all five genes have higher expression levels in the cold stimulation stages compared to the physiological after-ripening stage, but have varying levels of expression among the different cold stages. Genes showing significant differential expression between different stages of cold stimulation can be further explored as candidate genes for cold tolerance using functional genomics approaches.
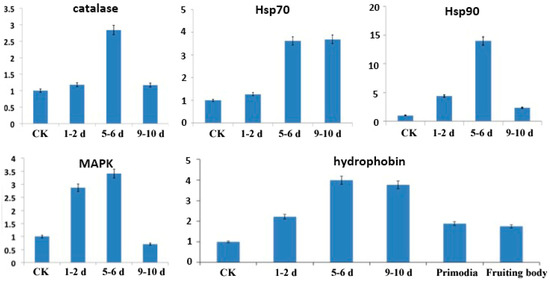
Figure 4.
qRT-PCR analysis of selected genes in mycelia during the physiological after-ripening stage (control (CK) = 25 °C) and different cold stimulation stages (1–2 d, 5–6 d, and 9–10 d at −3 °C). Transcript levels of five genes (PoH2-type hydrophobin, MAPK (mitogen-activated protein kinase hog1), catalase, Hsp70 (heat shock protein 70) and Hsp90 (heat shock protein 90)) that appeared to be up-regulated in all cold stimulation stages compared to the physiological after-ripening stage. The X-axis represents relative quantification of transcript levels of the three selected genes, and the Y-axis represents the control and the three sampling times after cold stimulation.
Overall, all above results suggest that de novo transcriptome sequencing for identifying cold-responsive genes in Bailinggu provides a good estimation of gene expression trends and levels in response to temporal variation in cold stimulation conditions. Our methods and results could be helpful in further studies of Bailinggu or in similar studies in the context of other moshrooms.
2.10. SSR Mining and Identification
Bailinggu’s genome has not been sequenced until now. So, our transcript/EST-based molecular markers are an important resource for genetic diversity analysis, genetic map construction, and molecular marker-assisted selection in breeding. Overall, 607 transcripts/EST-SSR loci were identified and primers were developed (Table S3). Among them, TNR is the most commonly repeated motif with a frequency of 63.76%, followed by DNR (30.64%), TTNR (3.79%), PNR (1.32%) and HNR (0.49%). TNR has generally been detected with the highest frequency in mushroom and crops, including Pleurotus ostreatus, Boletus edulis, Coprinopsis cinerea, Schizophyllum commune [77], Auricularia polytricha [22], Lentinula edodes [78], maize, rice [79], peanut [80], and Gossypium hirsutum [81].
To obtain high-quality EST-SSR primer pairs and test their polymorphism, we selected 100 primer pairs including 22 DNR, 68 TNR, 8 TTNR, and 2 PNR. Ninety-five out of the 100 SSR primer pairs generated a product in at least one of the tested wild collected strains. Ninety-two primer pairs resulted in polymorphic products for all genotypes of wild collected strains and were distinguishable from each other. The results of five-selected primer pairs used for polymorphism analysis were shown in Figure 5. The TNR primer pairs were the most abundant polymorphism of these primer pairs. Five primer pairs—1 TNR, 3 DNR, and 1 TTNR—failed to generate a product in all tested genotypes. Therefore, the overall amplification rate was 95% and the polymorphism rate was 92%. These 92 primers can be used for subsequent population genetics diversity, genetic linkage, and QTL analysis of agronomic traits for Bailinggu.

Figure 5.
Amplification products obtained from PCR using EST-SSR primers and DNA from 10 Bailinggu wild strains using non-denaturing PAGE. EST-SSR primers are blgSSR22, blgSSR51, blgSSR65, and blgSSR74. Lines 1–10 are wild Bailinggu strains collected from Xinjiang Autonomous Region in China.
3. Methods
3.1. Mushroom Tissue Source
The commercial strain and mycelia samples of Pleurotus eryngii subsp. tuoliensis used in this work were kindly provided by Hengdaxing, a year-round cultivation mushroom factory in Beijing, China. Mycelium was grown at 25 °C in cultivation bottles containing 780 g medium (25% corncob, 35% sawdust, 24% wheat bran, 10% maize powder, 4.5% soybean meal) in the dark for 60 days up to the physiological after-ripening stage, which time the substrate was fully colonized in the bottles. Then, the mature mycelia are grown at −3 °C for 1, 2, 5, 6, 9 and 10 d. We randomly selected mature mycelia samples from the different cultivation stages and divided them into two groups, the control sample (the physiological after-ripening stage grown at 25 °C for 60 d) and the plus cold stage samples that were grown at −2–3 °C for 1, 2, 5, 6, 9 and 10 d. All samples were collected and stored in liquid nitrogen in separate bottles and labeled according to number of days under 25 °C and cold stress.
