Metabolites of Siamenoside I and Their Distributions in Rats
Abstract
:1. Introduction
2. Results
2.1. Profiling the Metabolites of Siamenoside I in Different Biosamples by HPLC-ESI-IT-TOF-MSn
2.2. Identification of the Metabolites of Siamenoside I in Different Biosamples by HPLC-ESI-IT-TOF-MSn
| No. | tR (min) | Meas. (Da) | Pred.(Da) | Err. (ppm) | DBE 2 | Formula | Identification | Reactions |
|---|---|---|---|---|---|---|---|---|
| M0 1 | 24.966 | 1169.5959 | 1169.5961 | 0.17 | 9 | C54H92O24 | siamenoside I | − |
| M1 | 25.698 | 1285.6444 | 1285.6434 | 0.78 | 10 | C60H102O29 | mogroside V isomer | +Glc |
| M2 | 26.043 | 1285.6441 | 1285.6434 | 0.54 | 10 | C60H102O29 | mogroside V isomer | +Glc |
| M3 1 | 25.345 | 1169.5905 | 1169.5961 | −0.09 | 9 | C54H92O24 | mogroside IVA | isomerization |
| M4 1 | 25.960 | 1169.5923 | 1169.5961 | −3.25 | 9 | C54H92O24 | mogroside IVE | isomerization |
| M5 | 26.696 | 1169.5950 | 1169.5961 | −0.94 | 9 | C54H92O24 | mogroside IV isomer | isomerization |
| M6 | 25.923 | 1167.5730 | 1167.5804 | −2.23 | 10 | C54H90O24 | dehydrogenated siamenoside I | −2H |
| M7 | 25.983 | 1153.5957 | 1153.6011 | −4.25 | 9 | C54H92O23 | deoxygenated siamenoside I | −O |
| M8 1 | 26.803 | 1007.5425 | 1007.5432 | −0.69 | 8 | C48H82O19 | mogroside III | −Glc |
| M9 1 | 27.173 | 1007.5387 | 1007.5432 | −4.47 | 8 | C48H82O19 | mogroside IIIE | −Glc |
| M10 1 | 30.217 | 1007.5376 | 1007.5432 | −5.56 | 8 | C48H82O19 | mogroside IIIA1 | −Glc |
| M11 | 30.525 | 1007.5395 | 1007.5432 | −3.67 | 8 | C48H82O19 | mogroside III isomer | −Glc |
| M12 | 30.940 | 1007.5385 | 1007.5432 | −4.66 | 8 | C48H82O19 | mogroside III isomer | −Glc |
| M13 | 26.927 | 1005.5269 | 1005.5276 | 0.70 | 9 | C48H80O19 | dehydrogenated mogroside III isomer | −Glc − 2H |
| M14 | 27.728 | 991.5441 | 991.5483 | −4.24 | 8 | C48H82O18 | deoxygenated mogroside III isomer | −Glc − O |
| M15 1 | 29.365 | 845.4881 | 845.4904 | −2.72 | 7 | C42H72O14 | mogroside IIE | −2Glc |
| M16 | 30.648 | 845.4944 | 845.4904 | 4.73 | 7 | C42H72O14 | mogroside II isomer | −2Glc |
| M17 1 | 31.775 | 845.4910 | 845.4904 | 0.71 | 7 | C42H72O14 | mogroside IIA2 | −2Glc |
| M18 | 33.298 | 845.4884 | 845.4904 | −2.37 | 7 | C42H72O14 | mogroside II isomer | −2Glc |
| M19 | 33.905 | 845.4918 | 845.4904 | 1.66 | 7 | C42H72O14 | mogroside II isomer | −2Glc |
| M20 1 | 29.908 | 843.4737 | 843.4748 | −1.30 | 8 | C42H70O14 | 11-oxomogroside IIE | −2Glc − 2H |
| M21 | 33.604 | 843.4726 | 843.4748 | −2.61 | 8 | C42H70O14 | dehydrogenated mogroside II isomer | −2Glc − 2H |
| M22 | 34.028 | 829.4946 | 829.4955 | −1.08 | 7 | C42H72O13 | deoxygenated mogroside II isomer | −2Glc − O |
| M23 | 34.813 | 827.4780 | 827.4798 | −2.18 | 8 | C42H70O13 | dehydrogenated deoxygenated mogroside II isomer | −2Glc − 2H − O |
| M24 | 34.997 | 683.4348 | 683.4376 | −1.76 | 6 | C36H62O9 | mogroside IA1 | −3Glc |
