Cross-Talk of Phosphorylation and Prolyl Isomerization of the C-terminal Domain of RNA Polymerase II
Abstract
:1. Introduction
2. Covalent Modification of CTD
3. Non-Covalent Modification on CTD
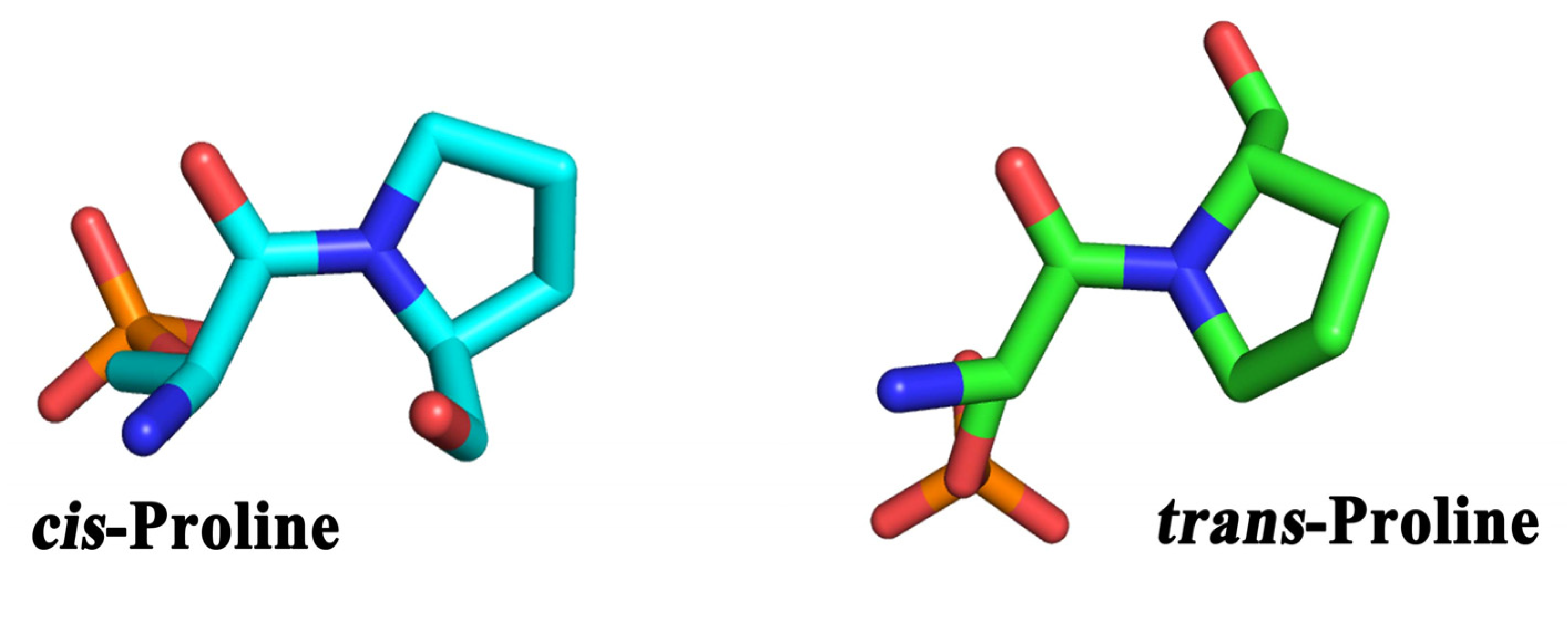
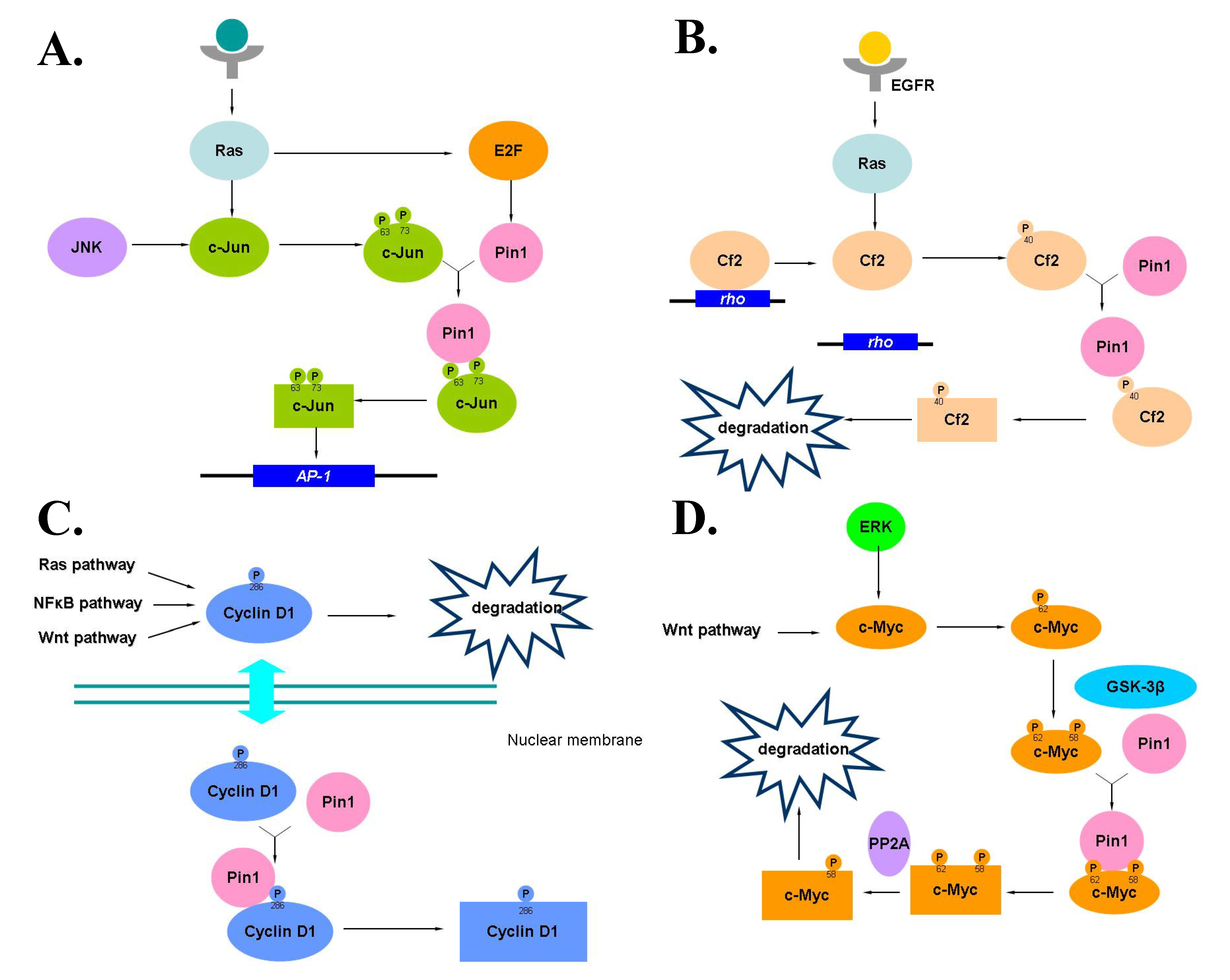
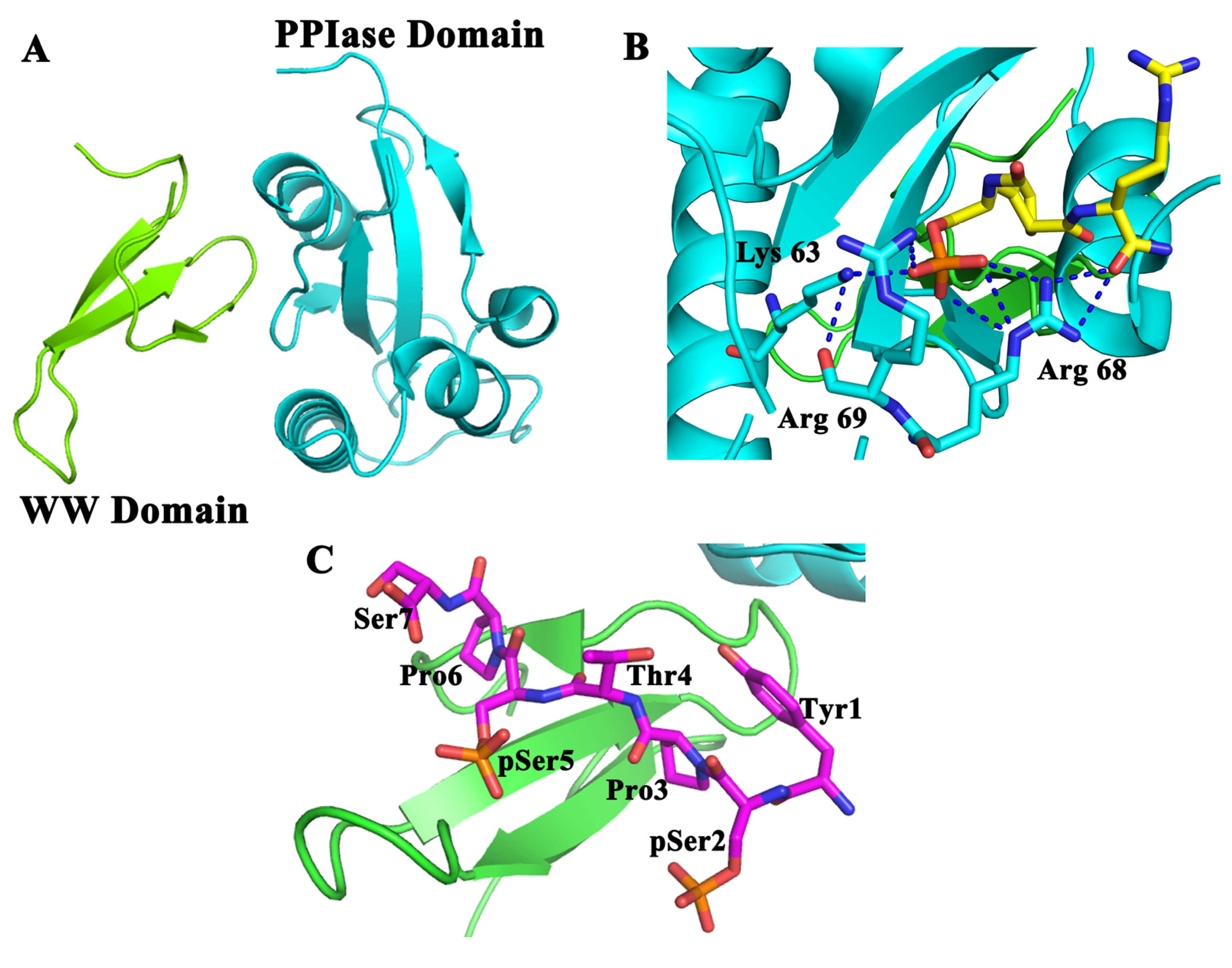
4. Erasers, CTD Phosphatases
4.1. Ssu72
4.2. Scp1
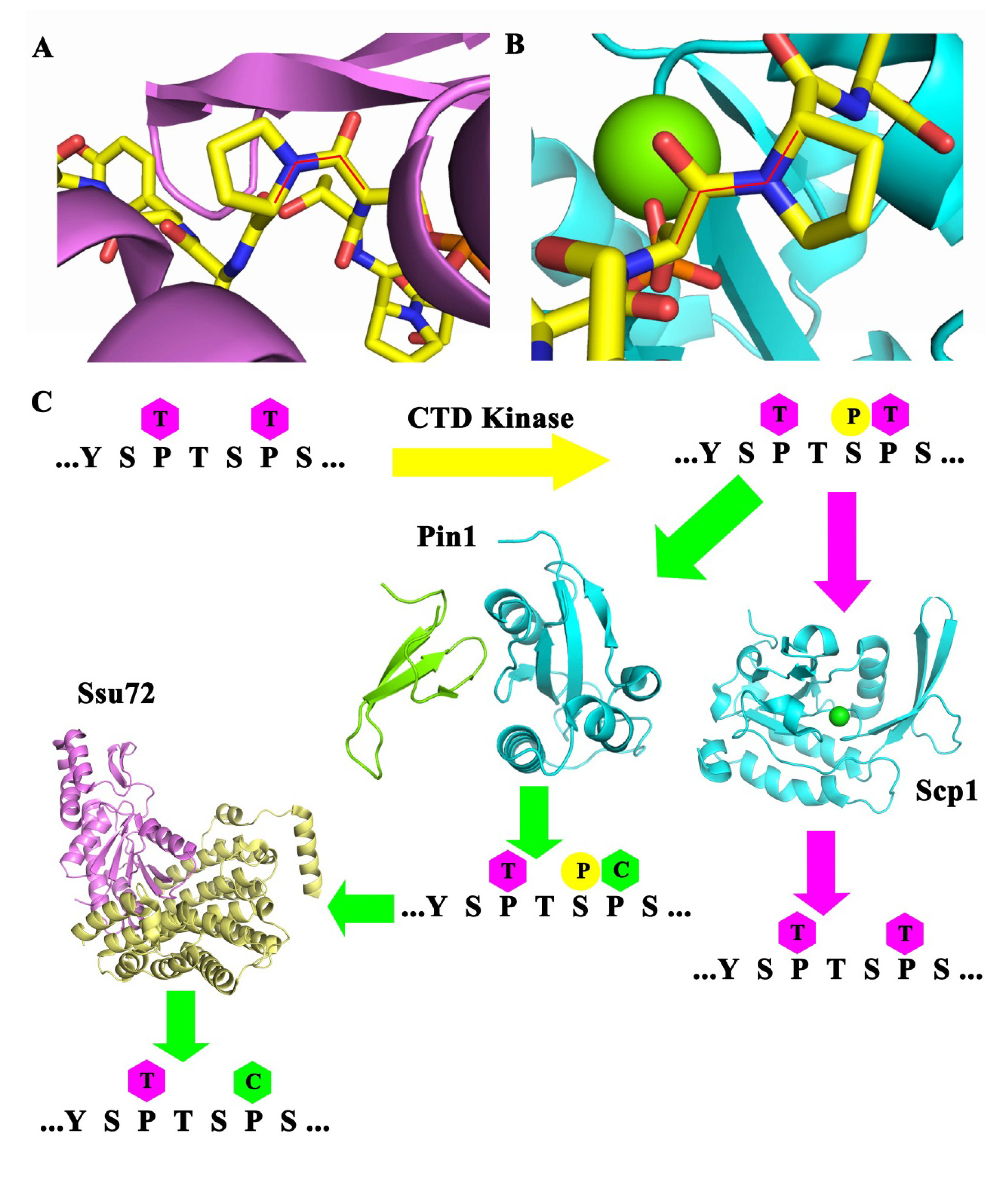
4.3. Fcp1
4.4. Rtr1
5. Writers, CTD Kinases
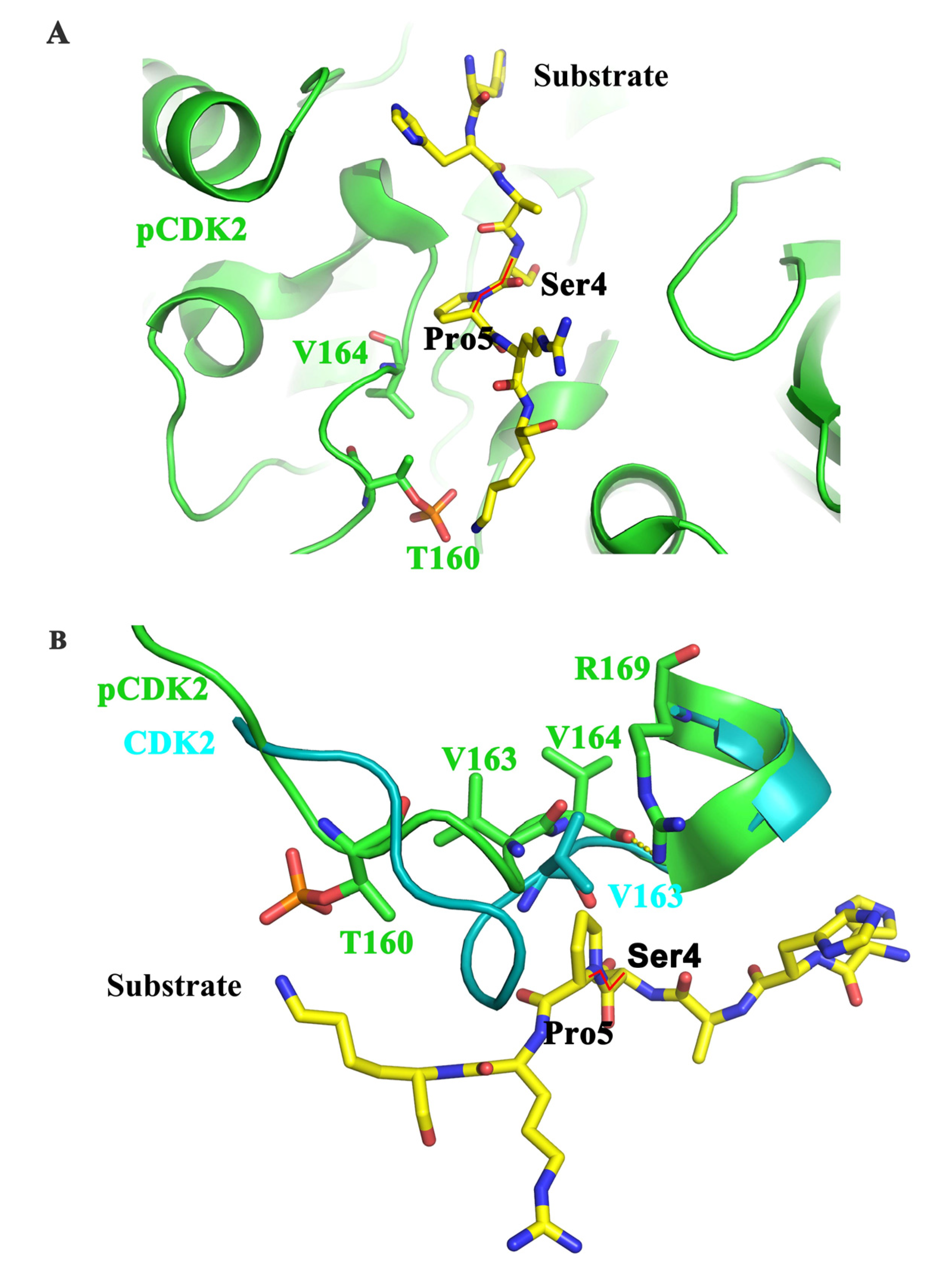
5.1. CDK7
5.2. CDK9
5.3. CDK12
5.4. CDK8
6. Readers, CTD Binding Proteins that Regulate Transcription
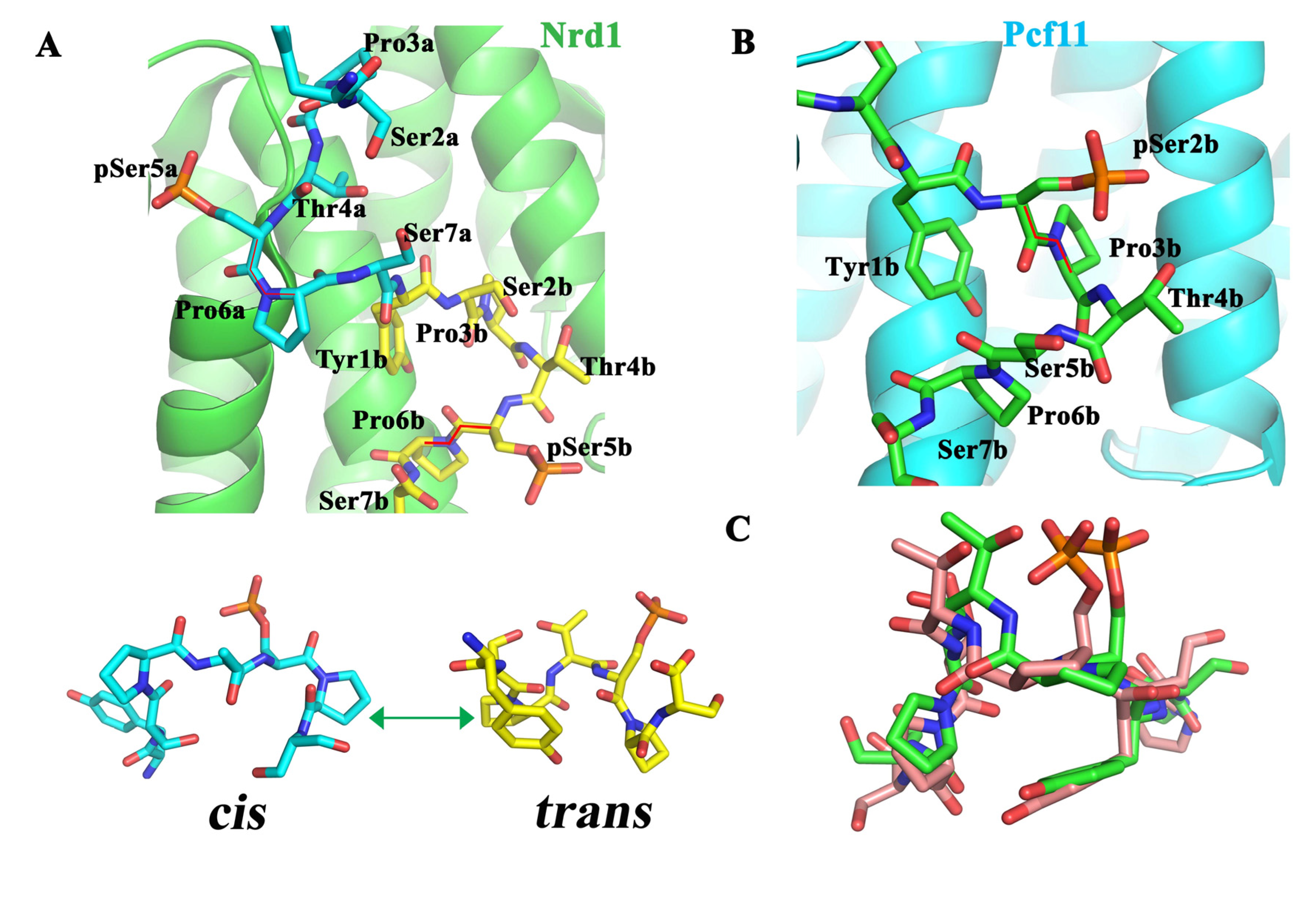
6.1. Nrd1
6.2. Pcf11
6.3. Capping Enzymes
6.4. Rtt103
6.5. SCAF8
7. Methods to Study the Prolyl Isomeric Specificity
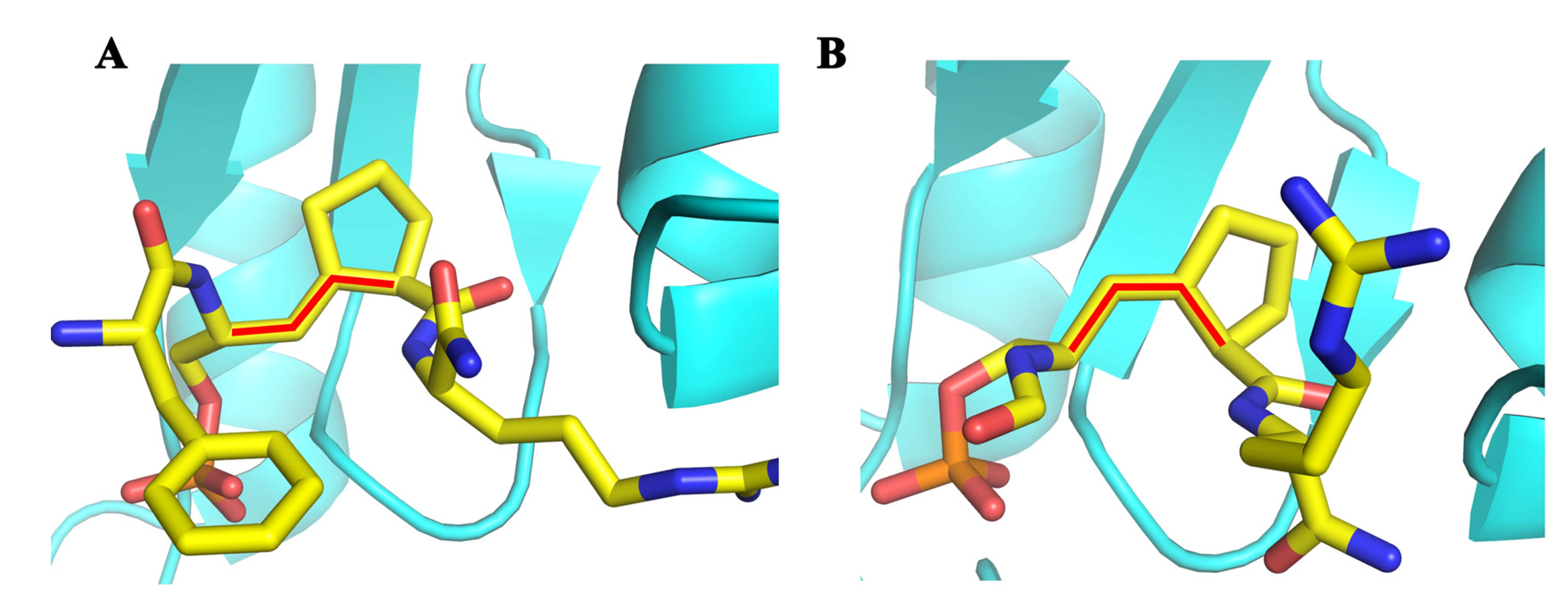
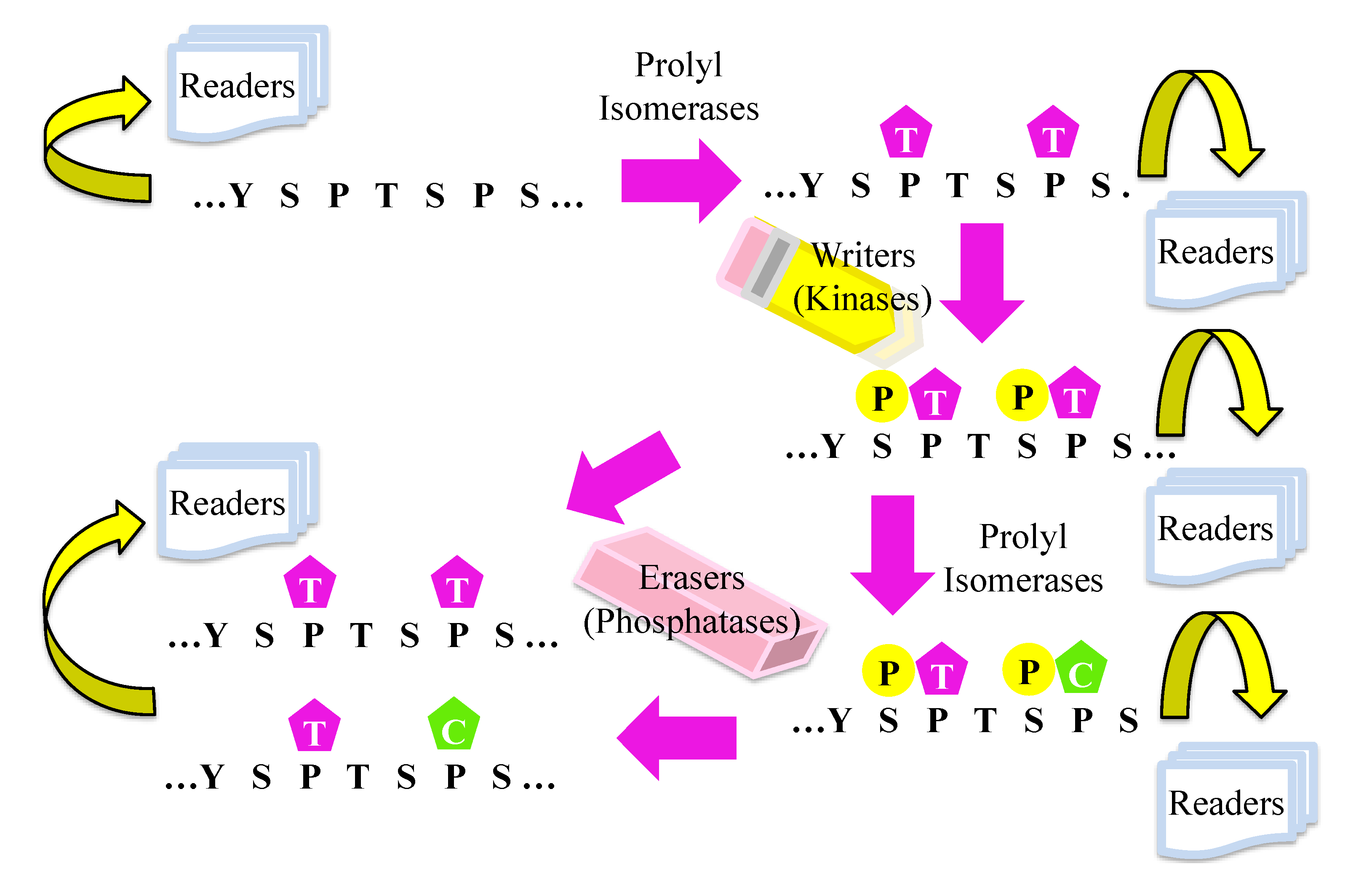
8. Hypothetical Model for the Effect of Prolyl Isomerization on Transcription
Acknowledgments
Conflicts of Interest
References
- Kornberg, R.D. Eukaryotic transcriptional control. Trends Cell Biol. 1999, 9, M46–M49. [Google Scholar] [CrossRef]
- Sims, R.J.; Mandal, S.S.; Reinberg, D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 2004, 16, 263–271. [Google Scholar] [CrossRef]
- Buratowski, S. The CTD code. Nat. Struct. Biol. 2003, 10, 679–680. [Google Scholar] [CrossRef]
- Maniatis, T.; Reed, R. An extensive network of coupling among gene expression machines. Nature 2002, 416, 499–506. [Google Scholar] [CrossRef]
- Bentley, D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 1999, 11, 347–351. [Google Scholar] [CrossRef]
- Hirose, Y.; Manley, J.L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000, 14, 1415–1429. [Google Scholar]
- Corden, J.L. Tails of RNA polymerase II. Trends Biochem. Sci. 1990, 15, 383–387. [Google Scholar] [CrossRef]
