Abstract
Lipo-alkaloid is a kind of C19-norditerpenoid alkaloid usually found in Aconitum species. Structurally, they contain an aconitane skeleton and one or two fatty acid moieties of 3–25 carbon chains with 1–6 unsaturated degrees. Analysis of the lipo-alkaloids in roots of Aconitum carmichaelii resulted in the isolation of six known pure lipo-alkaloids (A1–A6) and a lipo-alkaloid mixture (A7). The mixture shared the same aconitane skeleton of 14-benzoylmesaconine, but their side chains were determined to be 9-hydroxy-octadecadienoic acid, 13-hydroxy-octadecadienoic acid and 10-hydroxy-octadecadienoic acid, respectively, by MS/MS analysis after alkaline hydrolysis. To our knowledge, this is the first time of the reporting of the oxygenated fatty acids as the side chains in naturally-occurring lipo-alkaloids. In order to identify more lipo-alkaloids, a compound database was established based on various combinations between the aconitane skeleton and the fatty acid chain, and then, the identification of lipo-alkaloids was conducted using the database, UHPLC-Q-TOF-MS and MS/MS. Finally, 148 lipo-alkaloids were identified from A. carmichaelii after intensive MS/MS analysis, including 93 potential new compounds and 38 compounds with oxygenated fatty acid moieties.
1. Introduction
Lipo-alkaloid is a kind of C19-norditerpenoid alkaloid usually found in Aconitum species. Structurally, they consist of an aconitane skeleton and one or two fatty acid moieties of 3–25 carbon chains with 1–6 unsaturated degrees [1]. So far, more than 200 lipo-alkaloids have been reported from plants [1], semisynthesis [2] and biotransformations [3,4,5]. Because naturally-occurring lipo-alkaloids are so close structurally, it is very difficult to purify them from the mixture. Their characterization is mainly conducted by high sensitive mass spectrometry (MS) or liquid chromatography-mass spectrometry (LC-MS) techniques, such as ion trap-MS [6], MALDI-TOF-MS [7], ESI-Fourier Transform Ion Cyclotron Resonance FTICR-MS [8], LC-ESI-Ion TrapIT-MS [8,9,10], and LC-Linear Ion TrapLTQ-Orbitrap-MS [11]. To our knowledge, only one pure lipo-alkaloid, 8-O-azeloyl-14-benzoylaconine, was isolated and elucidated by NMR from plants [12]. In this research, the separation of naturally-occurring lipo-alkaloids was optimized using column chromatography (CC), and six pure lipo-alkaloids (A1–A6) were obtained from a natural source for the first time. At the same time, a mixture of oxygenated fatty acid-containing lipo-alkaloids (A7) was also obtained, and the structures were determined by MS/MS analysis after alkaline hydrolysis. Furthermore, a compound database of lipo-alkaloids was established based on the basic skeletons and fatty acids groups, and by the combination of the compound database, UHPLC-Q-TOF-MS and MS/MS analysis, 148 lipo-alkaloids, including 93 potential new ones, were identified from A. carmichaelii.
2. Results and Discussion
2.1. Isolation and Structural Elucidation of Lipo-Alkaloids A1–A7
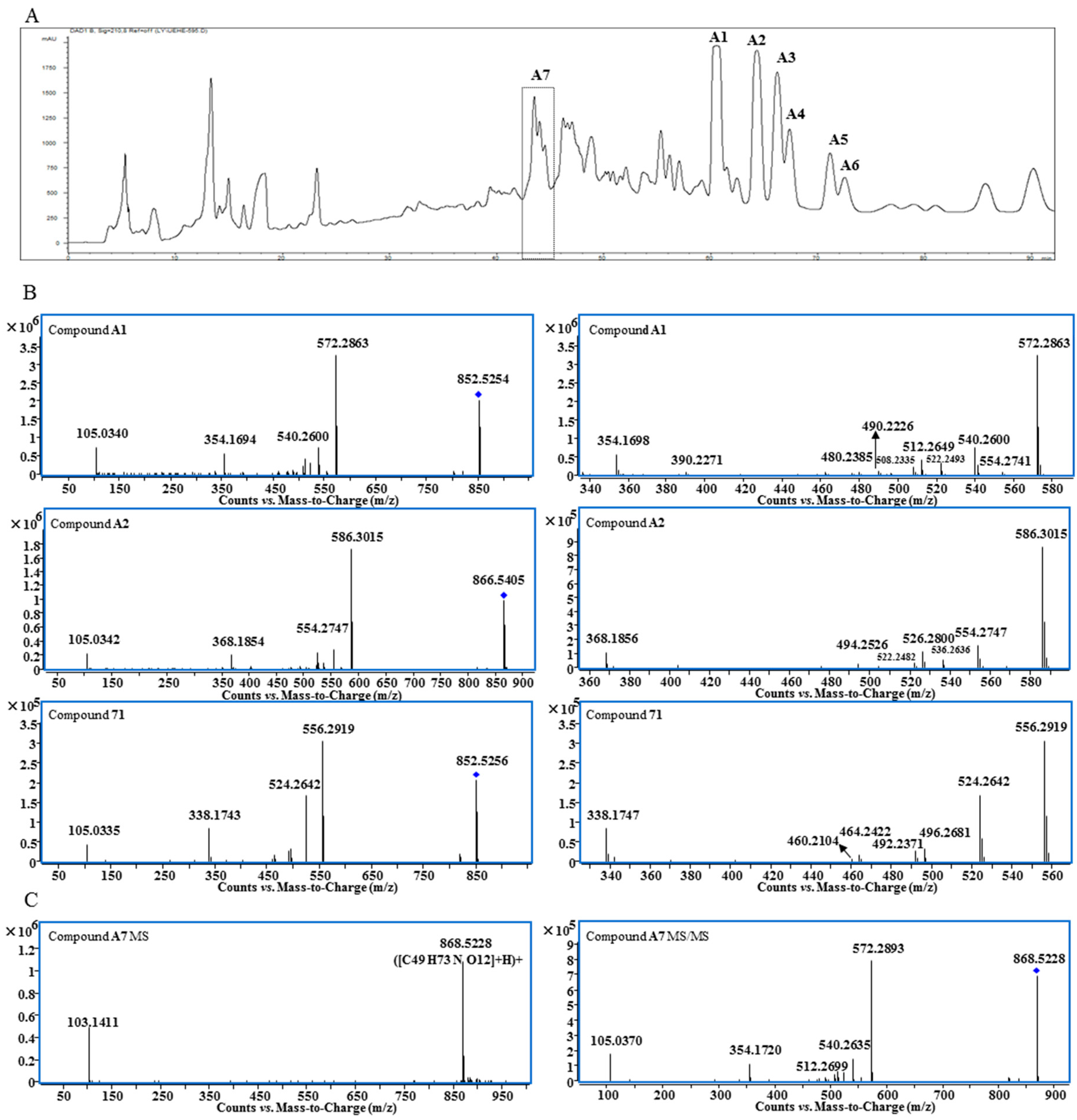
Structurally, lipo-alkaloids usually contain one or two long fatty acid moieties; therefore, they have low polarity and can be extracted by n-hexane. After column chromatography separation on silica gel and ODS, a lipo-alkaloids-rich fraction was obtained. Compared to water and methanol as mobile phases, 0.01% diethylamine in water and methanol gave better separation of lipo-alkaloids on a preparative HPLC C18 column (Figure 1A). Seven compounds (A1–A7) were determined to be lipo-alkaloids by UHPLC-Q-TOF-MS and NMR techniques.
