Ag+ as a More Effective Elicitor for Production of Tanshinones than Phenolic Acids in Salvia miltiorrhiza Hairy Roots
Abstract
:1. Introduction
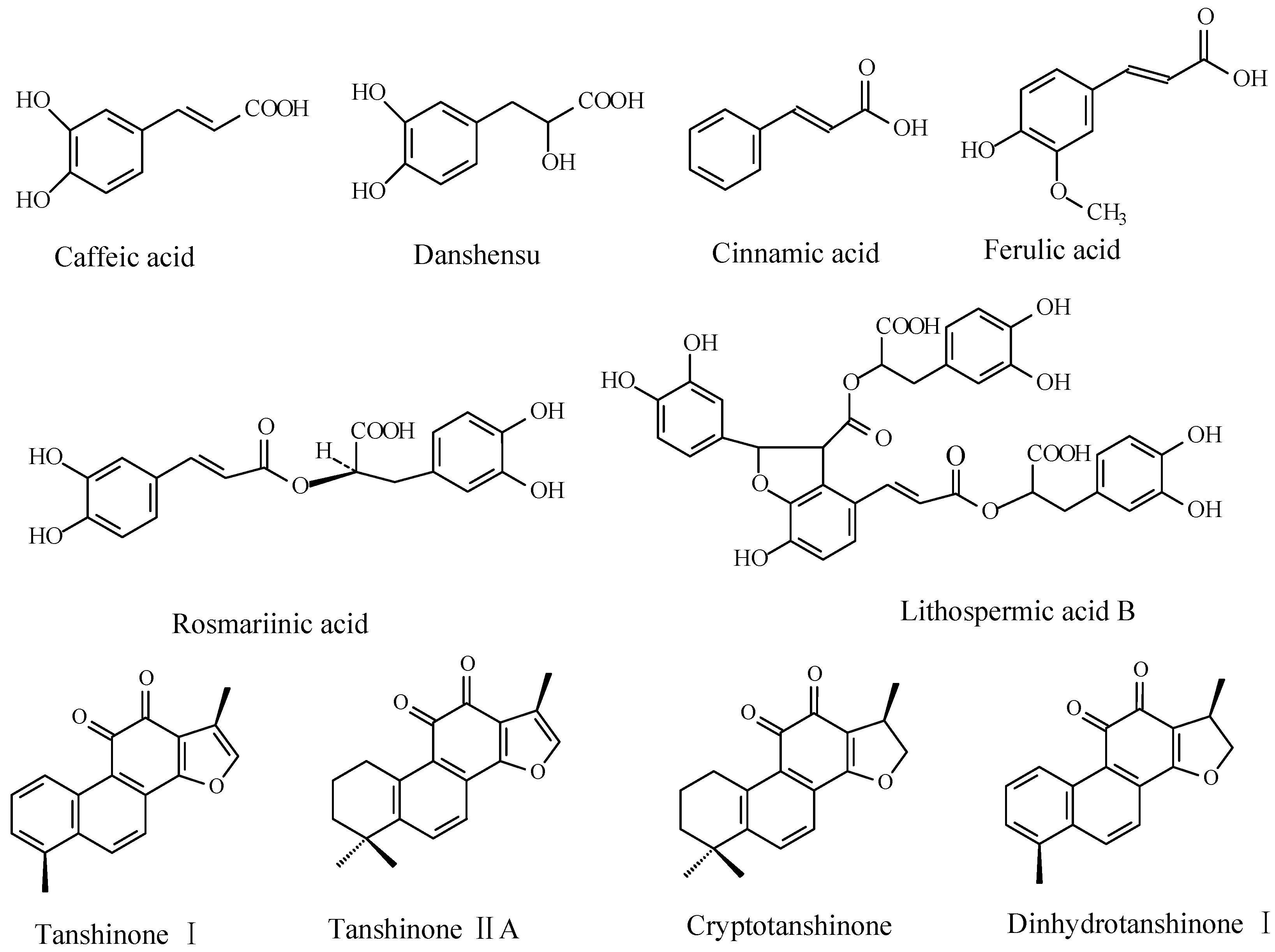
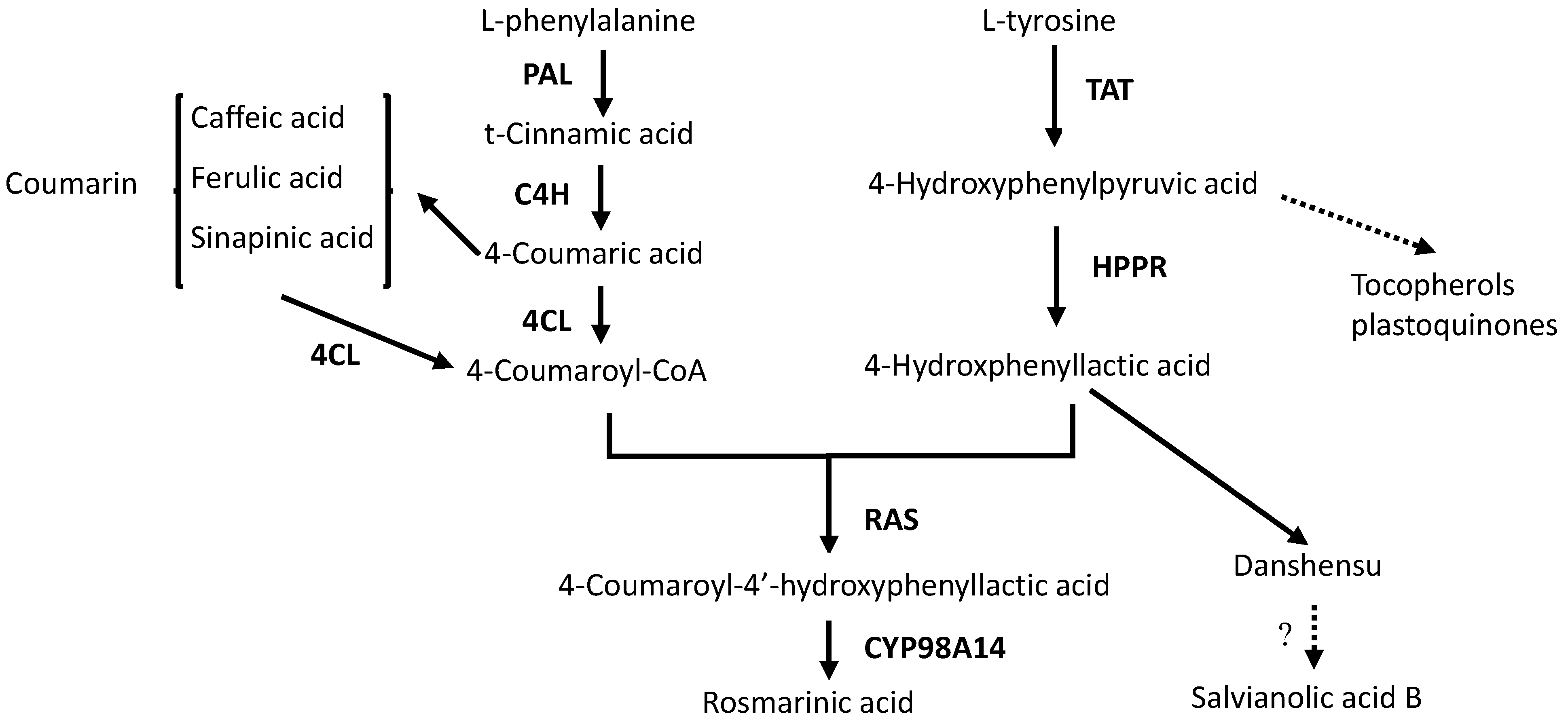
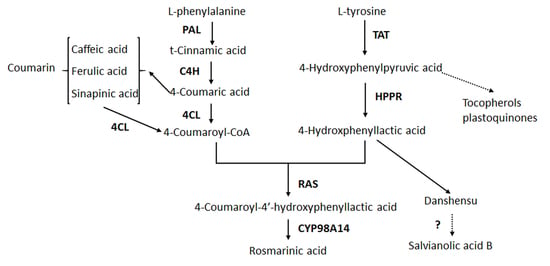
2. Results and Discussion
2.1. Effects of Ag+ Elicitor on Cell Growth of S. miltiorrhiza Hairy Roots
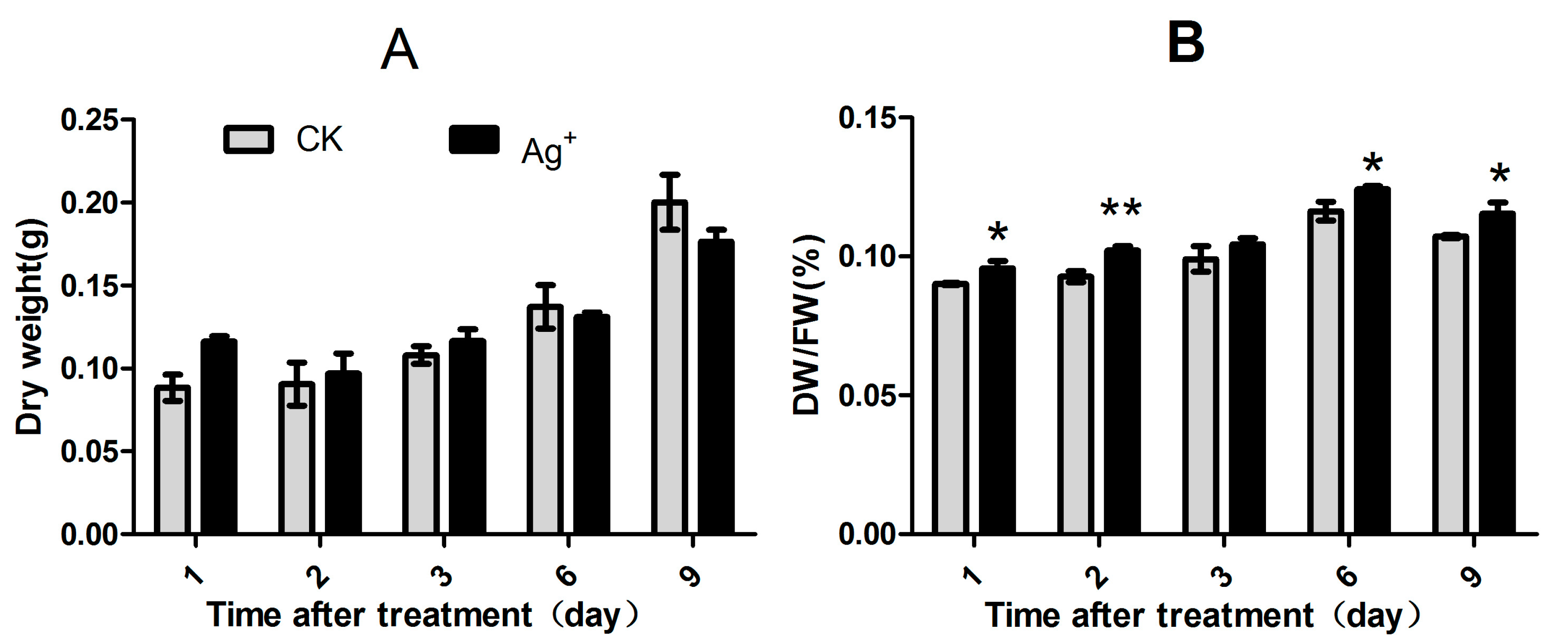
2.2. Effects of Ag+ Elicitor on Accumulation of Phenolic Acids in S. miltiorrhiza Hairy Roots
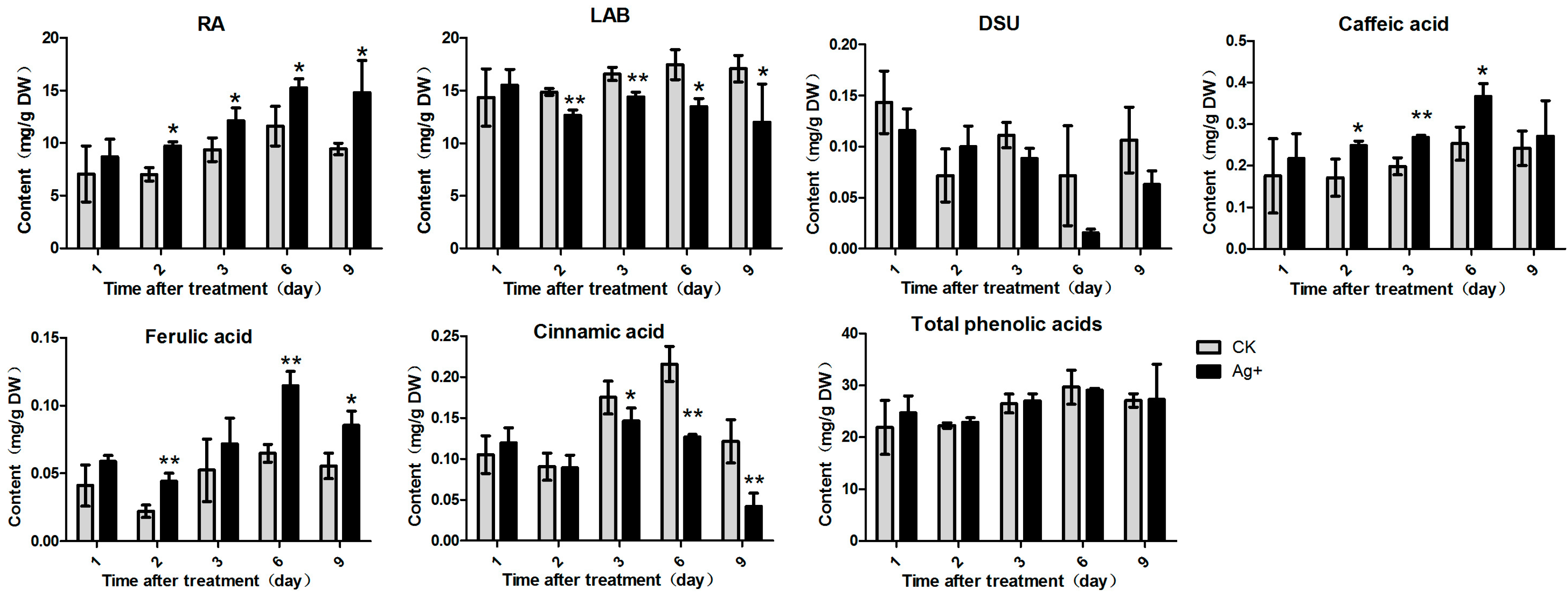
2.3. Effects of Ag+ Elicitor on Tanshinones Accumulation in S. miltiorrhiza Hairy Roots
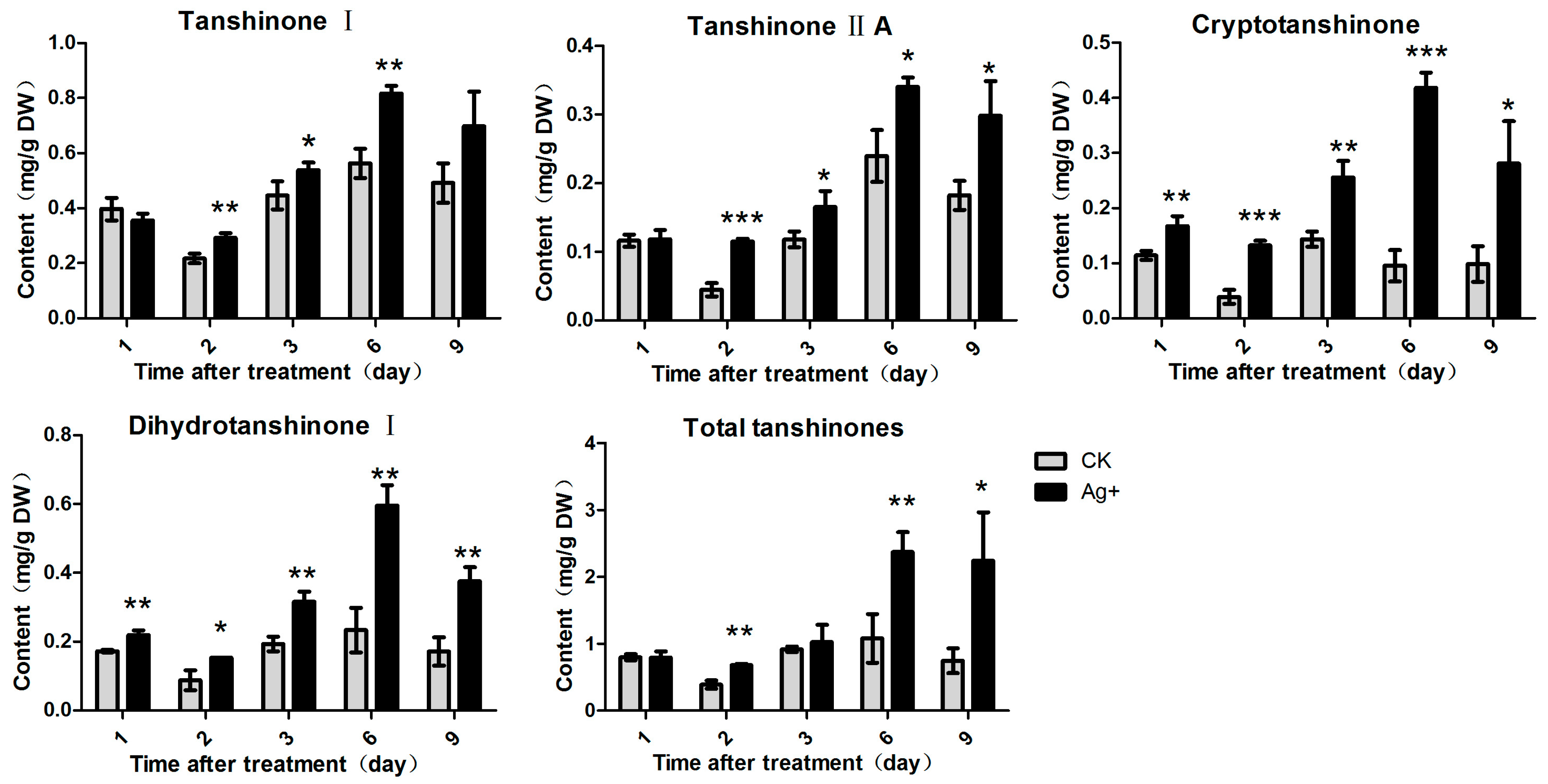
2.4. Effects of Ag+ Elicitor on Genes Expressions Involved in the Two Parallel Pathways of LAB Biosynthesis
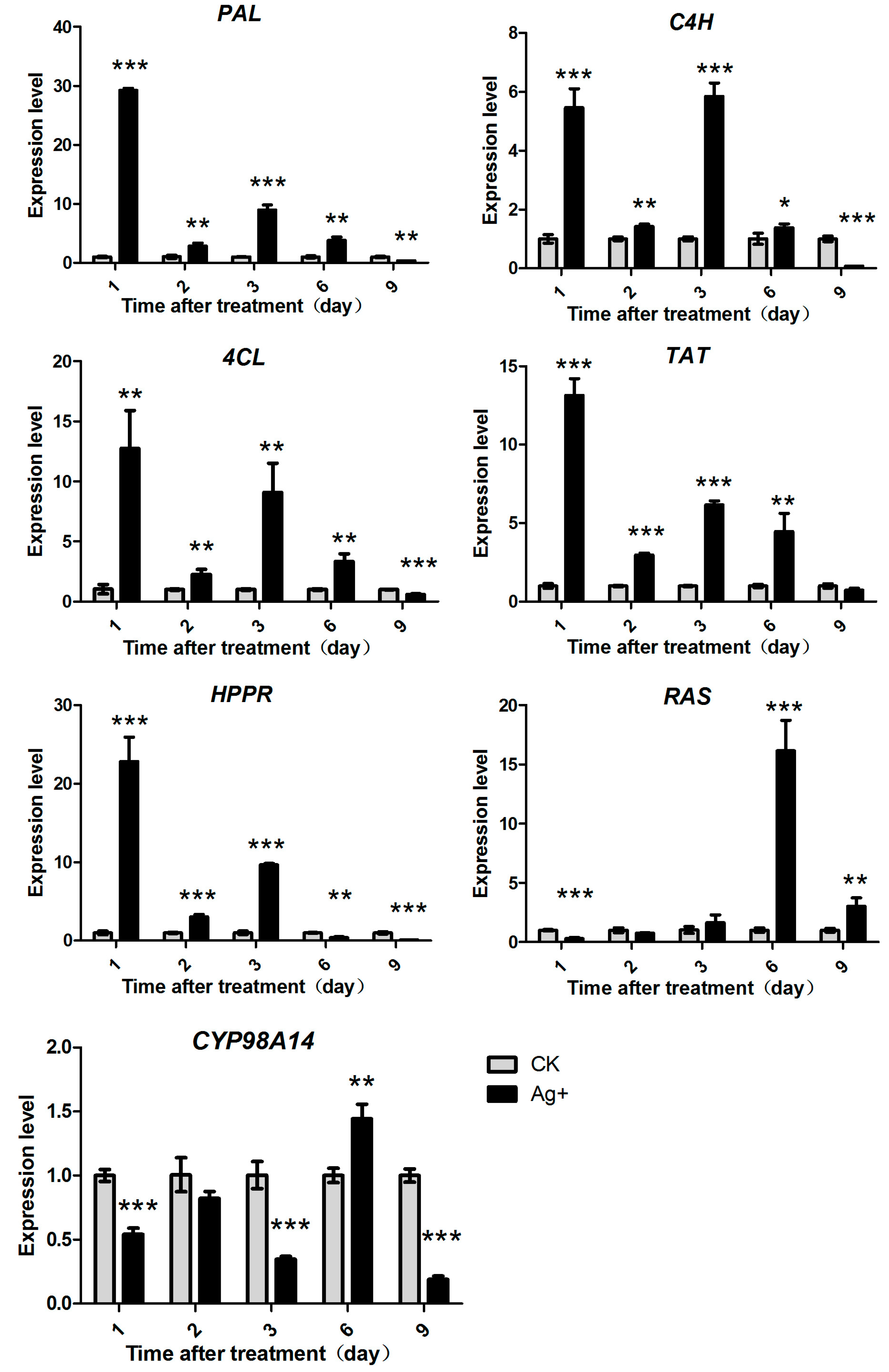
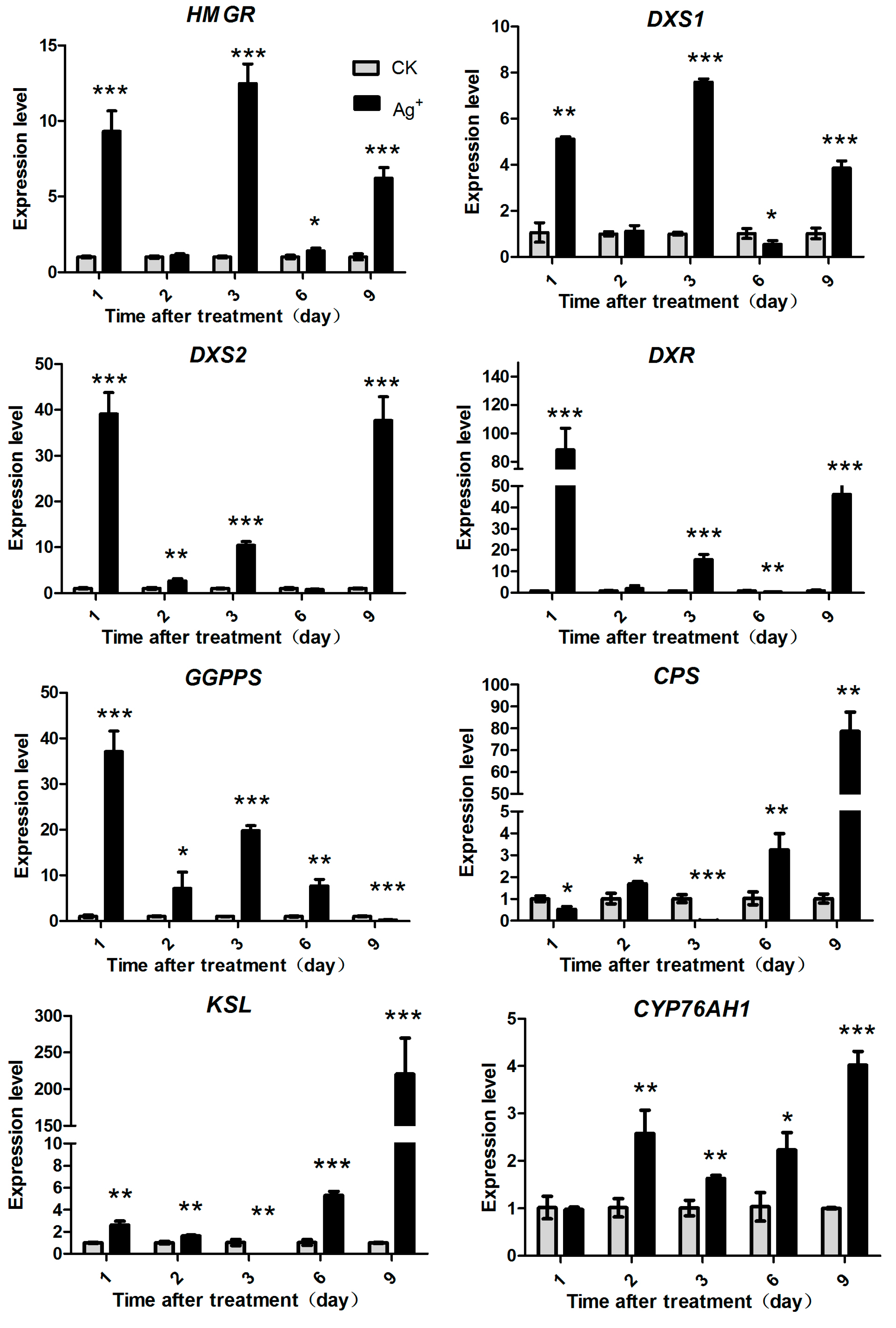
2.5. Effects of Ag+ Elicitor on Genes Expression Involved in Pathways of Tanshinones Biosynthesis
2.6. Discussion
3. Experimental Section
3.1. Hairy Roots Culture
3.2. Preparation of Ag+ Elicitor
3.3. Metabolite Extraction and HPLC Analysis
3.4. RNA Isolation and Real-Time Quantitative PCR Analysis
| Gene | Sense Primer (5’ to 3’) | Reverse Prime (5’ to 3’) | Genbank ID |
|---|---|---|---|
| PAL | GGCGGCGATTGAGAGCAGGA | ATCAGCAGATAGGAAGAGGAGCACC | GQ249111 |
| 4CL | TCGCCAAATACGACCTTTCC | TGCTTCAGTCATCCCATACCC | AY237164 |
| C4H | CCAGGAGTCCAAATAACAGAGCC | GAGCCACCAAGCGTTCACCAA | EF377337 |
| TAT | TTCAACGGCTACGCTCCAACT | AAACGGACAATGCTATCTCAAT | DQ334606 |
| HPPR | GACTCCAGAAACAACCCACATT | CCCAGACGACCCTCCACAAGA | DQ099741 |
| CYP98A14 | CTAAGGAGGTGCTGAAGGAG | GTGGAGTCGTTGTAGATGGA | HQ316179 |
| RAS | CGCCCTAGTTGAGTTCTACCCTTACGC | TCGGATAGGTGGTGCTCGTTTGC | FJ906696 |