- Corden, J.L.; Cadena, D.L.; Ahearn, J.M., Jr.; Dahmus, M.E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl. Acad. Sci. USA 1985, 82, 7934–7938. [Google Scholar] [CrossRef]
- Allison, L.A.; Moyle, M.; Shales, M.; Ingles, C.J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 1985, 42, 599–610. [Google Scholar] [CrossRef]
- Li, W.B.; Bzik, D.J.; Gu, H.M.; Tanaka, M.; Fox, B.A.; Inselburg, J. An enlarged largest subunit of Plasmodium falciparum RNA polymerase II defines conserved and variable RNA polymerase domains. Nucleic Acids Res. 1989, 17, 9621–9636. [Google Scholar] [CrossRef]
- Chapman, R.D.; Conrad, M.; Eick, D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol. Cell Biol. 2005, 25, 7665–7674. [Google Scholar] [CrossRef]
- Corden, J.L. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem. Rev. 2013, 113, 8423–8455. [Google Scholar] [CrossRef]
- Dahmus, M.E. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol. 1994, 48, 143–179. [Google Scholar]
- Dahmus, M.E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996, 271, 19009–19012. [Google Scholar]
- Cadena, D.L.; Dahmus, M.E. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J. Biol. Chem. 1987, 262, 12468–12474. [Google Scholar]
- Fuda, N.J.; Ardehali, M.B.; Lis, J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 2009, 461, 186–192. [Google Scholar] [CrossRef]
- Heidemann, M.; Hintermair, C.; Voss, K.; Eick, D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta 2013, 1829, 55–62. [Google Scholar]
- Hintermair, C.; Heidemann, M.; Koch, F.; Descostes, N.; Gut, M.; Gut, I.; Fenouil, R.; Ferrier, P.; Flatley, A.; Kremmer, E.; et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012, 31, 2784–2797. [Google Scholar] [CrossRef]