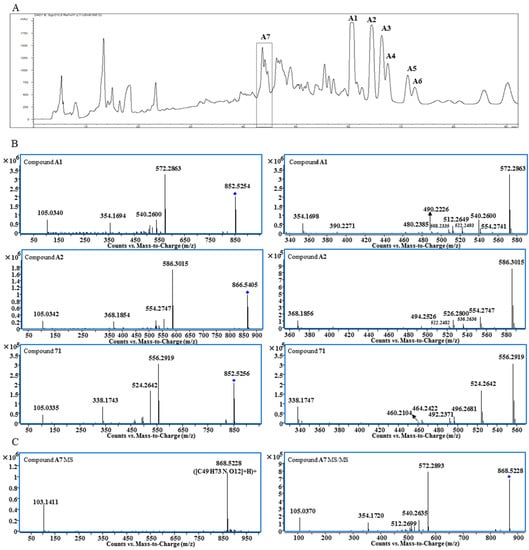
Figure 1.
Preparative HPLC chromatogram of Fraction D4-1-8 (A); MS/MS and expanded MS/MS spectra of Compounds A1, A2 and 71 (B); MS and MS/MS spectra of Compound A7 (C).
The molecular formula of Compound A1 was determined as C49H73NO11 based on the quasi-molecular ion at m/z 852.5254. The base peak at m/z 572.2863 in the MS/MS spectrum is produced from the neutral loss of C18H32O2 (Figure 1B), indicating the presence of long chain fatty acid moiety. Other fragmentation ions at m/z 540.2600, 522.2493, 512.2649, 508.2335, 490.2226, 480.2385 and 390.2271 were related to the neutral losses of MeOH, H2O, CO and BzOH (Table 1), while the fragmentation ion at m/z 105.0340 was assigned to the benzoyl group. Therefore, the basic skeleton of A1 should be 14-benzoylmesaconitine (BMA) [13], and this was supported by its 1H- and 13C-NMR spectra (Table S1). 1H-NMR also displayed the unconjugated vinylic protons at δ 5.376 (4H, m, H-9″, 10″, 12″, 13″), bis-allylic protons at δ 2.802 (2H, m, H-11″), allylic protons at δ 2.045 (4H, m, H-8″, 14″), acylated methylene at δ 1.450 (1H, m, H-2″a) and 1.816 (1H, m, H-2″b), methylene at δ 1.380 (8H, m, H-7″, 15″, 16″, 17″), 1.177 (2H, m, H-6″), 1.026 (2H, m, H-5″), 1.179 (1H, m, H-3″a), 1.025 (1H, m, H-3″b) and 0.885 (2H, m, H-4″) and a methyl group at δ 0.915 (3H, t, H-18″); thus, the fatty acid moiety was determined to be the linoleoyl group. Comparing to the 1H-NMR spectra of free fatty acids, significant up-field shifting was observed for H-2″, H-3″ and H-4″; moreover, the signals of two protons of the methylene at positions 2″ and 3″ were un-equivalent, which should be induced by the 14-benzoyl group. Finally, Compound A1 was determined to be 8-O-linoleoyl-14-benzoylmesaconine.

Table 1.
MS/MS characteristic fragmentation ions of aconitane skeletons in lipo-alkaloids a.
The molecular formulae of Compounds A3 and A4 were C47H73NO11 and C49H75NO11, which were two carbons less and two hydrogens more than that of Compound A1. The same MS/MS spectra (Table S2) as A1 indicated that A3 and A4 should be BMA derivatives. Meanwhile, the neutral losses of C16H32O2 and C18H34O2 in MS/MS spectra and 1H-NMR data (Table S1) suggested that the side chains for A3 and A4 were palmitic acid and oleic acid, respectively. Therefore, their structures were determined to be 8-O-palmitoyl-14-benzoylmesaconine (A3) and 8-O-oleoyl-14-benzoylmesaconine (A4).
The molecular formula of Peak A2 was determined as C50H75NO11 from the high resolution MS, which was one carbon and two hydrogens more than that of A1. The pattern of fragmentation ions of A2 was very similar to that of A1 with most of the peaks moved 14 Da (CH2) to the right side (Figure 1B and Table 1). The 1H-NMR spectrum of A2 showed the presence of NCH2CH3 (Table S1); therefore, the basic skeleton should be 14-benzoylaconine (BA), and it was determined to be 8-O-linoleoyl-14-benzoylaconine. With a similar comparison, Compounds A5 (C48H75NO11) and A6 (C50H77NO11) were determined to be 8-O-palmitoyl-14-benzoylaconine and 8-O-oleoyl-14-benzoylaconine, respectively, based on their MS/MS and 1H-NMR spectra.