| HMGR | GCAACATCGTCTCCGCCGTCTACA | GATGGTGGCCAGCAGCCTGGAGTT | FJ747636 |
| DXS1 | CGACCAGGTAGTGCACGACG | TCATCTGAAGGAGCCATCACCAC | EU670744 |
| DXS2 | TTGGAGATTGGGAAGGGAAGGAT | AGGCTTGCAGAATCTCGCATCAG | FJ643618 |
| DXR | GAGAATCTACTGCTCCGAGA | CTGGTCGTAGTGGATGATCT | DQ991431 |
| GGPPS | GGGGCTATTTTGGGAGGTGGAA | CAGCAGCTTGGGATACGTGGTC | FJ178784 |
| CPS | GAGGGAGAGGTGAGGAAGGAA | AGGGAACAAAAGTTGAAAAGG | EU003997 |
| KSL | CATGTCGAACAAGGACGTA | AATCATCCAAGGTTAGTGCC | EF635966 |
| CYP76AH1 | CAGGAGGTGAACGGCTATCT | GTTATGAACCAGAGTCGCAGTAG | JX422213 |
4. Conclusions
Acknowledgments
Author Contributions
Abbreviation
| ABA | abscisic acid |
| C4H | cinnamic acid 4-hydroxylase |
| CPS | copalyl diphosphate synthase |
| CT | cryptotanshinone |
| CYP98A14 | cytochrome P450-dependent monooxygenase |
| CYP76AH1 | cytochrome P450-dependent monooxygenase |
| DMAPP | dimethylallyl diphosphate |
| DTI I | dihydrotanshinone I |
| DXR | 1-deoxy-d-xylulose 5-phosphate reductoisomerase |
| DXS | 1-deoxy-d-xylulose 5-phosphate synthase |
| DW | dry weight |
| EB | ethidium bromide |
| FW | fresh weight |
| GA | gibberellins acid |
| GGPP | geranylgeranyl diphosphate |
| GGPPS | Geranylgeranyl diphosphate synthase |
| HMGR | 3-hydroxy-3-methylglutaryl CoA reductase |
| HPLC | high performance liquid chromatography |
| HPPR | 4-hydroxyphenylpyruvate reductase |
| IPP | isopentenl diphosphate |
| KSL | kaurene synthase-like |
| LAB | salvianolic acid B |
| MeJA | methyl jasmonate |
| MEP | methylerythritol phosphate |
| MVA | mevalonate |
| PAL | Phenylalanine ammonia-lyase |
| PDA | photodiode array detector |
| PEG | polyethylene glycol |
| RA | rosmarinic acid |
| RAS | rosmarinic acid synthase |
| RNAi | RNA interference |
| RT-qPCR | quantitative real-time PCR |
| SA | salicylic acid |