| M25 | 37.507 | 683.4366 | 683.4376 | 1.46 | 6 | C36H62O9 | mogroside IE1 | −3Glc |
| M26 | 36.120 | 681.4196 | 681.4219 | −3.38 | 7 | C36H60O9 | dehydrogenated mogroside I isomer | −3Glc − 2H |
| M27 | 38.860 | 681.4215 | 681.4219 | −0.59 | 7 | C36H60O9 | dehydrogenated mogroside I isomer | −3Glc − 2H |
| M28 | 45.978 | 521.3834 | 521.3848 | −2.69 | 5 | C30H52O4 | mogrol isomer | −4Glc |
| M29 1 | 46.467 | 521.3838 | 521.3848 | −1.92 | 5 | C30H52O4 | mogrol | −4Glc |
| M30 | 52.478 | 519.3676 | 519.3691 | −2.89 | 6 | C30H50O4 | dehydrogenated mogrol | −4Glc − 2H |
| M31 | 52.953 | 519.3687 | 519.3691 | −0.77 | 6 | C30H50O4 | dehydrogenated mogrol | −4Glc − 2H |
| M32 | 23.007 | 553.3721 | 553.3746 | −4.52 | 5 | C30H52O6 | dihydroxylated mogrol | −4Glc + 2O |
| M33 | 26.528 | 553.3699 | 553.3746 | −8.49 | 5 | C30H52O6 | dihydroxylated mogrol | −4Glc + 2O |
| M34 | 27.482 | 553.3717 | 553.3746 | −5.24 | 5 | C30H52O6 | dihydroxylated mogrol | −4Glc + 2O |
| M35 | 28.152 | 553.3740 | 553.3746 | −1.08 | 5 | C30H52O6 | dihydroxylated mogrol | −4Glc + 2O |
| M36 | 26.475 | 551.3559 | 551.3589 | −5.40 | 6 | C30H50O6 | dehydrogenated dihydroxylated mogrol | −4Glc − 2H + 2O |
| M37 | 27.050 | 551.3557 | 551.3589 | −5.80 | 6 | C30H50O6 | dehydrogenated dihydroxylated mogrol | −4Glc − 2H + 2O |
| M38 | 29.487 | 551.3566 | 551.3589 | −4.17 | 6 | C30H50O6 | dehydrogenated dihydroxylated mogrol | −4Glc − 2H + 2O |
| M39 | 30.887 | 551.3558 | 551.3589 | −5.62 | 6 | C30H50O6 | dehydrogenated dihydroxylated mogrol | −4Glc − 2H + 2O |
| M40 | 31.537 | 551.3561 | 551.3589 | −5.04 | 6 | C30H50O6 | dehydrogenated dihydroxylated mogrol | −4Glc − 2H + 2O |
| M41 | 33.122 | 551.3549 | 551.3589 | −7.25 | 6 | C30H50O6 | dehydrogenated dihydroxylated mogrol | −4Glc − 2H + 2O |
| M42 | 16.305 | 569.3655 | 569.3695 | −7.03 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M43 | 16.728 | 569.3671 | 569.3695 | −4.22 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M44 | 17.280 | 569.3660 | 569.3695 | −6.15 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M45 | 18.140 | 569.3674 | 569.3695 | −3.69 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M46 | 18.923 | 569.3675 | 569.3695 | −3.51 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M47 | 21.464 | 569.3675 | 569.3695 | −3.51 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M48 | 21.755 | 569.3674 | 569.3695 | −3.69 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M49 | 22.121 | 569.3666 | 569.3695 | −5.09 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M50 | 22.531 | 569.3667 | 569.3695 | −7.03 | 5 | C30H52O7 | trihydroxylated mogrol | −4Glc + 3O |