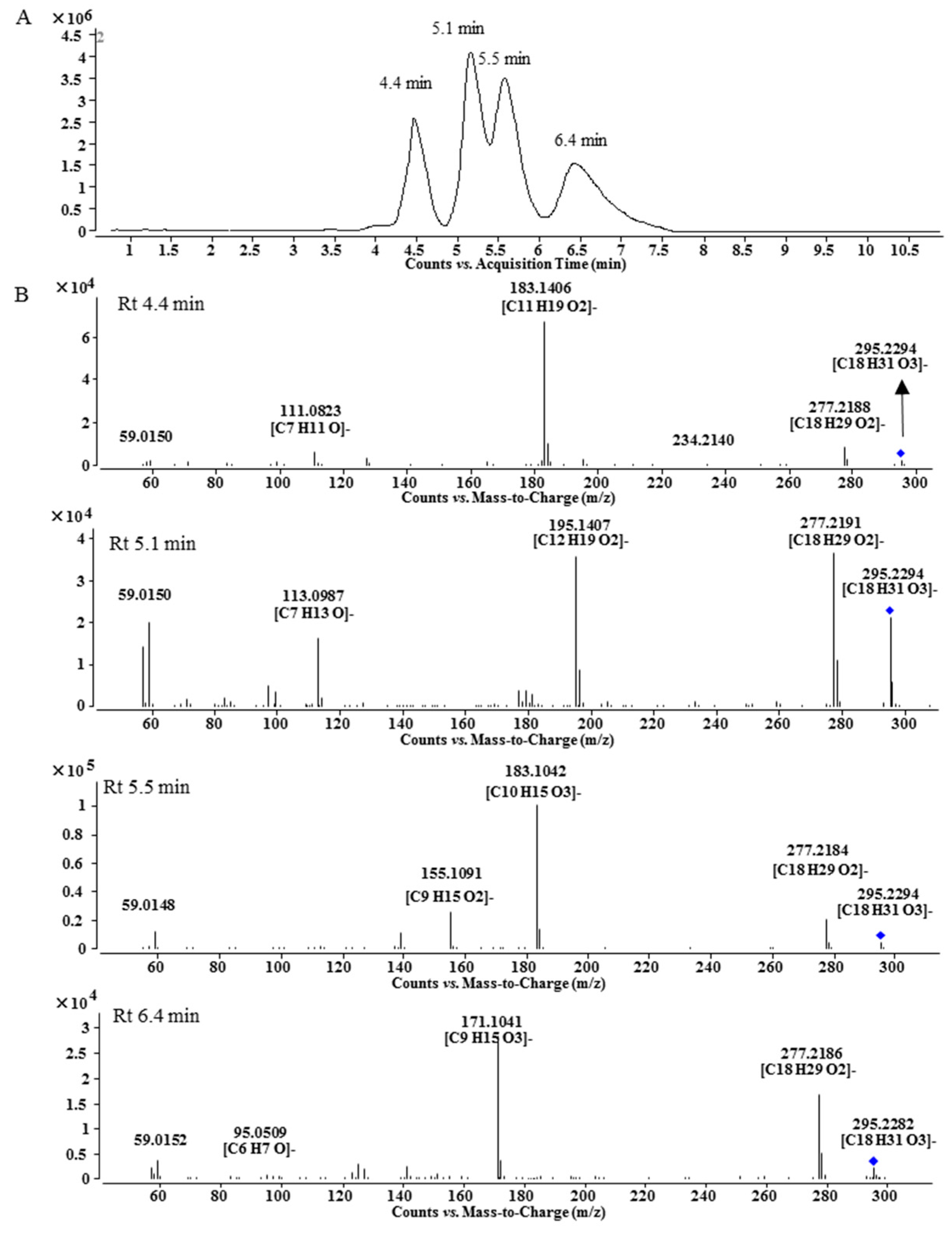
Peak A7 was obtained as a single peak in the UHPLC-MS chromatogram and with a quasi-molecular ion at m/z 868.5228, which is in agreement with the molecular formula of C49H73NO12 that is one oxygen more than that of A1. The MS/MS spectrum showed that A7 was a BMA derivative with C18H32O3 as the side chain (Figure 1C). However, the 1H-NMR spectrum of A7 was unexpectedly complicated. Except for BMA signals, the unconjugated vinyl protons at δ 5.371 (4H, m), bis-allylic methylene at δ 2.790, allylic methylene at δ 2.051 and methylene at around δ 1.3, there were three proton signals at δ 4.413, 4.189 and 4.127 with unproportioned integrations to other signals. Therefore, A7 might be a mixture containing different side chains. After alkaline hydrolysis of A7, the released fatty acids were analyzed by UHPLC-MS and MS/MS, and the results are shown in Figure 2. At least four peaks were detected at 4.4, 5.1, 5.5 and 6.4 min with the same m/z at 295.23 (C18H32O3), but their MS/MS spectra were significantly different (Figure 2B). In the MS/MS spectrum of the peak at 5.1 min, the fragmentation ions at m/z 277.2191 ([M − H − H2O]−) and 195.1407 ([M − H − C6H12O]−) indicate the presence of a hydroxyl group at C-13 [14] and the unconjugated vinyl bonds should be between C-2 and C-12, i.e., 13-hydroxyoctadecadienoic acid. The fragmentation ions in the MS/MS spectrum for the third peak at 5.5 min were related to the neutral losses of H2O, C8H16 and CO, just like the MS/MS spectrum of 10-hydroxyoctadecadienoic acid [15]. The fourth peak was identified as 9-hydroxyoctadecadienoic acid based on the ions of [M − H − H2O]− and [M − H − C9H16]− [14]. For the first peak, the fragmentation ions from the neutral losses of H2O and C5H12O were observed. The cleavage of hydroxyl group usually gives an unsaturated carbonyl group, which is different from the neutral loss of C5H12O, so the fatty acid at 4.4 min needs further investigation. Finally, Peak A7 was determined as the mixture of the oxygenated fatty acids-containing BMA derivative by the combination of 1H-NMR and MS/MS analysis after alkaline hydrolysis.
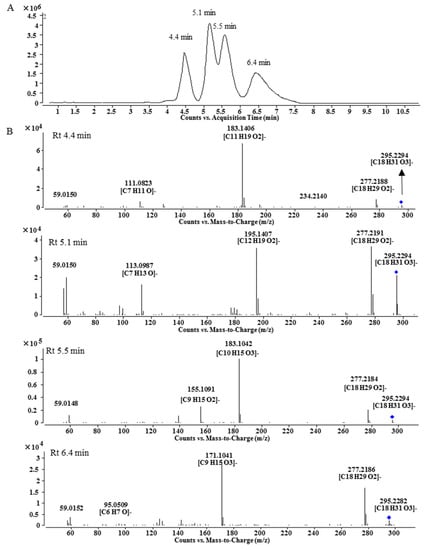
Figure 2.
LC-MS chromatogram (A) and MS/MS spectra (B) of fatty acids released by alkaline hydrolysis.
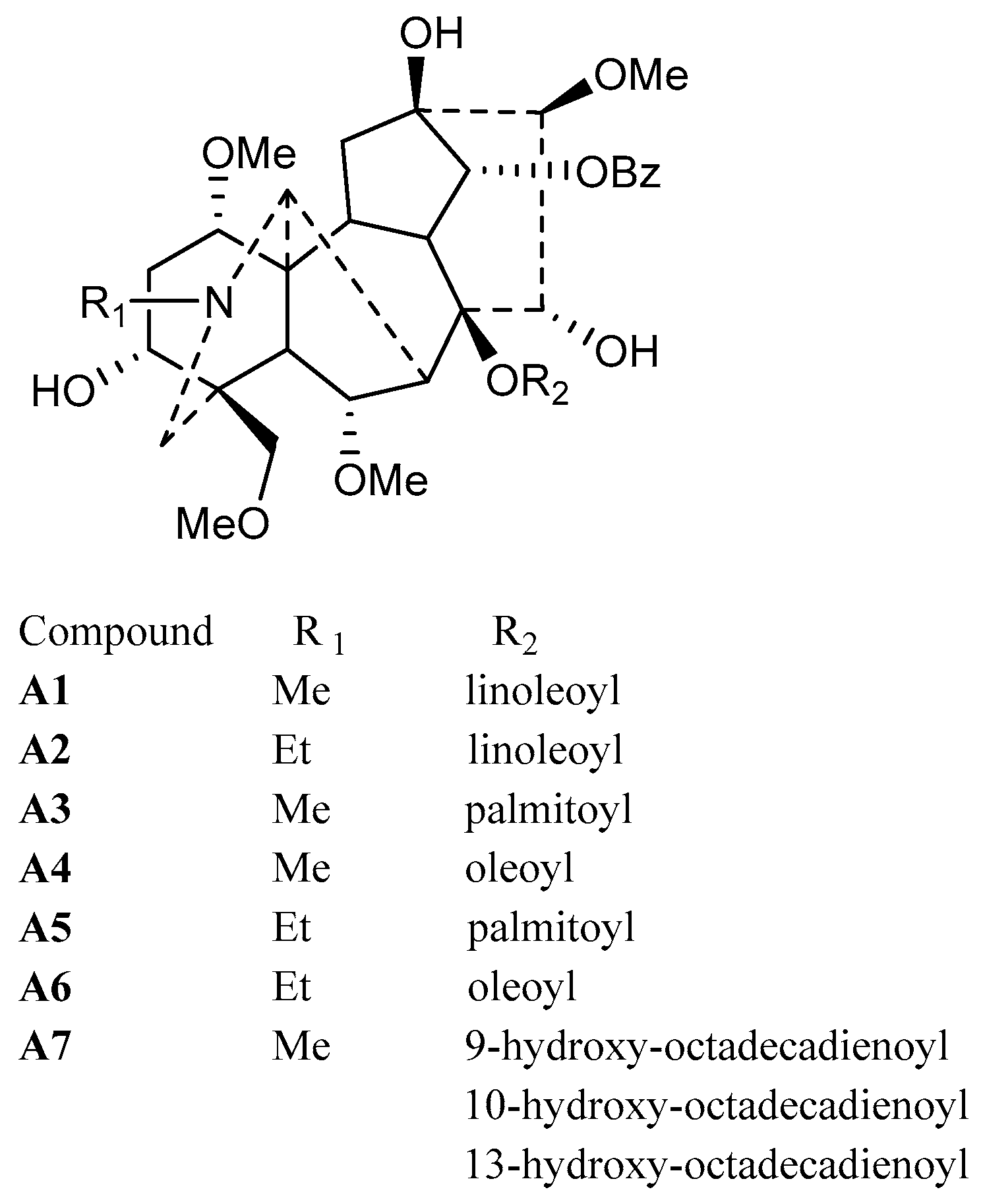
As a result, six pure lipo-alkaloids were obtained by optimized column chromatography and characterized as 8-O-linoleoyl-14-benzoylmesaconine (A1), 8-O-linoleoyl-14-benzoylaconine (A2), 8-O-palmitoyl-14-benzoylmesaconine (A3), 8-O-oleoyl-14-benzoylmesaconine (A4), 8-O-palmitoyl-14-benzoylaconine (A5) and 8-O-oleoyl-14-benzoylaconine (A6), respectively. Although these compounds were identified by LC-MS and/or semi-synthetic methods before, this is the first time that they have been purified from nature. Besides, oxygenated fatty acid-containing lipo-alkaloids were firstly obtained, and they shared the same aconitane skeleton of 14-benzoylmesaconine, while their side chains were determined to be 9-hydroxy-octadecadienoic acid, 13-hydroxy-octadecadienoic acid and 10-hydroxy-octadecadienoic acid, respectively. Their structures are shown in Figure 3.