| Salvia miltiorrhiza | Salvia miltiorrhiza |
| TAT | tyrosine aminotransferase |
| T I | tanshinone I |
| T II A | tanshinone II A |
| YE | yeast extract |
| 4CL | hydroxycinnamate coenzyme A ligase |
Conflicts of Interest
References
- Cheng, H.T.; Li, X.L.; Li, X.; Li, Y.; Wang, L.; Xue, M. Simultaneous quantification of selected compounds from Salvia herbs by HPLC method and their application. Food Chem. 2012, 130, 1031–1035. [Google Scholar]
- Liu, A.H.; Li, L.; Xu, M.; Lin, Y.H.; Guo, H.Z.; Guo, D.A. Simultaneous quantification of six major phenolic acids in the roots of Salvia miltiorrhiza and four related traditional Chinese medicinal preparations by HPLC-DAD method. J. Pharm. Biomed. Anal. 2006, 41, 48–56. [Google Scholar]
- Shi, Z.H.; He, J.T.; Yao, T.T.; Chang, W.B.; Zhao, M.P. Simultaneous determination of cryptotanshinone, tanshinone I and tanshinone II A in traditional chinese medicinal preparations containing Radix Salvia miltiorrhiza by HPLC. J. Pharm. Biomed. Anal. 2005, 37, 481–486. [Google Scholar]
- Zhou, Y.Q.; Li, W.Z.; Xu, L.; Chen, L.Y. In Salvia miltiorrhiza, phenolic acids possess protective properties against amyloid β-induced cytotoxicity, and tanshinones act as acetylcholinesterase inhibitors. Environ. Toxicol. Pharmacol. 2011, 31, 443–452. [Google Scholar]
- Chen, X.P.; Guo, J.J.; Bao, J.L.; Lu, J.J.; Wang, Y.T. The anticancer properties of Salvia miltiorrhiza Bunge (danshen): A systematic review. Med. Res. Rev. 2014, 34, 768–794. [Google Scholar]
- Wang, X.; Lee, W.Y.W.; Zhou, X.L.; Or, P.M.Y.; Yeung, J.H.K. A pharmacodynamics-pharmacokinetic (PD-PK) study on the effects of Danshen (Salvia miltiorrhiza) on midazolam, a model CYP3A probe substrate, in the rat. Phytomedicine 2010, 17, 876–883. [Google Scholar]