| M51 | 11.410 | 567.3489 | 567.3539 | −8.80 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M52 | 18.482 | 567.3463 | 567.3539 | −13.4 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M53 | 19.893 | 567.3534 | 567.3539 | −2.64 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M54 | 20.977 | 567.3494 | 567.3539 | −0.88 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M55 | 21.761 | 567.3512 | 567.3539 | −7.93 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M56 | 22.365 | 567.3507 | 567.3539 | −4.76 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M57 | 24.123 | 567.3497 | 567.3539 | −5.64 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M58 | 24.478 | 567.3508 | 567.3539 | −7.23 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M59 | 25.380 | 567.3499 | 567.3539 | −5.46 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M60 | 27.050 | 567.3498 | 567.3539 | −7.05 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M61 | 27.728 | 567.3520 | 567.3539 | −7.23 | 6 | C30H50O7 | dehydrogenated trihydroxylated mogrol | −4Glc − 2H + 3O |
| M62 | 24.412 | 565.3351 | 565.3382 | −5.48 | 7 | C30H48O7 | didehydrogenated trihydroxylated mogrol | −4Glc − 4H + 3O |
| M63 | 26.052 | 565.3357 | 565.3382 | −4.42 | 7 | C30H48O7 | didehydrogenated trihydroxylated mogrol | −4Glc − 4H + 3O |
| M64 | 28.810 | 565.3368 | 565.3382 | −2.48 | 7 | C30H48O7 | didehydrogenated trihydroxylated mogrol | −4Glc − 4H + 3O |
| M65 | 30.340 | 565.3347 | 565.3382 | −6.19 | 7 | C30H48O7 | didehydrogenated trihydroxylated mogrol | −4Glc − 4H + 3O |
| M66 | 13.872 | 585.3613 | 585.3644 | −5.30 | 5 | C30H52O8 | tetrahydroxylated mogrol | −4Glc + 4O |
| M67 | 14.242 | 585.3601 | 585.3644 | −7.35 | 5 | C30H52O8 | tetrahydroxylated mogrol | −4Glc + 4O |
| M68 | 14.603 | 585.3608 | 585.3644 | −6.15 | 5 | C30H52O8 | tetrahydroxylated mogrol | −4Glc + 4O |
| M69 | 18.307 | 585.3603 | 585.3644 | −4.95 | 5 | C30H52O8 | tetrahydroxylated mogrol | −4Glc + 4O |
| M70 | 19.408 | 585.3613 | 585.3644 | −5.30 | 5 | C30H52O8 | tetrahydroxylated mogrol | −4Glc + 4O |
| M71 | 15.573 | 583.3444 | 583.3488 | −7.54 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M72 | 15.997 | 583.3454 | 583.3488 | −8.54 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M73 | 20.492 | 583.3454 | 583.3488 | −6.00 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M74 | 20.800 | 583.3465 | 583.3488 | −3.94 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M75 | 21.453 | 583.3465 | 583.3488 | −3.94 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M76 | 22.895 | 583.3452 | 583.3488 | −6.17 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M77 | 24.710 | 583.3447 | 583.3488 | −7.03 | 6 | C30H50O8 | dehydrogenated tetrahydroxylated mogrol | −4Glc − 2H + 4O |