3.2. Library Preparation and RNA-Seq
For comparative transcriptomic analysis, we selected 3 bottles from 7 stages respectively, including the control sample (the physiological after-ripening stage grown at 25 °C for 60 d) and the cold stage samples that were grown at −3 °C for 1, 2, 5, 6, 9 and 10 d. Total RNA was extracted from mycelia obtained from each bottles, then equal amounts total RNA from each bottles of one stage mix together. When referring to the 1–2, 5–6 and 9–10 days, equal amounts total RNA obtained from 1 d and 2 d cold samples were merged to represent the first stage of cold stimulation (1–2 d). At the same, the 5 and 6 d cold mycelia samples were merged to represent the second cold stage that the minimum number of days required for primordia initiation (5–6 d). The 9 and 10 d cold mycelia samples were merged to represent the third cold stage (9–10 d) that the minimum number of days required for budding consistency and high production in factory-based Bailinggu cultivation.
Using TRIzol reagent (Life technologies, New York, NY, USA), total RNA was extracted from mycelia obtained from six culture bottles each for the control and the three cold stage samples. The total RNA was further evaluated for integrity and quality using an Agilent Technologies 2100 Bioanalyzer (Santa Clara, CA, USA). The four cDNA libraries were constructed and a paired-end sequencing strategy (on a Illumina HiSeq 2000 platform) was performed by Beijing Institute of Genomics, Chinese Academy of Sicences (Beijing, China) using the manufacturer’s standard protocol. Data have been deposited in the National Center for Biotechnology Information (NCBI) database under the accession number SRR2080100.
3.3. De novo Transcriptome Assembly and Homology Search
Using an in-house Perl script, we first filtered and trimmed the adapters, low-quality sequences, and duplicate sequences of the raw reads to obtain clean data. Then, using Trinity’s [82] standard protocol (http://trinityrnaseq.sf.net), all clean reads from the four libraries were de novo assembled to yield transcripts and unigenes.
These genes were assigned putative gene descriptions following Basic Local Alignment Search Tool X (BLASTX) alignment to the Non-Redundant (NR) protein database of NCBI, Gene Ontology (GO) (http://wego.genomics.org.cn) database, Interproscan database, Clusters of Orthologous Groups (COGs) in the COG database (http://www.ncbi.nlm.nih.gov/COG) [82], and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) pathway annotations.
3.4. Identification of Differentially Expressed Genes
Differential gene expression (DEG) analysis was calculated using RSEM (RNA-Seq by Expectation Maximization) [83] and R package EBseq [84] and DEGseq [85] to compare the control and the three cold stage libraries. Functional analysis of the differentially expressed genes (DEGs) was carried out using Web Gene Ontology Annotation Plotting (WEGO) [86] and KEGG pathways. The significance of gene expression differences was assessed using the threshold of false discovery rate p-value ≤ 0.05 and |log2 ratio| ≥ 1.
3.5. Identification of Transcription Factors
Transcription factors (TFs) were identified from the lists of significant DEGs using InterPro terms for conserved domains via the pipeline of Fungal Transcription Factor Database (FTFD) (http://ftfd.snu.ac.kr/) and Plant Transcription Factor Database (Plant TFDB) (http://planttfdb.cbi.pku.edu.cn/) with an E-value cut-off of <10−5 [87,88]. The TFs were then used to classify DEGs according to the gene family information. TFs believed to be associated with cold stimulation were selected for further investigation.
3.6. Quantitative Real Time PCR
The quantitative real time PCR (qRT-PCR) was used to assess the results of RNA-seq analysis. Samples of mature mycelia from the different cold stages (1–2 d, 5–6 d and 9–10 d at −3 °C) and control (untreated, at 25 °C) were randomly selected from Hengdaxing mushroom factory in Beijing. Total RNA extraction from these samples used the same procedures described above. Approximately 1 μg of total RNA of each sample was subjected to reverse transcription using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). The qRT-PCR was performed with SYBR® Green Supermix (Thermo Scientific) on the Stratagene Mx3005P (Agilent Technologies) thermal cycler. Each reaction contained 1 μL of the first-strand cDNA as a template and 10 μM of each primer in a total volume reaction of 20 μL. The amplification program was performed under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The gene-specific primers were used for qRT-PCR are shown in Table S4. Two biological and three technical repeats were performed for each sample. The actin gene (GenBank accession number: AY772706) was used as an internal reference gene for normalization of data. Relative gene expression level was calculated using the 2−ΔΔCt method [89].