- Kai, G.Y.; Hao, X.L.; Cui, L.J.; Ni, X.L.; Zekria, D.; Wu, J.Y. Metabolic engineering and biotechnological approaches for production of bioactive diterpene tanshinones in Salvia miltiorrhiza. Biotechnol. Adv. 2014. [Google Scholar] [CrossRef]
- Zhang, S.C.; Yan, Y.; Wang, B.Q.; Liang, Z.S.; Liu, Y.; Liu, F.H.; Qi, Z.H. Selective responses of enzymes in the two parallel pathways of rosmarinic acid biosynthetic pathway to elicitors in Salvia miltiorrhiza hairy root cultures. J. Biosci. Bioeng. 2014, 117, 645–651. [Google Scholar]
- Di, P.; Zhang, L.; Chen, J.F.; Tan, H.X.; Xiao, Y.; Dong, X.; Zhou, X.; Chen, W.S. 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 2013, 8, 1537–1548. [Google Scholar]
- Yang, D.F.; Sheng, D.F.; Duan, Q.M.; Liang, X.; Liang, Z.S.; Liu, Y. PEG and ABA trigger the burst of reactive oxygen species to increase tanshinone production in Salvia miltiorrhiza hairy roots. J. Plant Growth Regul. 2012, 31, 579–587. [Google Scholar]
- Xiao, Y.; Gao, S.H.; Di, P.; Chen, J.F.; Chen, W.S.; Zhang, L. Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol. Plant 2009, 137, 1–9. [Google Scholar]
- Yan, Q.; Shi, M.; Ng, J.; Wu, J.Y. Elicitor-induced rosmarinic acid accumulation and secondary metabolism enzyme activities in Salvia miltiorrhiza hairy roots. Plant Sci. 2006, 170, 853–858. [Google Scholar]
- Liang, Z.S.; Ma, Y.N.; Xu, T.; Cui, B.M.; Liu, Y.; Guo, Z.X.; Yang, D.F. Effects of Abscisic Acid, Gibberellin, Ethylene and Their Interactions on Production of Phenolic Acids in Salvia miltiorrhiza Bunge Hairy Roots. PLoS One 2013, 8, e72806. [Google Scholar]
- Jiao, M.L; Cao, R.R.; Chen, H.Y.; Hao, W.F.; Dong, J.E. Effects of salicylic acid on synthesis of rosmarinic acid and related enzymes in the suspension cultures of Salvia miltiorrhiza. Sheng Wu Gong Cheng Xue Bao 2012, 28, 320–328. [Google Scholar]