- Egloff, S.; O’Reilly, D.; Chapman, R.D.; Taylor, A.; Tanzhaus, K.; Pitts, L.; Eick, D.; Murphy, S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007, 318, 1777–1779. [Google Scholar] [CrossRef]
- Chapman, R.D.; Heidemann, M.; Albert, T.K.; Mailhammer, R.; Flatley, A.; Meisterernst, M.; Kremmer, E.; Eick, D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 2007, 318, 1780–1782. [Google Scholar] [CrossRef]
- Mayer, A.; Heidemann, M.; Lidschreiber, M.; Schreieck, A.; Sun, M.; Hintermair, C.; Kremmer, E.; Eick, D.; Cramer, P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012, 336, 1723–1725. [Google Scholar] [CrossRef]
- Hsin, J.P.; Sheth, A.; Manley, J.L. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3' end processing. Science 2011, 334, 683–686. [Google Scholar] [CrossRef]
- Sims, R.J., III; Rojas, L.A.; Beck, D.; Bonasio, R.; Schuller, R.; Drury, W.J., III; Eick, D.; Reinberg, D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 2011, 332, 99–103. [Google Scholar] [CrossRef]
- Liou, Y.C.; Zhou, X.Z.; Lu, K.P. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 2011, 36, 501–514. [Google Scholar] [CrossRef]
- Lu, K.P.; Finn, G.; Lee, T.H.; Nicholson, L.K. Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 2007, 3, 619–629. [Google Scholar] [CrossRef]
- Payne, J.M.; Dahmus, M.E. Partial purification and characterization of two distinct protein kinases that differentially phosphorylate the carboxyl-terminal domain of RNA polymerase subunit IIa. J. Biol. Chem. 1993, 268, 80–87. [Google Scholar]
- Eberhardt, E.S.; Panisik, N., Jr.; Raines, R.T. Inductive Effects on the Energetics of Prolyl Peptide Bond Isomerization: Implications for Collagen Folding and Stability. J. Am. Chem. Soc. 1996, 118, 12261–12266. [Google Scholar]