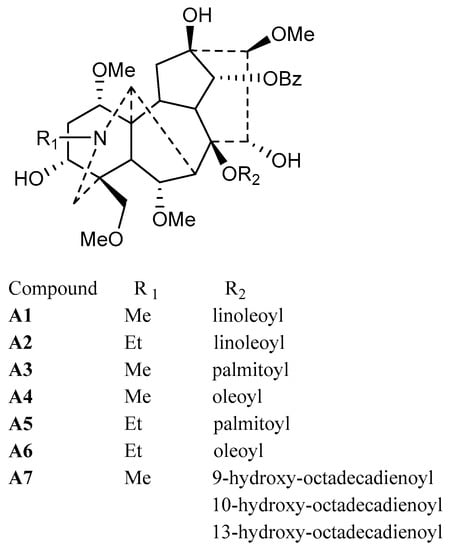
Figure 3.
Structures of isolated lipo-alkaloids (A1–A7).
2.2. Establishment of Lipo-Alkaloids Database
In the structures of the previously-reported naturally-occurring lipo-alkaloids [1,11], there were 13 basic aconitine skeletons and 51 fatty acid chains. Oxygenated fatty acids are widely present in plants; thus, it is possible that these fatty acids connect with aconitane skeletons to form the lipo-alkaloids, as reported above. All possible lipo-alkaloids can be hypothesized by the following formula:
MFlipo-alkaloid = MFbasic skeleton + MFfatty acid − H2O
Herein, MFlipo-alkaloid and MFbasic skeleton are the molecular formulae of potential lipo-alkaloids and the 13 reported aconitane skeletons, while MFfatty acid is the molecular formula of possible fatty acids, which have 3–25 carbons and 2–6 oxygen atoms with the unsaturated degrees from 1–7. Then, the names and molecular formulae of the hypothesized lipo-alkaloids were input into an Agilent MassHunter database file to establish an in-house lipo-alkaloids database with a total of 484 molecular formulae.
2.3. Determination of Lipo-Alkaloids by Combination of Database, UHPLC-Q-TOF-MS and MS/MS Analysis
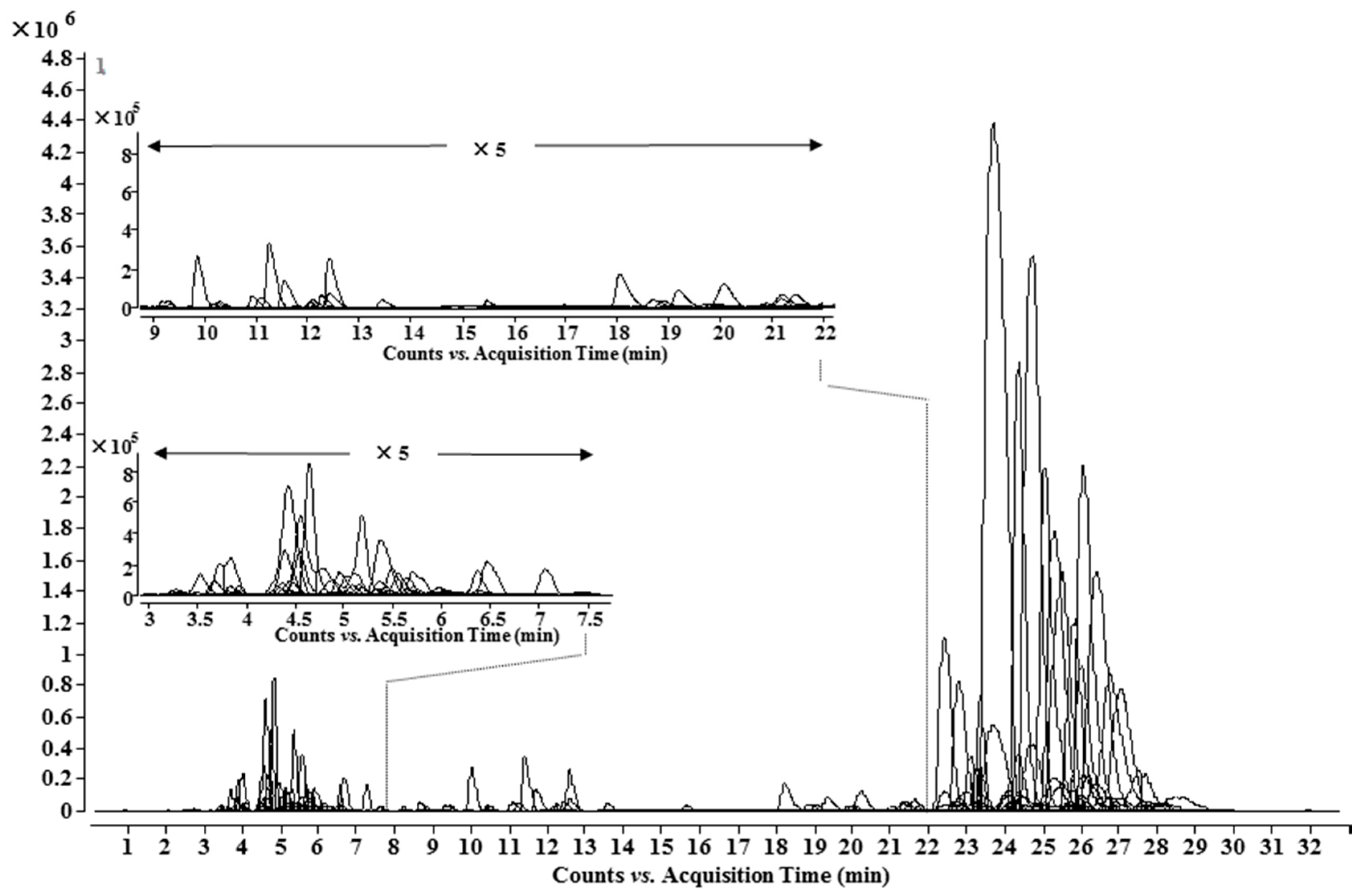
Based on information from the lipo-alkaloids database, UHPLC-Q-TOF-MS and MS/MS analysis, 148 lipo-alkaloids, including 93 potential new ones, were determined (Table 2 and Figure 4). Among them, 21 molecular formulae were found to have at least two isomers with different MS/MS spectra and/or retention times, e.g., Compounds 71 and 102, 79 and 122, 62, 72 and 109. Compounds 71 and 102 had the same molecular formula of C49H73NO11. Compound 102 has the same retention time, MS and MS/MS spectra as Compound A1, 8-O-linoleoyl-14-benzoylmesaconine. The MS/MS base peak of 71 was at m/z 556.2919, which was one oxygen less than that of A1; other fragmentation ions were also one oxygen less than the corresponding ions in A1, and no ion related to neutral loss of CO was observed (Table 1). Therefore, the basic skeleton of 71 was identified as 14-benzyolhypaconine (BHA). The fatty acid in 71 was determined to have a formula of C18H32O3, corresponding to a hydroxyoctadecadienoic acid as in Compound A7. Similarly, Compounds 79 and 122 have the same molecular formula of C49H75NO11 with two more hydrogens than 71 and A1. Compound 122 was unambiguously determined as 8-O-oleoyl-14-benzoylmesaconine based on the same retention time, MS and MS/MS spectra as A4. The MS/MS spectrum of 79 suggested a basic skeleton of BHA, so the fatty acid should have two hydrogens more than that in 71, i.e., hydroxyoctadecenoic acid (C18H34O3) in 79. Compounds 61, 72 and 109 had one oxygen more than 79 and A4, and the fragmentation patterns of 61 and 72 were almost the same as those of 79 and A4; thus, the additional oxygen should substitute on the fatty acid side chains, which are C18H34O4 and C18H34O3, respectively. The MS/MS base peak ion of Compound 109 showed one oxygen more than that of 72, and other fragmentation ions were also 16 Da more than that of Compound 72; therefore, the basic skeleton should be 10-OH-BMA. Therefore, Compound 109 was identified as 8-O-oleoyl-10-hydroxy-14-benzoylmesaconine. Similarly, by comprehensive analysis of the MS/MS spectra, 22 other fatty acid chains, e.g., C18H30O3, C18H32O4, C18H36O3 and C23H46O2, were also determined, as shown in Table 3.