- Kai, G.Y.; Zhang, A.; Guo, Y.Y.; Li, L.; Cui, L.J.; Luo, X.Q.; Liu, C.; Xiao, J.B. Enhancing the production of tropane alkaloids in transgenic Anisodus acutangulus hairy root cultures by over-expressing tropinone reductase I and hyoscyamine-6β-hydroxylase. Mol. Biosyst. 2012, 8, 2883–2890. [Google Scholar]
- Song, J.; Wang, Z.Z. RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J. Plant Res. 2011, 124, 183–192. [Google Scholar]
- Kai, G.Y.; Xu, H.; Zhou, C.C.; Liao, P.; Xiao, J.B.; Luo, X.Q.; You, L.J.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar]
- Yang, D.F.; Du, X.H.; Liang, X.; Han, R.L.; Liang, Z.S.; Liu, Y.; Liu, F.H.; Zhao, J. Different roles of the mevalonate and methylerythritol phosphate pathways in cell growth and tanshinone production of Salvia miltiorrhiza hairy roots. PLoS One 2012, 7, e46797. [Google Scholar]
- Kai, G.Y.; Liao, P.; Zhang, T.; Zhou, W.; Wang, J.; Xu, H.; Liu, Y.Y.; Zhang, L. Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnol. Bioprocess Eng. 2010, 15, 236–245. [Google Scholar]
- Cheng, Q.Q.; Su, P.; Hu, Y.T.; He, Y.F.; Gao, W.; Huang, L.Q. RNA interference-mediated repression of SmCPS (copalyldiphosphate synthase) expression in hairy roots of Salvia miltiorrhiza causes a decrease of tanshinones and sheds light on the functional role of SmCPS. Biotechnol. Lett. 2014, 36, 363–369. [Google Scholar]
- Zhou, J.; Fang, L.; Wang, X.; Guo, L.P.; Huang, L.Q. La dramaticaly enhances the accumulation of tanshinones in Salvia miltiorrhiza hairy root cultures. Earth Sci. Res. 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Kai, G.Y.; Liao, P.; Xu, H.; Wang, J.; Zhou, C.C.; Zhou, W.; Qi, Y.P.; Xiao, J.B.; Wang, Y.L.; Zhang, L. Molecular mechanism of elicitor-induced tanshinone accumulation in Salvia miltiorrhiza hairy root cultures. Acta Physiol. Plant. 2012, 34, 1421–1433. [Google Scholar]