3.7. SSR Mining and Identification
The MIcroSAtellite (MISA) identification tool [90] was utilized to identify simple sequence repeats of the final genes. Criteria of default parameters for SSR primer development were dinucleotide repeats (DNR) ≥ 6, trinucleotide repeats (TNR) of ≥ 5, tetranucleotide repeats (TTNR) ≥ 5, pentanucleotide repeats (PNR) ≥ 5, and hexanucleotide repeats (HNR) ≥ 5. Primers were generated from the primer modelling software Primer3 (Version 2.3.5, Whitehead Institute, Cambridge, MA, USA). Then we removed the same potential SSR loci and the primers of the different isoforms of the same gene.
One hundred SSR primers were tested for amplification and polymorphism of Bailinggu. Ten wild collected strains of Bailinggu collected from Xinjiang Autonomous Region of China were evaluated in this work. All strains (NO. CCMJ2501-2510) are maintained in the Engineering Research Center of Chinese Ministry of Education for Edible and Medicinal Fungi of the Jilin Agriculture University, China. Genomic DNA was isolated from mycelia using the Plant DNA Mini Kit (KANGWEI, Beijing, China) following manufacturer instructions. Quality of isolated genomic DNA was assessed by the NanoDrop 2000c spectrophotometer (Thermo Scientific). Fifty SSR primers were synthesized at Sangon Biotech Co., Ltd. (Shanghai, China). PCR amplification reactions (20 μL total volume) contained 1 × buffer (Mg2+ free), 2 mM MgCl2, 0.2 mM dNTPs, 0.2 μM primers, 0.5 units of DNA Polymerase (Thermo Scientific), and 1.0 ng genomic DNA. PCR was performed as follows: denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final step at 72 °C for 8 min. PCR products were mixed with a volume of loading buffer and then subjected to 8% polyacrylamide gel for 1.5 hours.
4. Conclusions
In this study, we presented comprehensive profiles of the transcriptome of Pleurotus eryngii subsp. tuoliensis (in the mycelium tissue) and their changes during the cold stimulation process using RNA-Seq. By comparing different gene expression profiles, we revealed candidate genes and complex regulatory networks that play key roles in signal transduction, cell growth, and metabolite biosynthesis in response to cold stimulation. A large number of genes involved in diverse biological or molecular pathways were identified during the cold stimulation process, including the following: (1) genes involved in cold signal sensors or transduction; (2) genes encoding cold-regulated proteins associated with cell wall and membrane; (3) antioxidant enzymatic defense system genes; (4) genes associated with soluble sugars and protein biosynthesis and metabolism; and (5) stress-responsive transcription factor genes. Our results also demonstrated that a series of complex regulatory networks are triggered in Bailinggu during cold stimulation process. Our study provides new insights into the molecular mechanisms regulating Bailinggu mycelium tissue response to cold stimulation. Our study can also serve as a valuable resource for future, relevant genetic research associated with cold stimulation in edible mushrooms. Furthermore, numerous SSR loci were predicted based on transcripts/EST and 607 SSR primers were designed; 92 out of the 100 detected SSR primer pairs generated a product in at least one of the tested wild collected strains. These loci and primers can be used for subsequent population genetics diversity, genetic linkage, and QTL analysis of agronomic traits for Bailinggu.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/5/560/s1.
Acknowledgments
This study was financially supported by National Basic Research Program of China (No. 2014CB138305), National Natural Science Foundation of China (31471926), Natural Science Foundation of Jilin Province (20140101149JC), and University S & T Innovation Platform of Jilin Province for Economic Fungi (#2014B-1). We are grateful to Yuning He and Dingxuan Huang for the sample preparation of Bailinggu. We are also grateful to Peng Wei provide wild strains of Bailinggu. We also thank Linda R. Klein for valuable advice on the manuscript.