| M78 | 20.615 | 581.3297 | 581.3331 | −5.85 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M79 | 21.815 | 581.3290 | 581.3331 | −7.05 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M80 | 23.007 | 581.3292 | 581.3331 | −6.71 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M81 | 23.433 | 581.3306 | 581.3331 | −4.03 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M82 | 23.988 | 581.3308 | 581.3331 | −3.96 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M83 | 25.682 | 581.3304 | 581.3331 | −4.64 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M84 | 25.990 | 581.3291 | 581.3331 | −4.00 | 7 | C30H48O8 | didehydrogenated tetrahydroxylated mogrol | −4Glc − 4H + 4O |
| M85 | 17.707 | 587.2977 | 587.2992 | −2.55 | 7 | C30H48O9 | didehydrogenated pentahydroxylated mogrol | −4Glc − 4H + 5O |
| M86 | 18.607 | 587.2983 | 587.2992 | −1.53 | 7 | C30H48O9 | didehydrogenated pentahydroxylated mogrol | −4Glc − 4H + 5O |
2.2.1. Metabolites Formed by Monoglycosylation (M1, M2)
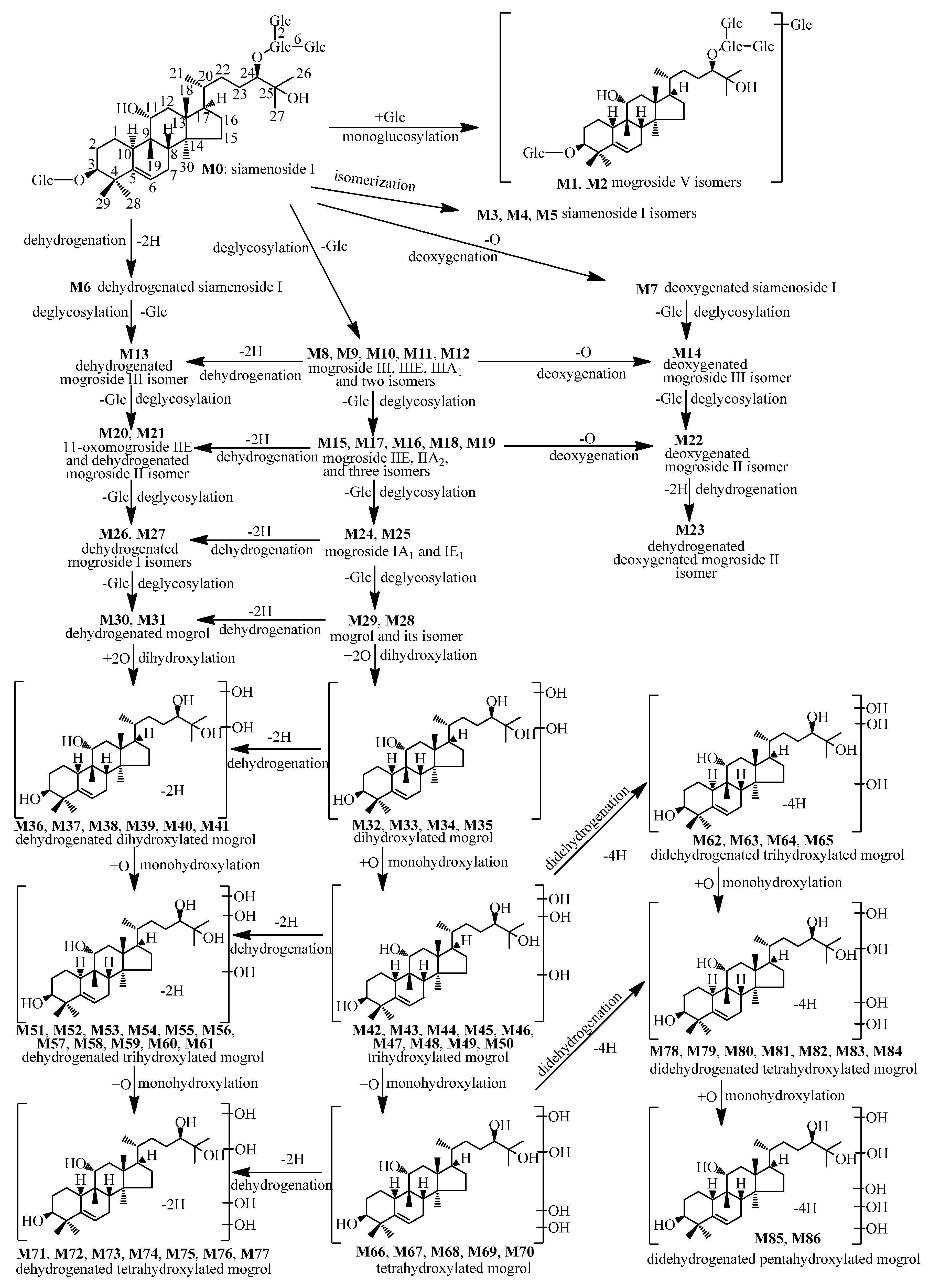
2.2.2. Metabolites Formed by Isomerization (M3–M5)
2.2.3. Metabolites Formed by Dehydrogenation (M6)
2.2.4. Metabolites Formed by Deoxygenation (M7)
2.2.5. Metabolites Formed by Deglucosylation (M8–M12)
2.2.6. Metabolites Formed by Deglucosylation and Dehydrogenation (M13)
2.2.7. Metabolites Formed by Deglucosylation and Deoxygenation (M14)
2.2.8. Metabolites Formed By Dideglucosylation (M15–M19)
2.2.9. Metabolites Formed by Dideglucosylation and Dehydrogenation (M20–M21)
2.2.10. Metabolites Formed by Dideglucosylation and Deoxygenation (M22)
2.2.11. Metabolites Formed by Dideglucosylation, Dehydrogenation, and Deoxygenation (M23)
2.2.12. Metabolites Formed by Trideglucosylation (M24–M25)
2.2.13. Metabolites Formed by Trideglucosylation and Dehydrogenation (M26–M27)
2.2.14. Metabolites Formed by Tetradeglucosylation (M28–M29)
2.2.15. Metabolites Formed by Tetradeglucosylation and Dehydrogenation (M30–M31)
2.2.16. Metabolites Formed by Tetradeglucosylation and Dihydroxylation (M32–M35)
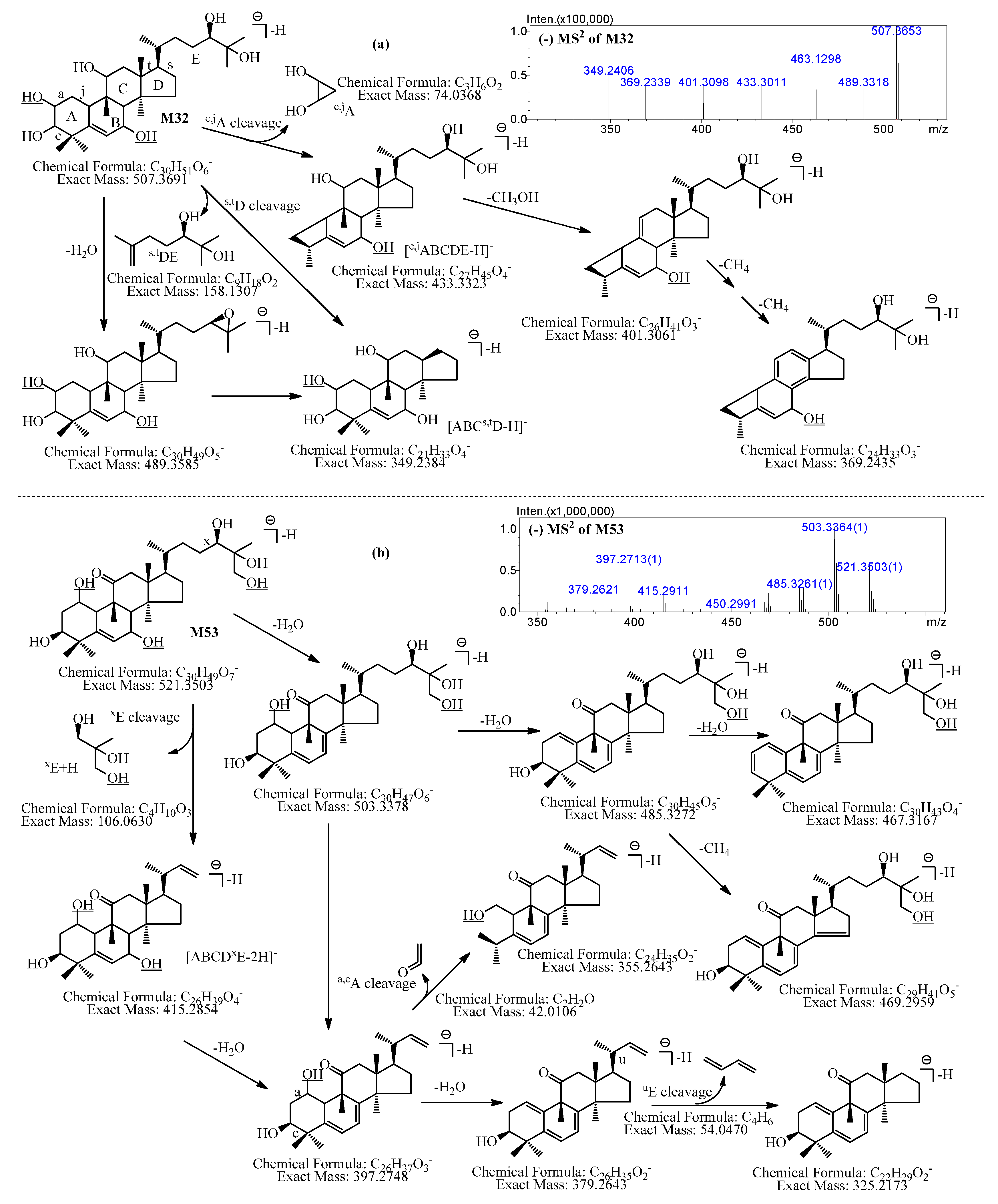
2.2.17. Metabolites Formed by Tetradeglucosylation, Dihydroxylation, and Dehydrogenation (M36–M41)
2.2.18. Metabolites Formed by Tetradeglucosylation and Trihydroxylation (M42–M50)
2.2.19. Metabolites Formed by Tetradeglucosylation, Trihydroxylation, and Dehydrogenation (M51–M61)
2.2.20. Metabolites Formed by Tetradeglucosylation, Trihydroxylation, and Didehydrogenation (M62–M65)
2.2.21. Metabolites Formed by Tetradeglucosylation and Tetrahydroxylation (M66–M70)
2.2.22. Metabolites Formed by Tetradeglucosylation, Tetrahydroxylation, and Dehydrogenation (M71–M77)
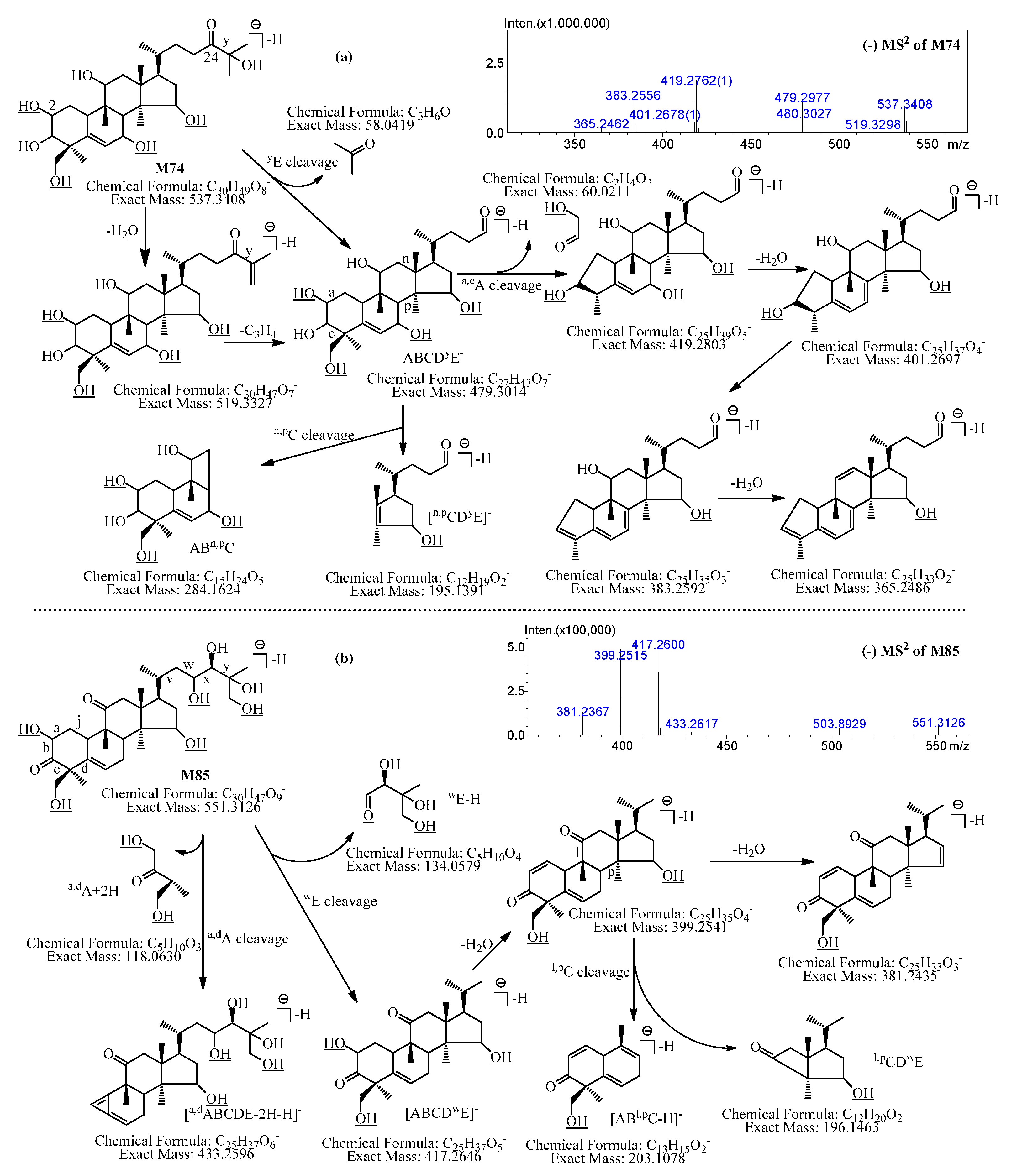
2.2.23. Metabolites Formed by Tetradeglucosylation, Tetrahydroxylation, and Didehydrogenation (M78–M84)
2.2.24. Metabolites Formed by Tetradeglucosylation, Pentahydroxylation, and Didehydrogenation (M85–M86)
2.3. Distribution of the Metabolites of Siamenoside I in Rats