- Hao, X.L.; Shi, M.; Cui, L.J.; Xu, C.; Zhang, Y.J.; Kai, G.Y. Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 2014. [Google Scholar] [CrossRef]
- Shi, M.; Luo, X.Q.; Ju, G.H.; Yu, X.H.; Hao, X.L.; Huang, Q.; Xiao, J.B.; Cui, L.J.; Kai, G.Y. Increased accumulation of the cardio-cerebrovascular disease treatment drug tanshinone in Salvia miltiorrhiza hairy roots by the enzymes 3-hydroxy-3-methylglutaryl CoA reductase and 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Funct. Integr. Genomics 2014, 14, 603–615. [Google Scholar]
- Peng, X.; He, J.Y. The inhibitory effect of Ca2+ on the flavonoid production of Tetrastigma hemsleyanum suspension cells induced by metal elicitors. In Vitro Cell. Dev. Biol. Plant 2013, 49, 550–559. [Google Scholar]
- Cai, Z.Z.; Kastell, A.; Speiser, C.; Smetanska, I. Enhanced resveratrol production in Vitis vinifera cell suspension cultures by heavy metals without loss of cell viability. Appl. Biochem. Biotechnol. 2013, 171, 330–340. [Google Scholar]
- Zhang, C.H.; Wu, J.Y. Ethylene inhibitors enhance elicitor-induced paclitaxel production in suspension cultures of Taxus spp. Cells. Enzym. Microb. Technol. 2003, 32, 71–77. [Google Scholar]
- Chen, W.H.; Xu, C.M.; Zeng, J.L.; Zhao, B.; Wang, X.-D.; Wang, Y.C. Improvement of echinacoside and acteoside production by two-stage elicitation in cell suspension culture of Cistanche deserticola. World J. Microbiol. Biotechnol. 2007, 23, 1451–1458. [Google Scholar]