- Brandts, J.F.; Halvorson, H.R.; Brennan, M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 1975, 14, 4953–4963. [Google Scholar] [CrossRef]
- Brandts, J.F.; Lin, L.N. Proline isomerization studied with proteolytic enzymes. Meth. Enzymol. 1986, 131, 107–126. [Google Scholar]
- Cook, K.H.; Schmid, F.X.; Baldwin, R.L. Role of proline isomerization in folding of ribonuclease A at low temperatures. Proc. Natl. Acad. Sci. USA 1979, 76, 6157–6161. [Google Scholar] [CrossRef]
- Lu, K.P.; Hanes, S.D.; Hunter, T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 1996, 380, 544–547. [Google Scholar] [CrossRef]
- Zhou, X.Z.; Lu, P.J.; Wulf, G.; Lu, K.P. Phosphorylation-dependent prolyl isomerization: A novel signaling regulatory mechanism. Cell. Mol. Life Sci. 1999, 56, 788–806. [Google Scholar] [CrossRef]
- Lu, K.P. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 2003, 4, 175–180. [Google Scholar] [CrossRef]
- Shaw, P.E. Peptidyl-prolyl isomerases: A new twist to transcription. EMBO Rep. 2002, 3, 521–526. [Google Scholar] [CrossRef]
- Shaw, P.E. Peptidyl-prolyl cis/trans isomerases and transcription: Is there a twist in the tail? EMBO Rep. 2007, 8, 40–45. [Google Scholar] [CrossRef]
- Ranganathan, R.; Lu, K.P.; Hunter, T.; Noel, J.P. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 1997, 89, 875–886. [Google Scholar] [CrossRef]
- Zhang, Y.; Daum, S.; Wildemann, D.; Zhou, X.Z.; Verdecia, M.A.; Bowman, M.E.; Lucke, C.; Hunter, T.; Lu, K.P.; Fischer, G.; et al. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem. Biol. 2007, 2, 320–328. [Google Scholar] [CrossRef]
- Etzkorn, F.A. Pin1 flips Alzheimer’s switch. ACS Chem. Biol. 2006, 1, 214–216. [Google Scholar] [CrossRef]