Author Contributions
Y.L., Z.-W.Z. and S.-N.H. conceptualized the study and designed the research framework. Y.-P.F. performed the data analyses and wrote the manuscript. Y.L. de novo assembled and analysis the statistical data using DEGseq. C.-T.Y. analysis DEG unigenes using EBseq. Y.-T.D. and M.-Z.D. carried out the SSR marker experiments; Z.Z. was performed qRT-PCR. Y.L. supervised the study with help from N. B. R. P. C. and C. N. S. F. B. F and other projects. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| DEGs | Differentially expressed genes |
| MAPK | Mitogen-activated protein kinases |
| DNR | Dinucleotide repeats |
| TNR | Trinucleotide repeats |
| TTNR | Tetranucleotide repeats |
| PNR | Pentanucleotide repeats |
| HNR | Hexanucleotide repeats |
| MISA | MIcroSAtellite |
| TFs | Transcription factors |
| qRT-PCR | Quantitative real time PCR |
| P5CDH | δ-1-Pyrroline-5-carboxylate dehydrogenase |
| ROS | Reactive oxygen species |
References
- Zervakis, G.I.; Ntougias, S.; Gargano, M.L.; Besi, M.I.; Polemis, E.; Typas, M.A.; Venturella, G. A reappraisal of the Pleurotus eryngii complex—New species and taxonomic combinations based on the application of a polyphasic approach, and an identification key to Pleurotus taxa associated with Apiaceae plants. Fungal Biol. 2014, 118, 814–834. [Google Scholar]
- Zhao, M.; Huang, C.; Chen, Q.; Wu, X.; Qu, J.; Zhang, J. Genetic variability and population structure of the mushroom Pleurotus eryngii var. tuoliensis. PLoS ONE 2013, 8, e83253. [Google Scholar]
- Alam, N.; Yoon, K.N.; Lee, T.S. Evaluation of the antioxidant and antityrosinase activities of three extracts from Pleurotus nebrodensis fruiting bodies. Afr. J. Biotechnol. 2011, 10, 2978–2986. [Google Scholar]
- Kawai, G.; Babasaki, K.; Neda, H. Taxonomic position of a Chinese Pleurotus “Bai-Ling-Gu”, It belongs to Pleurotus eryngii (DC., Fr.) Quél. and evolved independently in China. Mycoscience 2008, 49, 75–87. [Google Scholar] [CrossRef]
- Chen, M.; Chen, Q.; Zhao, M.; Ming, Z. Screening and analysis of gene fragments related to fructification of Pleurotus eryngii var. tuoliensis. Biotechnology 2013, 23, 4–8. [Google Scholar]
- Liu, P.; Xu, Y.; She, D.; Sun, Y. Progress of study on Pleurotus nebrodensis. Seed 2007, 26, 55–58. [Google Scholar]
- Zhou, C.Q. Study on Basis Physiology and Key Technology of Culture of Pleurotus nerbrodensis. Ph.D. Thesis, Shandong Agricultural University, 2007. [Google Scholar]
- Zhou, C.Q.; Wang, X.; Li, Y. Extracellular enzyme production by Pleurotus nebrodensis at different developmental stages. Acta Edulis Fungi 2008, 15, 67–71. [Google Scholar]
- Ramírez, L.; Oguiza, J.A.; Pérez, G.; Lavín, J.L.; Omarini, A.; Santoyo, F.; Alfaro, M.; Castanera, R.; Parenti, A.; Muguerza, E.; et al. Genomics and transcriptomics characterization of genes expressed during postharvest at 4 °C by the edible basidiomycete Pleurotus ostreatus. Int. Microbiol. 2011, 14, 111–120. [Google Scholar] [PubMed]
- Zampieri, E.; Balestrini, R.; Kohler, A.; Abbà, S.; Martin, F.; Bonfante, P. The Perigord black truffle responds to cold temperature with an extensive reprogramming of its transcriptional activity. Fungal Genet. Biol. 2011, 48, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Chen, Z.; Xia, J.; Zhang, K.; Chen, X.; Zhou, Y.; Bo, W.; Song, S.; Deng, D.; Guo, X.; et al. Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in Cassava. BMC Plant Biol. 2014, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, Q.; Ma, C.; Zhang, Z.; Cao, H.; Kong, Y.; Yue, C.; Hao, X.Y.; Chen, L.; Ma, J.; et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 2013, 14, 415. [Google Scholar] [CrossRef] [PubMed]
- Morin, E.; Kohler, A.; Baker, A.R.; Foulongne-Oriol, M.; Lombard, V.; Nagy, L.G.; Ohm, R.A.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.L.; et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Jian, H.; Song, C.; Bao, D.; Shang, X.; Wu, D. Transcriptome analysis of candidate genes and signaling pathways associated with light-induced brown film formation in Lentinula edodes. Appl. Microbiol. Biotechnol. 2013, 97, 4977–4989. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Baek, J.H.; Lee, S.; Kim, C.; Rhee, H.; Kim, H.; Seo, J.S.; Park, H.R.; Yoon, D.E.; Nam, J.Y.; et al. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS ONE 2014, 9, e93560. [Google Scholar] [CrossRef] [PubMed]