- Xiao, Y.; Gao, S.H.; Di, P.; Chen, J.F.; Chen, W.S.; Zhang, L. Lithospermic acid b is more responsive to silver ions (Ag+) than rosmarinic acid in Salvia miltiorrhiza hairy root cultures. Biosci. Rep. 2010, 30, 33–40. [Google Scholar]
- Ge, X.C.; Wu, J.Y. Tanshinone production and isoprenoid pathways in salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Sci. 2005, 168, 487–491. [Google Scholar]
- Ma, P.D.; Liu, J.L.; Zhang, C.L.; Liang, Z.S. Regulation of water-soluble phenolic acid biosynthesis in Salvia miltiorrhiza Bunge. Appl. Biochem. Biotechnol. 2013, 170, 1253–1262. [Google Scholar]
- Pitta-Alvarez, S.I.P.; Spollansky, T.C.; Giulietti, A.M. The influence of different biotic and abiotic elicitors on the production and profile of tropane alkaloids in hairy root cultures of Brugmansia candida. Enzym. Microb. Technol. 2000, 26, 252–258. [Google Scholar]
- Zhao, J.L.; Zhou, L.G.; Wu, J.Y. Effects of biotic and abiotic elicitors on cell growth and tanshinone accumulation in Salvia miltiorrhiza cell cultures. Appl. Microbiol. Biotechnol. 2010, 87, 137–144. [Google Scholar]
- Guo, Y.X.; Zhang, D.J.; Wang, H.; Xiu, Z.L.; Wang, L.X.; Xiao, H.B. Hydrolytic kinetics of lithospermic acid b extracted from roots of Salvia miltiorrhiza. J. Pharm. Biomed. Anal. 2007, 43, 435–439. [Google Scholar]
- Song, J.; Ji, Y.Y.; Xu, K.; Wang, Z.Z. An integrated analysis of the rosmarinic acid-biosynthetic genes to uncover the regulation of rosmarinic acid pathway in Salvia miltiorrhiza. Acta Physiol. Plant. 2012, 34, 1501–1511. [Google Scholar]
- Xiao, Y.; Zhang, L.; Gao, S.H.; Saechao, S.; Di, P.; Chen, J.F.; Chen, W.S. The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. PLoS One 2011, 6, e29713. [Google Scholar]