- Wang, X.J.; Etzkorn, F.A. Peptidyl-prolyl isomerase inhibitors. Biopolymers 2006, 84, 125–146. [Google Scholar]
- Xu, G.G.; Etzkorn, F.A. Pin1 as an anticancer drug target. Drug News Perspect. 2009, 22, 399–407. [Google Scholar] [CrossRef]
- Xu, Y.X.; Hirose, Y.; Zhou, X.Z.; Lu, K.P.; Manley, J.L. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003, 17, 2765–2776. [Google Scholar] [CrossRef]
- Wu, X.; Wilcox, C.B.; Devasahayam, G.; Hackett, R.L.; Arevalo-Rodriguez, M.; Cardenas, M.E.; Heitman, J.; Hanes, S.D. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000, 19, 3727–3738. [Google Scholar] [CrossRef]
- Singh, N.; Ma, Z.; Gemmill, T.; Wu, X.; Defiglio, H.; Rossettini, A.; Rabeler, C.; Beane, O.; Morse, R.H.; Palumbo, M.J.; et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell 2009, 36, 255–266. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Ghazy, M.A.; Moore, C.; Hampsey, M. Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Mol. Cell Biol. 2009, 29, 2925–2934. [Google Scholar] [CrossRef]
- Verdecia, M.A.; Bowman, M.E.; Lu, K.P.; Hunter, T.; Noel, J.P. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 2000, 7, 639–643. [Google Scholar]
- Armache, K.J.; Mitterweger, S.; Meinhart, A.; Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005, 280, 7131–7134. [Google Scholar]
- Cramer, P.; Bushnell, D.A.; Kornberg, R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 2001, 292, 1863–1876. [Google Scholar] [CrossRef]
- Myers, J.K.; Morris, D.P.; Greenleaf, A.L.; Oas, T.G. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry 2001, 40, 8479–8486. [Google Scholar]
- Zhang, M.; Wang, X.J.; Chen, X.; Bowman, M.E.; Luo, Y.; Noel, J.P.; Ellington, A.D.; Etzkorn, F.A.; Zhang, Y. Structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. ACS Chem. Biol. 2012, 7, 1462–1470. [Google Scholar] [CrossRef]