Table 2.
Lipo-alkaloids identified from A. carmichaelii.
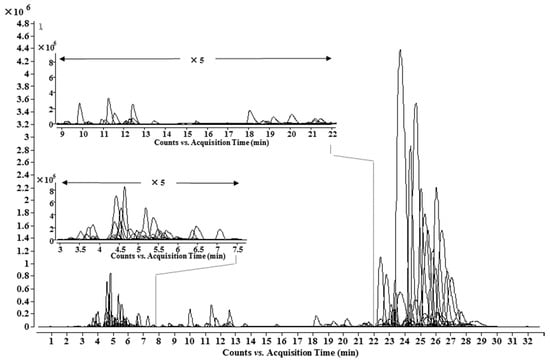
Figure 4.
UHPLC-MS chromatogram of the identified lipo-alkaloids in A. carmichaelii.

Table 3.
Fatty acid side chains in lipo-alkaloids.
In addition to 13 known aconitane skeletons included in the database, four new aconitane skeletons were found in the identified lipo-alkaloids for the first time (Figure 5). The elucidation of new skeletons is discussed below, while the MS/MS characterization of known skeletons is shown in Figure 6 and Table 1.
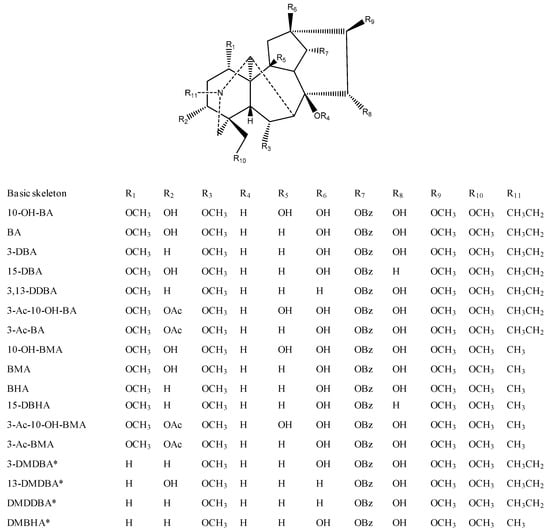
Figure 5.
Structures of aconitane skeletons of lipo-alkaloids. * Indicates the skeleton was detected for the first time.
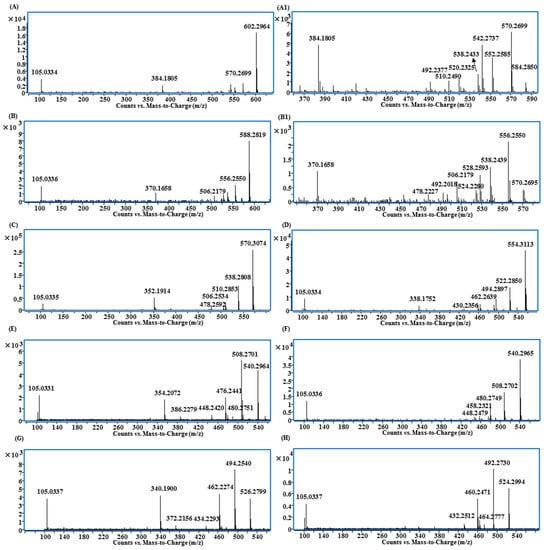
Figure 6.
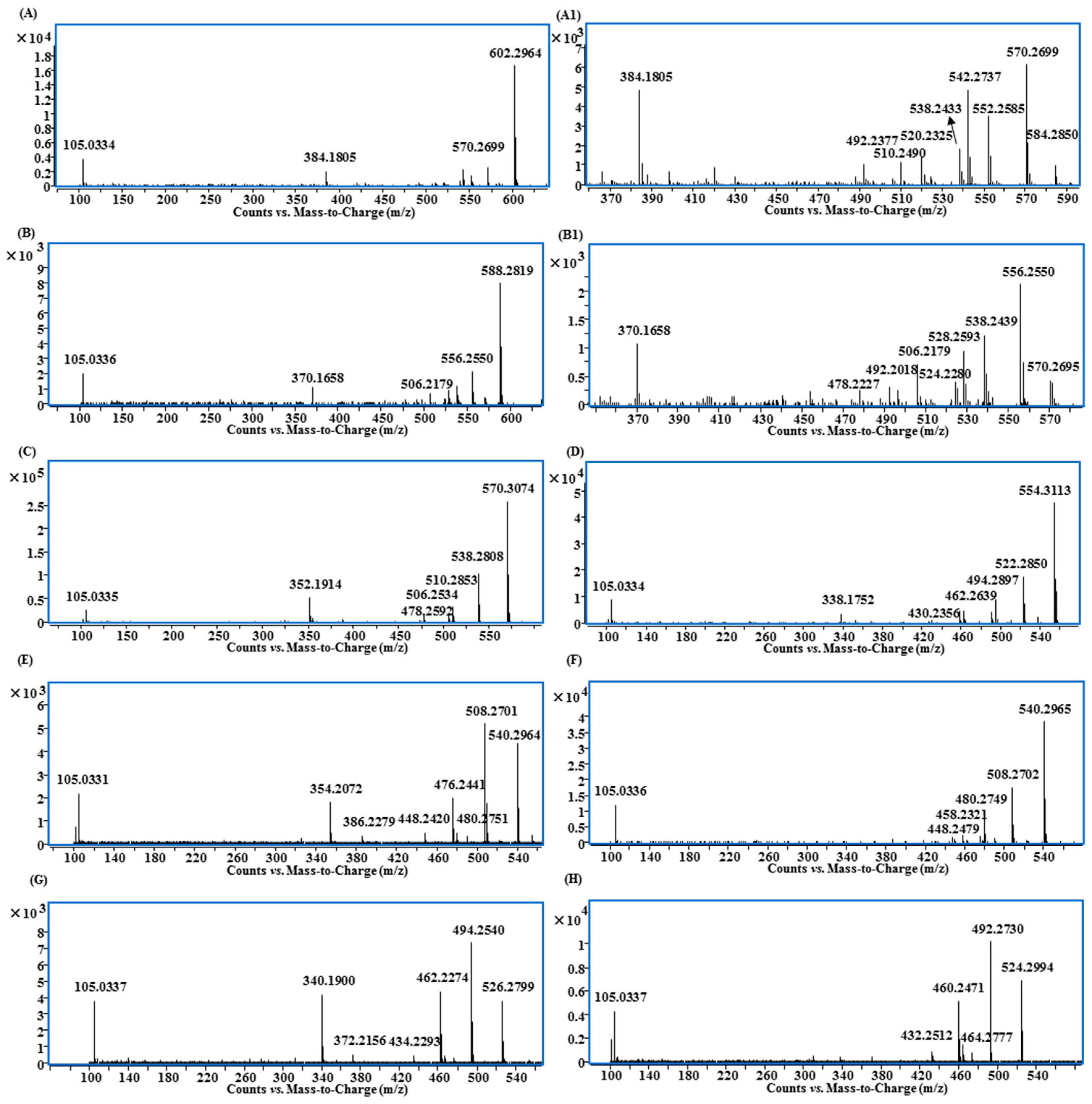
MS/MS spectra of lipo-alkaloids with the basic skeletons of: (A and A1) 10-OH-BA; (B and B1) 10-OH-BMA; (C) 3-DBA; (D) 3,13-DDBA; (E) 3-DMDBA; (F) 13-DMDBA; (G) DMBHA; (H) DMDDBA.