- Veneault-Fourrey, C.; Commun, C.; Kohler, A.; Morin, E.; Balestrini, R.; Plett, J.; Danchin, E.; Coutinho, P.; Wiebenga, A.; de Vries, R.P.; et al. Genomic and transcriptomic analysis of Laccaria bicolor CAZome reveals insights into polysaccharides remodelling during symbiosis establishment. Fungal Genet. Biol. 2014, 72, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ouyang, B.; Zhang, J.; Wang, T.; Li, H.; Zhang, Y.; Yu, C.; Ye, Z. Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLoS ONE 2012, 7, e50785. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, J.; Liu, Q.; Guo, W.; Zhao, T.; Ma, Q.; Wang, G. Transcriptome sequencing and identification of cold tolerance genes in Hardy corylus species (C. heterophylla fisch) floral buds. PLoS ONE 2014, 9, e108604. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zou, Z.; Wang, S.; Gong, M. Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas, L. PLoS ONE 2013, 8, e82817. [Google Scholar] [CrossRef] [PubMed]
- Die, J.V.; Rowland, L.J. Elucidating cold acclimation pathway in blueberry by transcriptome profiling. Environ. Exp. Bot. 2014, 106, 87–98. [Google Scholar]
- Li, Y.H.; Zhang, W.; Li, Y. Transcriptomic Analysis of Flower Blooming in Jasminum sambac through De Novo RNA Sequencing. Molecules 2015, 20, 10734–10747. [Google Scholar]
- Zhou, Y.; Chen, L.; Fan, X.; Bian, Y. De novo assembly of Auricularia polytricha transcriptome using Illumina sequencing for gene discovery and SSR marker identification. PLoS ONE 2014, 9, e91740. [Google Scholar]
- Pang, T.; Ye, C.Y.; Xia, X.; Yin, W. De novo sequencing and transcriptome analysis of the desert shrub, Ammopiptanthus mongolicus, during cold acclimation using Illumina/Solexa. BMC Genomics 2013, 14, 488. [Google Scholar] [PubMed]
- Liang, M.; Yang, X.; Li, H.; Su, S.; Yi, H.; Chai, L.; Deng, X. De novo transcriptome assembly of Pummelo and molecular marker development. PLoS ONE 2015, 10, e0120615. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Wang, Y.; Wu, S.; Wang, J.; Tian, X.; Pei, X. De novo assembly of transcriptome sequencing in Caragana korshinskii Kom. and Characterization of EST-SSR markers. PLoS ONE 2015, 10, e0115805. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.M.; Zhou, T.; Li, Z.H.; Zhao, G.F. Characterization of Global Transcriptome Using Illumina Paired-End Sequencing and Development of EST-SSR Markers in Two Species of Gynostemma (Cucurbitaceae). Molecules 2015, 20, 21214–21231. [Google Scholar] [CrossRef] [PubMed]
- Foulongne-Oriol, M.; Lapalu, N.; Férandon, C.; Spataro, C.; Ferrer, N.; Amselem, J.; Savoie, J.M. The first set of expressed sequence tags (EST) from the medicinal mushroom Agaricus subrufescens delivers resource for gene discovery and marker development. Appl. Microbiol. Biotechnol. 2014, 7879–7892. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Jiao, Y.; Wang, G.; Li, Y.; Jia, H.; Wu, H.; Chai, C.; Dong, X.; Guo, Y.; Zhang, L.; et al. Genetic diversity of male and female Chinese bayberry (Myrica rubra) populations and identification of sex-associated markers. BMC Genomics 2015, 16, 394. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Liu, X.; Huang, J.; Wang, Q.; Gu, J.; Lu, Y. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genomics 2014, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Uemura, M. Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J. 2003, 36, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Homma, T.; Kitagawa, E.; Momose, Y.; Sato, M.S.; Odani, M.; Shimizu, H.; Hasegawa-Mizusawa, M.; Matsumoto, R.; Mizukami, S.; et al. Genome-wide expression analysis of yeast response during exposure to 4 °C. Extremophiles 2006, 10, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Whiteford, J.R.; Spanu, P.D. Hydrophobins and the interactions between fungi and plants. Mol. Plant Pathol. 2002, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A. Hydrophobins, multipurpose proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef] [PubMed]