- Zhang, Y.; Kim, Y.; Genoud, N.; Gao, J.; Kelly, J.W.; Pfaff, S.L.; Gill, G.N.; Dixon, J.E.; Noel, J.P. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell 2006, 24, 759–770. [Google Scholar] [CrossRef]
- Xiang, K.; Nagaike, T.; Xiang, S.; Kilic, T.; Beh, M.M.; Manley, J.L.; Tong, L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 2010, 467, 729–733. [Google Scholar] [CrossRef]
- He, X.; Khan, A.U.; Cheng, H.; Pappas, D.L., Jr.; Hampsey, M.; Moore, C.L. Functional interactions between the transcription and mRNA 3' end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003, 17, 1030–1042. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; He, X.; Reyes-Reyes, M.; Moore, C.; Hampsey, M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell 2004, 14, 387–394. [Google Scholar] [CrossRef]
- Sun, Z.W.; Hampsey, M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell Biol. 1996, 16, 1557–1566. [Google Scholar]
- Reyes-Reyes, M.; Hampsey, M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol. Cell Biol. 2007, 27, 926–936. [Google Scholar] [CrossRef]
- Dichtl, B.; Blank, D.; Ohnacker, M.; Friedlein, A.; Roeder, D.; Langen, H.; Keller, W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 2002, 10, 1139–1150. [Google Scholar] [CrossRef]
- Ganem, C.; Devaux, F.; Torchet, C.; Jacq, C.; Quevillon-Cheruel, S.; Labesse, G.; Facca, C.; Faye, G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003, 22, 1588–1598. [Google Scholar] [CrossRef]
- Luo, Y.; Yogesha, S.D.; Cannon, J.R.; Yan, W.; Ellington, A.D.; Brodbelt, J.S.; Zhang, Y. Novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem. Biol. 2013, 8, 2042–2052. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Zhang, Y. Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 2011, 434, 435–444. [Google Scholar] [CrossRef]