| No. | Feces | Urine | Plasma | Heart 1 | Liver 1 | Spleen 1 | Lung 1 | Kidney 1 | Stomach 1 | Intestine 1 | Brain 1 | Muscle | TPA 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M0 | 9,489,561 | 45,194,235 | 929,711 | 655,725 | 1,151,491 | 15,473,808 | 1,201,5247 | 1,162,190 | 86,071,968 | ||||
| M1 | 1,144,657 | 9,555,900 | 2,001,769 | 1,487,067 | 680,786 | 14,870,179 | |||||||
| M2 | 15,997,951 | 531,345 | 16,529,296 | ||||||||||
| M3 | 760,580 | 847,869 | 1,307,673 | 1,655,097 | 2,264,310 | 177,956 | 7,013,485 | ||||||
| M4 | 545,593 | 5,114,666 | 2,993,972 | 1,200,234 | 2,991,375 | 1,454,847 | 2,022,516 | 66,665 | 16,389,868 | ||||
| M5 | 1,984,320 | 2,244,041 | 3,096,436 | 301,433 | 7,626,230 | ||||||||
| M6 | 1,370,701 | 24,868,935 | 522,375 | 216,179 | 460,499 | 5,658,908 | 2,342,276 | 8,964,568 | 44,404,441 | ||||
| M7 | 151,476 | 1,610,485 | 1,761,961 | ||||||||||
| M8 | 686,710 | 509,105 | 9,795,285 | 3,463,191 | 14,454,291 | ||||||||
| M9 | 5,499,380 | 4,564,759 | 247,245 | 3,396,211 | 10,439,099 | 1,349,085 | 621,654 | 17,082,221 | 23,756,273 | 15,351,670 | 7,796,211 | 90,103,808 | |
| M10 | 25,071,251 | 25,071,251 | |||||||||||
| M11 | 6,584,321 | 6,584,321 | |||||||||||
| M12 | 7,864,503 | 7,864,503 | |||||||||||
| M13 | 5,673,582 | 927,152 | 1,104,655 | 1,569,801 | 126,725 | 2,884,615 | 10,255,235 | 861,731 | 23,403,496 | ||||
| M14 | 4,700,498 | 1,594,595 | 2,186,093 | 8,481,186 | |||||||||
| M15 | 122,469,048 | 2,086,502 | 752,082 | 1,976,,058 | 9,533,756 | 34,065,738 | 170,883,184 | ||||||
| M16 | 10,029,179 | 10,029,179 | |||||||||||
| M17 | 14,873,427 | 3,964,569 | 18,837,996 | ||||||||||
| M18 | 131,377,697 | 2,775,395 | 2,221,834 | 1,382,313 | 6,051,761 | 868,285 | 144,677,285 | ||||||
| M19 | 45,423,120 | 394,065 | 8,102,455 | 562,370 | 54,482,010 | ||||||||
| M20 | 50,669,383 | 447,658 | 1,165,195 | 34,649,284 | 86,931,520 | ||||||||
| M21 | 50,269,416 | 885,941 | 128,120 | 905,385 | 52,188,862 | ||||||||
| M22 | 162,025,962 | 861,129 | 162,887,091 | ||||||||||
| M23 | 29,484,814 | 29,484,814 | |||||||||||
| M24 | 180,532,696 | 352,905 | 685,128 | 1,649,227 | 184,835,243 | ||||||||
| M25 | 896,803,169 | 4,472,844 | 29,155,815 | 486,612 | 21,253,177 | 297,909,967 | 1,250,081,584 | ||||||
| M26 | 48,856,108 | 1,376,742 | 598,621 | 50,831,471 | |||||||||
| M27 | 790,013,598 | 2,541,940 | 18,180,494 | 810,736,032 | |||||||||
| M28 | 92,475,813 | 1,666,710 | 1,426,867 | 95,569,390 | |||||||||
| M29 | 646,804,735 | 459,452 | 313,543 | 12,256,309 | 1,839,703 | 661,673,742 | |||||||
| M30 | 24,183,728 | 24,183,728 | |||||||||||
| M31 | 324,786,331 | 5,151,046 | 329,937,377 | ||||||||||
| M32 | 390,217,566 | 818,492 | 391,036,058 | ||||||||||
| M33 | 46,223,582 | 46,223,582 | |||||||||||
| M34 | 125,631,723 | 2,735,164 | 1,822,325 | 1,674,281 | 131,863,493 | ||||||||
| M35 | 488,223,880 | 1,363,338 | 8,760,094 | 4,522,744 | 502,870,056 | ||||||||
| M36 | 33,006,739 | 33,006,739 | |||||||||||
| M37 | 23,418,337 | 23,418,337 | |||||||||||