- Bayry, J.; Aimanianda, V.; Guijarro, J.I.; Sunde, M.; Latgé, J.P. Hydrophobins-unique fungal proteins. PLoS Pathog. 2012, 8, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Peñas, M.M.; Rust, B.; Larraya, L.M.; Ramírez, L.; Pisabarro, A.G. Differentially regulated, vegetative-mycelium-specific hydrophobins of the edible basidiomycete Pleurotus ostreatus. Appl. Environ. Microbiol. 2002, 68, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Ahn, I.P.; Rho, H.S.; Lee, Y.H. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 2005, 57, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Mgbeahuruike, A.C.; Kovalchuk, A.; Asiegbu, F.O. Comparative genomics and evolutionary analysis of hydrophobins from three species of wood-degrading fungi. Mycologia 2013, 105, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, S.; Picone, D.; Ercole, C.; Spadaccini, R.; Stefano, L.D.; Rea, I.; Giardina, P. Environmental conditions modulate the switch among different states of the hydrophobin VMH2 from Pleurotus ostreatus. Biomacromolecules 2012, 13, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Segers, G.C.; Hamada, W.; Oliver, R.P.; Spanu, P.D. Isolation and characterisation of five different hydrophobin-encoding cDNAs from the fungal tomato pathogen Cladosporium fulvum. Mol. Gen. Genet. 1999, 261, 644–652. [Google Scholar]
- Asgeirsdóttir, S.A.; Vries, O.M.; Wessels, J.G. Identification of three differentially expressed hydrophobins in Pleurotus ostreatus (oyster mushroom). Microbiology 1998, 144, 2961–2969. [Google Scholar]
- Moellering, E.R.; Muthan, B.; Benning, C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 2010, 330, 226–228. [Google Scholar]
- Uemura, M.; Steponkus, P. A contrast of the plasma membrane lipid composition of oat and rye leaves in relation to freezing tolerance. Plant Physiol. 1994, 104, 479–496. [Google Scholar] [PubMed]
- Gibson, S.; Arondel, V.; Iba, K.; Somerville, C. Cloning of a temperature-regulated gene encoding a chloroplast omega-3 desaturase from Arabidopsis thaliana. Plant Physiol. 1994, 106, 1615–1621. [Google Scholar] [CrossRef] [PubMed]
- Nishida, I.; Murata, N. Chilling sensitivity in plants and cyanobacteria, the crucial contribution of membrane lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 541–568. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Vargas, S.; Sánchez-García, A.; Martínez-Rivas, J.M.; Prieto, J.A.; Randez-Gil, F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl. Environ. Microbiol. 2007, 73, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, M.; Zhang, W.; Welti, R.; Wang, X. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2004, 22, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, S.; Zhao, Y.; Nozawa, Y. Tetrahymena thermophila and its mRNA expression during thermal. Cloning 1996, 34, 29–34. [Google Scholar]
- Testerink, C.; Munnik, T. Phosphatidic acid, A multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005, 10, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses, roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, R.E. Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 697–725. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2014, 56, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Townley, H.E.; Knight, M.R. Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 2002, 128, 1169–1172. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, R.; Li, M.; Li, X.; Wang, C.; Welti, R.; Wang, X. Differential degradation of extraplastidic and plastidic lipids during freezing and post-freezing recovery in Arabidopsis thaliana. J. Biol. Chem. 2008, 283, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Kojima, K.; Takano, Y.; Tanaka, C. Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot Cell 2005, 4, 1820–1828. [Google Scholar] [CrossRef] [PubMed]
- Estruch, F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 2000, 24, 469–486. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Schumaker, K.; Zhu, J.K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 2004, 55, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Gene regulation during cold acclimation in plants. Physiol. Plant 2006, 126, 52–61. [Google Scholar] [CrossRef]
- Hagiwara, D.; Suzuki, S.; Kamei, K.; Gonoi, T.; Kawamoto, S. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2014, 73, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Esselman, W.J. Inhibition of PTPs by H2O2 regulates the activation of distinct MAPK pathways. Free Radic. Biol. Med. 2002, 33, 1121–1132. [Google Scholar] [CrossRef]