- Werner-Allen, J.W.; Lee, C.J.; Liu, P.; Nicely, N.I.; Wang, S.; Greenleaf, A.L.; Zhou, P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 2011, 286, 5717–5726. [Google Scholar]
- Zhang, M.; Yogesha, S.D.; Mayfield, J.E.; Gill, G.N.; Zhang, Y. Viewing serine/threonine protein phosphatases through the eyes of drug designers. FEBS J. 2013, 280, 4739–4760. [Google Scholar] [CrossRef]
- Bataille, A.R.; Jeronimo, C.; Jacques, P.E.; Laramee, L.; Fortin, M.E.; Forest, A.; Bergeron, M.; Hanes, S.D.; Robert, F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell 2012, 45, 158–170. [Google Scholar] [CrossRef]
- Zhang, D.W.; Mosley, A.L.; Ramisetty, S.R.; Rodriguez-Molina, J.B.; Washburn, M.P.; Ansari, A.Z. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 2012, 287, 8541–8551. [Google Scholar]
- Xiang, K.; Manley, J.L.; Tong, L. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 2012, 26, 2265–2270. [Google Scholar] [CrossRef]
- Yeo, M.; Lin, P.S.; Dahmus, M.E.; Gill, G.N. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem 2003, 278, 26078–26085. [Google Scholar]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005, 307, 596–600. [Google Scholar] [CrossRef]
- Zhang, M.; Cho, E.J.; Burstein, G.; Siegel, D.; Zhang, Y. Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem. Biol. 2011, 6, 511–519. [Google Scholar] [CrossRef]
- Chambers, R.S.; Dahmus, M.E. Purification and characterization of a phosphatase from HeLa cells which dephosphorylates the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1994, 269, 26243–26248. [Google Scholar]
- Zhang, M.; Liu, J.; Kim, Y.; Dixon, J.E.; Pfaff, S.L.; Gill, G.N.; Noel, J.P.; Zhang, Y. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 2010, 19, 974–986. [Google Scholar]