Compounds 85, 103, 116 and 117 shared the same fragmentation patterns, in which m/z 540.30 ([M + H − FA]+), 508.27 ([M + H − FA − CH3OH]+), 480.28 ([M + H − FA − CH3OH − CO]+), 476.24 ([M + H − FA − 2CH3OH]+), 448.24 ([M + H − FA − 2CH3OH − CO]+), 354.21 ([M + H − FA − 2CH3OH − benzoic acid]+) and 105.03 ([C6H5CO]+) were the major ions (Figure 6E). Except for the ion at m/z 354.21, all other ions contained one OCH2 group less than that of 3-deoxy-14-benzoylaconine (3-DBA) derivatives (Figure 6C); therefore the basic skeleton was determined to be demethoxy-3-deoxy-14-benzoylaconine (3-DMDBA). The ion at m/z 354.21 was derived from the loss of two methanols rather than three methanols, which in turn further confirmed that the basic skeleton had one methoxyl group less than 3-DBA with 1-OCH3 or 6-OCH3 missing (Scheme S4).
The isomers of Compounds 103, 117 and 116 were observed at the retention times of 24.5 min (97), 26.0 (115) and 26.1 min (119), respectively. They had similar MS/MS fragmentation ions, which were significantly different from Compounds 103, 117 and 116. The ions of [M + H − FA]+, [M + H − FA − CH3OH]+, [M + H − FA − CH3OH − CO]+, [M + H − FA − 2CH3OH]+, [M + H − FA − 2CH3OH − CO]+ and [C6H5CO]+ were also observed, but the relative intensities of [M + H − FA]+ and [M + H − FA − CH3OH − CO]+ were higher than others (Figure 6F). More importantly, there was no evidence of the losses of FA + 2CH3OH + benzoic acid, so that the basic skeleton should be demethoxy-13-deoxy-14-benzoylaconine (13-DMDBA).
Compounds 51, 95 and 110 had the molecular formulae of C46H71NO9, C48H71NO9 and C48H73NO9, and their fragmentation ions were mainly generated from the neutral losses of FA, methanol and benzoic acid showing at m/z 526.28, 494.25, 462.23 and 340.19 (Figure 6G and Scheme S3). The fragmentation patterns were very similar to those of 3-DMDBA derivatives (Figure 6E), but with one CH2 less. The ions corresponding to the loss of two molecules of methanol at m/z 462.23 ([M + H − FA − 2CH3OH]+) and 340.19 ([M + H − FA − 2CH3OH − benzoic acid]+) indicated that the differences were the substitution groups on the N atom, and it should be N-CH3 rather than N-C2H5 in these three compounds, i.e., the basic skeleton should be demethoxy-14-benzoylhypaconine (DMBHA).
Compounds 105, 120, 131 and 137 shared the same fragment ions at m/z 524.30, 492.27, 464.28, 460.25 and 432.25 (Figure 6H). The fragmentation patterns were very similar to those compounds with 3-DMDBA as the basic skeleton (Figure 6E), but with one oxygen less. Because there was no ion produced from the loss of FA + methanol + benzoic acid, the absence of 13-OH was indicated; therefore, the basic skeleton should be demethoxy-3,13-dideoxy-14-benzoylaconine (DMDDBA).
Based on the finding of oxygenated fatty acids as the side chains of lipo-alkaloids, the possible lipo-alkaloids were predicted and included in an in-house database. By the combination of the database, UHPLC-MS and MS/MS analysis, not only more oxygenated fatty acid-containing lipo-alkaloids were determined, but also four aconitane skeletons not reported in lipo-alkaloids before were detected. Finally, 148 lipo-alkaloids, including 93 potential new ones, were identified (Table 2). Although most of previous reports showed that the contents of lipo-alkaloids usually increased after processing, no significant difference was detected when using heat reflux extraction or ultrasonic extraction in our preliminary research (data not shown).
2.4. MS/MS Characterizations of Aconitane Skeletons in Lipo-Alkaloids
In this study, we reported 13 aconitane skeletons (including four new ones) in the lipo-alkaloids with their main fragmentation ions from the neutral losses of MeOH, H2O, CO and BzOH (Figure 6 and Table 1). Based on structures and MS/MS spectra, the relationship between the substitutions and the fragmentation ions can be summarized as follows. (1) The ions produced from the neutral loss of MeOH have higher abundance, and the numbers of methoxy group substituted on aconitane skeleton usually are determined from the corresponding ions. For instance, ions with the loss of three molecules of methanol were detected for the aconitine skeletons with tetramethoxy substitution, while ions with neutral loss of two molecules of methanol were observed for the trimethoxy-substituted skeletons. Due to the higher bond energy between C18 and the methoxy group [22], it is difficult to detect the fragment ions from the loss of C18-OMe; (2) The ion corresponding to the neutral loss of BzOH should be a diagnostic ion of 13-OH-14-OBz. The ion of [M + H − FA − 3CH3OH − BzOH]+ was observed for the lipo-alkaloids with the basic skeletons of 3-Ac-BMA, 10-OH-BA, 10-OH-MA, BA, BMA, DBA and BHA, and the ion of [M + H-FA-2CH3OH-BzOH]+ was detected from the derivatives of 3-DMDBA and DMBHA, while no evident ion was found in MS/MS spectra of the 3,13-DDBA, 13-DMDBA and 3,13-DMDDBA derivatives (Table 1). Moreover, the aforementioned ion could also be used to determine the numbers of methoxy groups substituted on the aconitane skeletons; (3) The ions of the loss of CO indicates the presence of the 15-OH-16-OMe group [11], e.g., [M + H − FA − 2CH3OH − CO]+ was observed in all identified lipo-alkaloids. (4) The loss of H2O usually indicates the substitution of a hydroxyl group at C-3. There is no such ion observed in DBA, BHA, 3, 13-DDBA, 3-DMDBA, DMBHA and 3, 13-DMDDBA.
2.5. Fatty Acid Side Chains in Lipo-Alkaloids
Besides common long chain fatty acids, medium and long chain oxidized fatty acids were detected as the side chains of lipo-alkaloids in plants for the first time, e.g., C9H16O3, C18H30O3, C18H32O3, C18H32O4, C18H34O3, C18H34O4, C18H34O5, and so on (Table 3). These oxygenated fatty acids might occur as hydroxyl-, oxo-, epoxy-, hydroperoxy-type or diacid [23]. However, due to the limitation of LC-MS data, it is difficult to determine in which form they exist in the lipo-alkaloids. In this study, three oxygenated fatty acids in a lipo-alkaloids mixture were determined by 1H-NMR, alkaline hydrolysis and MS/MS analysis, but other oxygenated fatty acid groups could not be determined due to the limited amount of sample available. Considering the polarity and occurrence of fatty acids in nature, the most possible structures were proposed in Table 2 by searching the lipid maps [24] and comparing retention times of the lipo-alkaloids to common fatty acid side chains.
Plant oxylipins are involved in the stress responses, and some of them have anti-microbial and anti-insecticidal activities [25]. Some oxylipins, e.g., 2-hydroxyoleic acid (C18H34O3), were found to have anti-cancer activity [26], while some oxylipins have anti-inflammatory activity [27]. When these oxidized fatty acids connect to aconitane alkaloids to form the lipo-alkaloids, the bioactivity and toxicity of aconitane alkaloids might change. Thus, the occurrence, bioactivity and toxicity of these oxygenated fatty acid-containing lipo-alkaloids are worth further investigations.