| M38 | 46,101,388 | 46,101,388 | |||||||||||
| M39 | 27,487,975 | 27,487,975 | |||||||||||
| M40 | 20,088,773 | 20,088,773 | |||||||||||
| M41 | 27,408,608 | 27,408,608 | |||||||||||
| M42 | 12,435,301 | 12,435,301 | |||||||||||
| M43 | 77,259,589 | 77,259,589 | |||||||||||
| M44 | 284,733,513 | 284,733,513 | |||||||||||
| M45 | 44,372,830 | 44,372,830 | |||||||||||
| M46 | 89,188,731 | 89,188,731 | |||||||||||
| M47 | 108,520,771 | 108,520,771 | |||||||||||
| M48 | 30986,855 | 30,986,855 | |||||||||||
| M49 | 34,753,952 | 34,753,952 | |||||||||||
| M50 | 53,158,418 | 53,158,418 | |||||||||||
| M51 | 15,908,470 | 15,908,470 | |||||||||||
| M52 | 30,236,800 | 43,804,798 | 2,369,281 | 76,410,879 | |||||||||
| M53 | 463,948,611 | 463,948,611 | |||||||||||
| M54 | 21,448,937 | 21,448,937 | |||||||||||
| M55 | 79,454,258 | 79,454,258 | |||||||||||
| M56 | 79,252,367 | 79,252,367 | |||||||||||
| M57 | 39,700,636 | 39,700,636 | |||||||||||
| M58 | 86,848,220 | 86,848,220 | |||||||||||
| M59 | 17,421,558 | 17,421,558 | |||||||||||
| M60 | 21,097,294 | 21,097,294 | |||||||||||
| M61 | 88,863,196 | 88,863,196 | |||||||||||
| M62 | 38,437,245 | 38,437,245 | |||||||||||
| M63 | 102,677,796 | 102,677,796 | |||||||||||
| M64 | 35,880,113 | 35,880,113 | |||||||||||
| M65 | 23,867,787 | 1,449,639 | 25,317,426 | ||||||||||
| M66 | 13,548,270 | 13,548,270 | |||||||||||
| M67 | 5,583,766 | 5,583,766 | |||||||||||
| M68 | 5,640,311 | 5,640,311 | |||||||||||
| M69 | 9,156,984 | 9,156,984 | |||||||||||
| M70 | 10,312,539 | 10,312,539 | |||||||||||
| M71 | 13,648,742 | 13,648,742 | |||||||||||
| M72 | 4,644,757 | 4,644,757 | |||||||||||
| M73 | 54,507,901 | 54,507,901 | |||||||||||
| M74 | 77,998,658 | 77,998,658 | |||||||||||
| M75 | 39,341,209 | 39,341,209 | |||||||||||
| M76 | 12,303,039 | 12,303,039 | |||||||||||
| M77 | 7,437,147 | 7,437,147 | |||||||||||
| M78 | 30,479,610 | 30,479,610 | |||||||||||
| M79 | 45,861,423 | 45,861,423 | |||||||||||
| M80 | 18,882,764 | 18,882,764 | |||||||||||
| M81 | 285,519,682 | 1,729,889 | 287,249,571 | ||||||||||
| M82 | 19,376,253 | 19,376,253 | |||||||||||
| M83 | 36,030,998 | 26,693,032 | 62,724,030 | ||||||||||
| M84 | 14,114,954 | 14,114,954 | |||||||||||
| M85 | 30,984,346 | 27,085,013 | 58,069,359 | ||||||||||
| M86 | 50,436,190 | 50,436,190 | |||||||||||
| TPA 2 | 7,540,531,449 | 125,100,073 | 1,699,331 | 4,148,293 | 50,488,315 | 37,920,485 | 1,620,283 | 115,671,871 | 147,536,576 | 449,779,867 | 53,283,701 | 0 | 8,527,780,244 |
| Sum 3 | 83 | 19 | 2 | 2 | 7 | 7 | 3 | 13 | 21 | 19 | 14 | 0 | |
| Peak Area (A) | A ≥ 109 | 109 >A ≥ 108 | 108 > A ≥ 107 | 107 > A ≥ 106 | 106 > A ≥ 105 | 104 ≤ A < 105 | A = 0 | ||||||
| Color | |||||||||||||
3. Discussion
3.1. The Metabolic Pathways of Siamenoside I in Rats
3.2. Distribution of the Metabolites of Siamenoside I in Rats