- Schaap, P.J.; Müller, Y.; Sonnenberg, S.M.; Van Griensven, L.J.L.D.; Visser, J. The Agaricus bisporus pruA gene encodes a cytosolic Δ1-pyrroline-5-carboxylate dehydrogenase which is expressed in fruit bodies but not in gill tissue. Appl. Environ. Microbiol. 1997, 63, 57–62. [Google Scholar] [PubMed]
- Mazzucotelli, E.; Mastrangelo, A.M.; Crosatti, C.; Guerra, D.; Stanca, M.; Cattivelli, L. Abiotic stress response in plants, When post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008, 174, 420–431. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline, a multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, C.; Zhu, J.K. Molecular genetic analysis of cold-regulated gene transcription. Philos Trans. R. Soc. Lond. B. Biol. Sci. 2002, 357, 877–886. [Google Scholar]
- Kandror, O.; Bretschneider, N.; Kreydin, E.; Cavalieri, D.; Goldberg, A.L. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol. Cell. 2004, 13, 771–781. [Google Scholar]
- Thomashow, M.F. Plant cold acclimation, freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar]
- Mitsuda, N.; Ohme-Takagi, M. Functional analysis of transcription factors in arabidopsis. Plant Cell Physiol. 2009, 50, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Koizuka, N.; Martin, R.C.; Nonogaki, H. The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 2005, 44, 960–971. [Google Scholar] [CrossRef] [PubMed]
- De Leonardis, A.M.; Marone, D.; Mazzucotelli, E.; Neffar, F.; Rizza, F.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A.M. Durum wheat genes up-regulated in the early phases of cold stress are modulated by drought in a developmental and genotype dependent manner. Plant Sci. 2007, 172, 1005–1016. [Google Scholar] [CrossRef]
- Ohm, R.A.; Jong, J.F.; Bekker, C.; Wösten, H.B.; Lugones, L.G. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011, 81, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Cao, H.; Zhang, L.; Huang, P.; Lin, F. Systematic analysis of Zn2 Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the Rice blast fungus. PLoS Pathog. 2014, 10, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Bai, X.; Zhu, D.; Li, Y.; Ji, W.; Cai, H.; Wu, J.; Liu, B.; Zhu, Y. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress. Planta 2012, 235, 1141–1155. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guo, S.; Zhao, Y.; Chen, D.; Chong, K.; Xu, Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2010, 29, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, M.; Wang, H.; Bao, D. Distribution pattern analysis of SSR loci in the genome of Boletus edulis. Mycosystema 2015, 34, 204–214. [Google Scholar]
- Polat, E.; Ince, A.G.; Karaca, M.; Onus, N. Mining and utilization of mushroom ESTs for microsatellites. Conserv. Genet. 2010, 11, 1123–1126. [Google Scholar] [CrossRef]
- Kantety, R.V.; La Rota, M.; Matthews, D.E.; Sorrells, M.E. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol. Biol. 2002, 48, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Chen, X.; Hong, Y.; Liu, H.; Zhou, G.; Li, S.; Guo, B. Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea, L.) and Arachis wild species. BMC Plant Biol. 2009, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Wang, C.; Song, X.; Guo, W.; Gou, J.; Li, C.; Chen, X.; Zhang, T. Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor. Appl. Genet. 2006, 112, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Philip, D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Macmanes, M.D.; et al. De novo transcript sequence reconstruction from RNA-Seq, reference generation and analysis with Trinity. Nat. Protoc. 2014, 8, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.G.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. Ebseq: An empirical bayes hierarchical model for inference in rna-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO, A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J.; Jang, S.; Kim, S.; Kong, S.; Choi, J.; Ahn, K.; Kim, J.; Lee, S.; Kim, S.; et al. FTFD, An informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 2008, 24, 1024–1025. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan, protein domains identifier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fu, Y.; Ma, J.; Zhang, C.; Wang, P. Isolation and characterization of soybean chalcone reductase cDNA, which encodes the key enzyme for the biosynthesis of 4,2′,4′-trihydroxychalcone in legumes. Mol. Breed 2014, 34, 2139–2149. [Google Scholar]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).