- Ming, Q.L.; Su, C.Y.; Zheng, C.J.; Jia, M.; Zhang, Q.Y.; Zhang, H.; Rahman, K.; Han, T.; Qin, L. Elicitors from the endophytic fungus trichoderma atroviride promote Salvia miltiorrhiza hairy root growth and tanshinone biosynthesis. J. Exp. Bot. 2013, 64, 5687–5694. [Google Scholar]
- Zhang, C.H.; Yan, Q.; Cheuk, W.K.; Wu, J.Y. Enhancement of tanshinone production in Salvia miltiorrhiza hairy root culture by Ag+ elicitation and nutrient feeding. Plant. Med. 2004, 70, 147–151. [Google Scholar]
- Zhang, X.N.; Guo, J.; Shen, Y.; Huang, L.Q. Cloning and expression analysis of a new 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza (SmHMGR3). Zhongguo Zhong Yao Za Zhi 2012, 37, 2378–2382. [Google Scholar]
- Sample Availability: Samples of the compounds are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, B.; Yang, D.; Guo, W.; Liang, Z.; Yan, X.; Zhu, Y.; Liu, Y. Ag+ as a More Effective Elicitor for Production of Tanshinones than Phenolic Acids in Salvia miltiorrhiza Hairy Roots. Molecules 2015, 20, 309-324. https://doi.org/10.3390/molecules20010309
Xing B, Yang D, Guo W, Liang Z, Yan X, Zhu Y, Liu Y. Ag+ as a More Effective Elicitor for Production of Tanshinones than Phenolic Acids in Salvia miltiorrhiza Hairy Roots. Molecules. 2015; 20(1):309-324. https://doi.org/10.3390/molecules20010309
Chicago/Turabian StyleXing, Bingcong, Dongfeng Yang, Wanli Guo, Zongsuo Liang, Xijun Yan, Yonghong Zhu, and Yan Liu. 2015. "Ag+ as a More Effective Elicitor for Production of Tanshinones than Phenolic Acids in Salvia miltiorrhiza Hairy Roots" Molecules 20, no. 1: 309-324. https://doi.org/10.3390/molecules20010309
APA StyleXing, B., Yang, D., Guo, W., Liang, Z., Yan, X., Zhu, Y., & Liu, Y. (2015). Ag+ as a More Effective Elicitor for Production of Tanshinones than Phenolic Acids in Salvia miltiorrhiza Hairy Roots. Molecules, 20(1), 309-324. https://doi.org/10.3390/molecules20010309