3. Materials and Methods
3.1. Chemicals and Reagents
AR-grade n-hexane, dichloromethane, n-butanol, methanol and HPLC-grade methanol were obtained from Anaqua Chemicals Supply (Houston, TX, USA). MS-grade acetonitrile, methanol and water were purchased from J.T. Baker (Danville, PA, USA), and MS-grade formic acid was provided by Sigma-Aldrich Laboratories, Inc. (St. Louis, MO, USA). AR-grade potassium hydroxide, hydrochloric acid (37%), and diethylamine were purchased from Merck and Advanced Technology & Industrial CO. Ltd. (Hong Kong, China), respectively. Silica gel (75–150 mesh) and ODS (35–70 µm) were provided by Grace (Columbia, MD, USA).
3.2. Plant Materials
The roots of Aconitum carmichaelii Debx. (ChW-02) were obtained from Hehuachi Medicinal Materials Market in Chengdu, Sichuan Province of China, and authenticated by Ying Liu, Chengdu University. A voucher specimen was deposited in Macau University of Science and Technology.
3.3. Separation of Lipo-Alkaloids
The air-dried roots of A. carmichaelii (7.2 kg) were powdered and soaked in methanol (12 L) at room temperature for one week and then extracted with methanol at reflux 3 times (3 × 12 L, 1 h for each extraction). The combined methanol extracts were evaporated under vacuum to give 356 g of residue, which was suspended in distilled water (3 L) followed by the participation with n-hexane (3 × 3 L), ethyl acetate (3 × 3 L) and n-butanol (3 × 3 L), successively. The n-hexane extract (32 g) was subjected to silica gel CC (6 × 60 cm) using CH2Cl2/CH3OH as the eluate to provide four fractions (A–D). Fraction D (5 g) was further subjected to ODS CC (4.5 × 50 cm) using water-containing methanol (0%–100%) as the eluate to produce five subfractions (D1–5). Subfraction D4 (1 g) was divided into 10 parts by another ODS CC with an increasing gradient of water-containing methanol (40%–100%). Preparative HPLC separation of the eighth part (D4–8, 134 mg) on an ODS column (10 × 250 mm, 5 µm) produced 7 compounds, 8 mg A1, 6 mg A2, 3 mg A3, 9 mg A4, 7 mg A5, 3 mg A6 and 4 mg A7. The mobile phases were 0.01% diethylamine-containing water (A) and methanol (B) with the following gradient: 0–40 min, 70%–95 B%; 40–120 min, 95% B. The flow rate was 2 mL/min, and the detection wavelength was set at 230 nm. The structures were characterized by NMR and mass spectrometry.
3.4. Alkaline Hydrolysis of Peak A7
One milligram of A7 was dissolved in 400 µL of KOH-saturated methanol solution and then heated to 75 °C for 15 min and 60 min. The reaction solution was neutralized with 800 µL of 5 M HCl-MeOH and participated with ethyl acetate, respectively. The ethyl acetate layer was analyzed by UHPLC-Q-TOF-MS.
3.5. Preparation of Methanol Extracts of Herbal Sample
One gram of powdered herbal sample was extracted with 6 mL methanol for 60 min with the aid of an ultrasonicator and then centrifuged at 13,000 rpm for 10 min. The supernatant was collected and diluted 10-times, then followed by the acquisition of UHPLC-Q-TOF-MS data.
3.6. UHPLC-Q-TOF-MS Analysis
Agilent 1290 UHPLC system (UHPLC, Agilent Technologies, Santa Clara, CA, USA) consisting of an autosampler, thermostated column compartment and binary pump and equipped with an Agilent Eclipse C18 column (2.1 × 100 mm, 1.8 μm, Agilent Technologies) was applied for the separation of components. The mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). Method 1 was applied for the determination of lipo-alkaloids, and the mobile phase gradient was set as follows, 0–0.5 min, 20% B; 0.5–30 min, 20%–98% B; 30–33 min 98% B; 33–33.1 min, 98%–20% B, and then maintained for 2 min. Method 2 was used for the analysis of fatty acids, and the gradient was 0–11 min, 25% B, 11–11.1 min, 25%–95% B, and then maintained for 2 min. The flow rate was 0.3 mL/min, and the injection volume was 2 µL. The mass spectrometry was conducted on a 6550 UHD Accurate-Mass Q-TOF/MS system (Agilent Technologies) with a dual Agilent Jet Stream electrospray ion source (dual AJS ESI). The mass parameters were optimized using the standards of aconitine, mesaconitine and hypaconitine and set as follows: dry gas temperature and flow were 250 °C and 15 L/min; sheath gas temperature and flow were 300 °C and 11 L/min; nebulizer at 20 psi; the capillary and nozzle voltages were 4000 and 500 V, respectively. The fragmentor was 380 V, and the collision cell energies were set at 50 eV for lipo-alkaloids in positive mode and 30 eV for fatty acids in negative mode, respectively.
3.7. Establishment of the Lipo-Alkaloids Database
Based on the possible fatty acid chains and known aconitane skeletons reported in Aconitum plants, the possible lipo-alkaloids were hypothesized and input into Agilent MassHunter database file (“Compound Formula Database”) to establish an in-house lipo-alkaloids database. Then, the potential lipo-alkaloids in A. carmichaelii were extracted using the function of “Find Compounds by Formula (FBF)” and determined by MS/MS analysis.
4. Conclusions
In this study, the separation method of lipo-alkaloids was optimized, and using this method, oxygenated fatty acids-containing lipo-alkaloids were obtained for the first time. A lipo-alkaloids database was established based on the known basic aconitane skeletons and possible fatty acid side chains. By using the database, potential lipo-alkaloids were first extracted from UHPLC-Q-TOF-MS, and then, the structures were determined from the comprehensive analysis and deduction of MS/MS spectra, resulting in successful identification of 148 lipo-alkaloids. Among them, 38 compounds contain medium or long chain oxidized fatty acids as side chains that were not reported previously. The combination of database and LC-MS dramatically speeds up the finding of potential new compounds and is confirmed to be a powerful tool in the study of natural product chemistry. The new finding of oxygenated fatty acids as side chains of lipo-alkaloids provides a kind of possible structures, which accounts for the bioactivities of A. carmichaelii, a widely-used traditional medicine.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/4/437/s1.
Acknowledgments
This work was supported by the Macao Science and Technology Development Fund (086/2013/A3).