3.3. The Proposed in Vivo Process of Siamenoside I in Rats
3.4. Bioactivities of the Metabolites of Siamenoside I
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animals
4.3. Instruments
4.4. Animal Experiments and Sample Collection
4.5. Sample Preparation
4.5.1. Blood Samples
4.5.2. Urine Samples
4.5.3. Feces Samples
4.5.4. Organ and Skeletal Muscle Samples
4.6. LC-MSn Conditions
4.7. Strategy for Profiling and Identification of the Metabolites of Siamenoside I in Biosamples
4.8. Preliminary Evaluation of the Relative Contents of Siamenoside I and Its Metabolites in Biosamples
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| EBV-EA | Epstein-Barr virus early antigen |
| TPA | 12-O-tetradecanoylphorbol-13-acetate |
| DMBA | 9,10-Dimethyl-1,2-benzanthracene |
| HPLC-ESI-IT-TOF-MSn | High-performance liquid chromatography-electrospray ionization-ion trap-time of flight-multistage mass spectrometry |
| Glc | Glucosyl group |
| NI | Negative ion |
| DAD-ELSD | Diode array detector coupled with evaporative light scattering detector |
| NS | Normal saline |
| BPC | Base peak chromatogram |
| EIC | Extracted ion chromatogram |
References
- Li, C.; Lin, L.M.; Sui, F.; Wang, Z.M.; Huo, H.R.; Dai, L.; Jiang, T.L. Chemistry and pharmacology of Siraitia grosvenorii: A review. Chin. J. Nat. Med. 2014, 12, 89–102. [Google Scholar] [CrossRef]
- Jin, J.K.; Lee, J.H. Phytochemical and pharmacological aspects of Siraitia grosvenorii, luo han kuo. Orient. Pharm. Exp. Med. 2012, 12, 233–239. [Google Scholar] [CrossRef]
- Kasai, R.; Nie, R.L.; Nashi, K.; Ohtani, K.; Zhou, J.; Tao, G.D.; Tanaka, O. Sweet cucurbitane glycosides from fruits of Siraitia siamensis (chi-zi luo-han-guo), a Chinese folk medicine. Agric. Biol. Chem. 1989, 53, 3347–3349. [Google Scholar] [CrossRef]
- Matsumoto, K.; Kasai, R.; Ohtani, K.; Tanaka, O. Minor cucurbitane glycosides from fruits of Siraitia grosvenori (Cucurbitaceae). Chem. Pharm. Bull. 1990, 38, 2030–2032. [Google Scholar] [CrossRef]
- Ukiya, M.; Akihisa, T.; Tokuda, H.; Toriumi, M.; Mukainaka, T.; Banno, N.; Kimura, Y.; Hasegawa, J.; Nishino, H. Inhibitory effects of cucurbitane glycosides and other triterpenoids from the fruit of Momordica grosvenori on Epstein-Barr virus early antigen induced by tumor promoter 12-O-tetradecanoylphorbol-13-acetate. J. Agric. Food Chem. 2002, 50, 6710–6715. [Google Scholar] [CrossRef] [PubMed]
- Konoshima, T. Inhibitory effects of sweet glycosides from fruits of Siraitia grosvenori on two stage carcinogenesis. Food Style 21 2004, 8, 77–81. [Google Scholar]
- Suzuki, Y.A.; Murata, Y.; Inui, H.; Sugiura, M.; Nakano, Y. Triterpene glycosides of Siraitia grosvenori inhibit rat intestinal maltase and suppress the rise in blood glucose level after a single oral Administration of maltose in rats. J. Agric. Food Chem. 2005, 53, 2941–2946. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.W.; Zhang, J.Y.; Xu, W. Biotransformation of mogroside III by human intestinal bacteria. J. Peking Univ. (Health Sci.) 2007, 39, 657–662. [Google Scholar]