Author Contributions
Liang Liu and Na Li conceived of and designed the experiments. Ying Liang, Jian-Lin Wu, Guanyu Yan and Na Li performed the experiments and analyzed the data. Ying Liu collected and authenticated the herbs. Elaine Lai-Han Leung, Hua Zhou, Zhongqiu Liu and Na Li wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviation
The following abbreviations are used in this manuscript:
| UHPLC-Q-TOF-MS | ultra-high performance liquid chromatography-quadrupole-time of flight-mass spectrometry |
| BA | 14-benzoylaconine |
| BMA | 14-benzoylmesaconine |
| BHA | 14-benzoylhypaconine |
| 10-OH-BA | 10-hydroxy-14-benzoylaconine |
| 3-DBA | 3-deoxy-14-benzoylaconine |
| 15-DBA | 15-deoxy-14-benzoylaconine |
| 3,13-DDBA | 3,13-dideoxy-14-benzoylaconine |
| 10-OH-BMA | 10-hydroxy-14-benzoylmesaconine |
| 15-DBHA | 15-deoxy-14-benzoylhypaconine |
| 3-Ac-10-OH-BA | 3-acetyl-10-hydroxy-14-benzoylaconine |
| 3-Ac-10-OH-BMA | 3-acetyl-10-hydroxy-14-benzoylmesaconine |
| 3-Ac-BA | 3-acetyl-14-benzoylaconine |
| 3-Ac-BMA | 3-acetyl-14-benzoylmesaconine |
| 3-DMDBA | demethoxy-3-deoxy-14-benzoylaconine |
| 13-DMDBA | demethoxy-13-deoxy-14-benzoylaconine |
| DMDDBA | demethoxy-3,13-dideoxy-14-benzoylaconine |
| DMBHA | demethoxy-14-benzoylhypaconine |
References
- Borcsa, B.; Csupor, D.; Forgo, P.; Widowitz, U.; Bauer, R.; Hohmann, J. Aconitum lipo-alkaloids-semisynthetic products of the traditional medicine. Nat. Prod. Commun. 2011, 6, 527–536. [Google Scholar] [PubMed]
- Chodoeva, A.; Bosc, J.J.; Guillon, J.; Costet, P.; Decendit, A.; Merillon, J.M.; Leger, J.M.; Jarry, C.; Robert, J. Hemisynthesis and antiproliferative properties of mono-[O-(14-benzoylaconine-8-yl)]esters and bis-[O-(14-benzoylaconine-8-yl)]esters. Eur. J. Med. Chem. 2012, 54, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Song, F.R.; Yue, H.; Guo, X.H.; Li, H.L.; Liu, Z.Q.; Liu, S.Y. Biotransformation of Deoxyaconitine of Metabolite of Aconitine by Human Intestinal Bacteria and Electrospray Ionization Tandem Mass Spectrometry. Chem. J. Chin. Univ. 2007, 28, 2051–2055. [Google Scholar]
- Zhao, Y.F.; Song, F.R.; Guo, X.H.; Liu, S.Y. Studies on the biotransformation of aconitine in human intestinal bacteria using soft-ionization mass spectrometry. Chem. J. Chin. Univ. 2008, 29, 55–59. [Google Scholar] [CrossRef]
- Zhao, Y.F.; Song, F.R.; Wang, X.Y.; Guo, X.H.; Liu, Z.Q.; Liu, S.Y. Studies on the biotransformation of 16-O-demethylaconitine and electrospray ionization tandem mass spectrometry. Acta Chim. Sin. 2008, 66, 525–530. [Google Scholar]
- Wang, Y.; Liu, Z.; Song, F.; Liu, S. Electrospray ionization tandem mass spectrometric study of the aconitines in the roots of aconite. Rapid Commun. Mass Spectrom. 2002, 16, 2075–2082. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liang, Z.; Zhao, Z.; Cai, Z. Direct analysis of alkaloid profiling in plant tissue by using matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 2007, 42, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Pi, Z.; Song, F.; Liu, Z.; Cai, Z.; Liu, S. Studies on the aconitine-type alkaloids in the roots of Aconitum carmichaeli Debx. by HPLC/ESIMS/MS(n). Talanta 2009, 77, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Song, F.; Cui, M.; Liu, S. Simultaneous determination of lipo-alkaloids extracted from Aconitum carmiechaeli using electrospray ionization mass spectrometry and multiple tandem mass spectrometry. Planta Med. 1999, 65, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Csupor, D.; Wenzig, E.M.; Zupko, I.; Wolkart, K.; Hohmann, J.; Bauer, R. Qualitative and quantitative analysis of aconitine-type and lipo-alkaloids of Aconitum carmichaelii roots. J. Chromatogr. A 2009, 1216, 2079–2086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, Z.H.; Qiu, X.H.; Yang, Y.M.; Zhu da, Y.; Xu, W. Neutral fragment filtering for rapid identification of new diester-diterpenoid alkaloids in roots of Aconitum carmichaeli by ultra-high-pressure liquid chromatography coupled with linear ion trap-orbitrap mass spectrometry. PLoS ONE 2012, 7, e52352. [Google Scholar] [CrossRef] [PubMed]
- Chodoeva, A.; Bosc, J.J.; Guillon, J.; Decendit, A.; Petraud, M.; Absalon, C.; Vitry, C.; Jarry, C.; Robert, J. 8-O-Azeloyl-14-benzoylaconine: A new alkaloid from the roots of Aconitum karacolicum Rapcs and its antiproliferative activities. Bioorg. Med. Chem. 2005, 13, 6493–6501. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.P.; Chen, Q.H. The C19-diterpenoid alkaloids. Alkaloids. Chem. Biol. 2010, 69, 1–577. [Google Scholar] [PubMed]
- Levison, B.S.; Zhang, R.; Wang, Z.; Fu, X.; DiDonato, J.A.; Hazen, S.L. Quantification of fatty acid oxidation products using online high-performance liquid chromatography tandem mass spectrometry. Free Radic. Biol. Med. 2013, 59, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Garscha, U.; Oliw, E.H. Steric analysis of 8-hydroxy- and 10-hydroxyoctadecadienoic acids and dihydroxyoctadecadienoic acids formed from 8R-hydroperoxyoctadecadienoic acid by hydroperoxide isomerases. Anal. Biochem. 2007, 367, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, F.; Xu, Q.; Liu, Z.; Liu, S. Characterization of aconitine-type alkaloids in the flowers of Aconitum kusnezoffii by electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 2003, 38, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Pi, Z.; Wang, X.; Song, F.; Liu, S. HPLC/ESI-MSn and ESI-MS studies on the Aconitum alkaloids in three Chinese medicinal herbs. J. Sep. Sci. 2010, 33, 2898–2906. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Pi, Z.; Song, F.; Liu, Z.; Liu, S. Studies on the Components of Aconitum carmichaeli by Using HPLC/ESI-MSn. Acta Chem. Sin. 2008, 66, 211–215. [Google Scholar]
- Yue, H.; Pi, Z.F.; Zhao, Y.F.; Song, F.R.; Liu, Z.Q.; Liu, S.Y. Investigation of Aconitine-type Alkaloids from Processed Tuber of Aconitum carmiechaeli by HPLC-ESI-MS/MSn. Chem. Res. Chin. Univ. 2007, 23, 625–627. [Google Scholar] [CrossRef]
- Bai, Y.; Desai, H.K.; Pelletier, S.W. Long-chain fatty acid esters of some norditerpenoid alkaloids. J. Nat. Prod. 1994, 57, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Liu, Z.Q.; Wang, Y.; Song, F.R.; Xu, Q.X.; Liu, N. Synthesis of Aconitum Alkaloid Type Fatty Acid Ester. Patent CN 1,580,047 A, 16 February 1995. [Google Scholar]
- hen, L.H.; Jian, L.J.; Su, Z.M.; Qiu, Y.Q.; Wang, Y.; Liu, S.Y. ESI-MSn Behavior and Quantum Chemistry Calculation of Stability of Fragment Ions of Diester-diterpenoid Alkaloids (DDA). Chem. J. Chin. Univ. 2005, 26, 2340–2344. [Google Scholar]
- Gobel, C.; Feussner, I. Methods for the analysis of oxylipins in plants. Phytochemistry 2009, 70, 1485–1503. [Google Scholar] [CrossRef] [PubMed]
- Lipidomics Gateway. Available online: http://www.lipidmaps.org (accessed on 21 March 2016).
- Blee, E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002, 7, 315–322. [Google Scholar] [CrossRef]
- Martinez, J.; Vogler, O.; Casas, J.; Barcelo, F.; Alemany, R.; Prades, J.; Nagy, T.; Baamonde, C.; Kasprzyk, P.G.; Teres, S.; et al. Membrane structure modulation, protein kinase C alpha activation, and anticancer activity of minerval. Mol. Pharmacol. 2005, 67, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.G.; Park, Y.M.; Lu, Y.; Chang, H.W.; Na, M.; Lee, S.H. Inhibition of prostaglandin D(2) production by trihydroxy fatty acids isolated from Ulmus davidiana var. Japonica. Phytother. Res. 2013, 27, 1376–1380. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).