- Murata, Y.; Ogawa, T.; Suzuki, Y.A.; Yoshikawa, S.; Inui, H.; Sugiura, M.; Nakano, Y. Digestion and absorption of Siraitia grosvenori triterpenoids in the rat. Biosci. Biotechnol. Biochem. 2010, 74, 673–676. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, D.P.; Huang, Z.C.; Lu, F.L.; Wang, L.; Huang, Y.L.; Wang, R.F.; Liu, G.X.; Shang, M.Y.; Cai, S.Q. Exploring in vitro, in vivo metabolism of mogroside V and distribution of its metabolites in rats by HPLC-ESI-IT-TOF-MSn. J. Pharm. Biomed. Anal. 2015, 115, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Zang, X.Y.; Wang, D.; Shang, M.Y.; Wang, X.; Chui, D.H.; Cai, S.Q. The profiling and identification of the metabolites of (+)-catechin and study on their distribution in rats by HPLC-DAD-ESI-IT-TOF-MSn technique. Biomed. Chromatogr. 2014, 28, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Li, D.P.; Ikeda, T.; Nohara, T.; Liu, J.L.; Wen, Y.X.; Sakamoto, T.; Nonaka, G. Cucurbitane glycosides from unripe fruits of Siraitia grosvenori. Chem. Pharm. Bull. 2007, 55, 1082–1086. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Lu, F.L.; Wang, L.; Chen, B.; Xu, F.; Cai, S.Q.; Li, D.P. Methodology of enzymic hydrolysis of fifty percent mogroside V. Guihaia 2015, 35, 812–816. [Google Scholar]
- Liang, J.; Xu, F.; Zhang, Y.Z.; Huang, S.; Zang, X.Y.; Zhao, X.; Zhang, L.; Shang, M.Y.; Yang, D.H.; Wang, X.; et al. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: A novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 2013, 83, 108–121. [Google Scholar] [PubMed]
- Sample Availability: Samples of the compounds siamenoside I, mogroside V are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.-R.; Xu, F.; Li, D.-P.; Lu, F.-L.; Liu, G.-X.; Wang, L.; Shang, M.-Y.; Huang, Y.-L.; Cai, S.-Q. Metabolites of Siamenoside I and Their Distributions in Rats. Molecules 2016, 21, 176. https://doi.org/10.3390/molecules21020176
Yang X-R, Xu F, Li D-P, Lu F-L, Liu G-X, Wang L, Shang M-Y, Huang Y-L, Cai S-Q. Metabolites of Siamenoside I and Their Distributions in Rats. Molecules. 2016; 21(2):176. https://doi.org/10.3390/molecules21020176
Chicago/Turabian StyleYang, Xue-Rong, Feng Xu, Dian-Peng Li, Feng-Lai Lu, Guang-Xue Liu, Lei Wang, Ming-Ying Shang, Yong-Lin Huang, and Shao-Qing Cai. 2016. "Metabolites of Siamenoside I and Their Distributions in Rats" Molecules 21, no. 2: 176. https://doi.org/10.3390/molecules21020176
APA StyleYang, X.-R., Xu, F., Li, D.-P., Lu, F.-L., Liu, G.-X., Wang, L., Shang, M.-Y., Huang, Y.-L., & Cai, S.-Q. (2016). Metabolites of Siamenoside I and Their Distributions in Rats. Molecules, 21(2), 176. https://doi.org/10.3390/molecules21020176