- Zhang, X.; Morera, S.; Bates, P.A.; Whitehead, P.C.; Coffer, A.I.; Hainbucher, K.; Nash, R.A.; Sternberg, M.J.; Lindahl, T.; Freemont, P.S. Structure of an XRCC1 BRCT domain: A new protein-protein interaction module. EMBO J. 1998, 17, 6404–6411. [Google Scholar] [CrossRef]
- Kobor, M.S.; Archambault, J.; Lester, W.; Holstege, F.C.; Gileadi, O.; Jansma, D.B.; Jennings, E.G.; Kouyoumdjian, F.; Davidson, A.R.; Young, R.A.; et al. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 1999, 4, 55–62. [Google Scholar] [CrossRef]
- Kobor, M.S.; Simon, L.D.; Omichinski, J.; Zhong, G.; Archambault, J.; Greenblatt, J. A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol. Cell Biol. 2000, 20, 7438–7449. [Google Scholar] [CrossRef]
- Ghosh, A.; Shuman, S.; Lima, C.D. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol. Cell 2008, 32, 478–490. [Google Scholar] [CrossRef]
- Hausmann, S.; Shuman, S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J. Biol. Chem. 2002, 277, 21213–21220. [Google Scholar] [CrossRef]
- Cho, H.; Kim, T.K.; Mancebo, H.; Lane, W.S.; Flores, O.; Reinberg, D. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 1999, 13, 1540–1552. [Google Scholar] [CrossRef]
- Kops, O.; Zhou, X.Z.; Lu, K.P. Pin1 modulates the dephosphorylation of the RNA polymerase II C-terminal domain by yeast Fcp1. FEBS Lett. 2002, 513, 305–311. [Google Scholar] [CrossRef]
- Palancade, B.; Marshall, N.F.; Tremeau-Bravard, A.; Bensaude, O.; Dahmus, M.E.; Dubois, M.-F. Dephosphorylation of RNA polymerase II by CTD-phosphatase FCP1 is inhibited by phospho-CTD associating proteins. J. Mol. Biol. 2004, 335, 415–424. [Google Scholar] [CrossRef]
- Gibney, P.A.; Fries, T.; Bailer, S.M.; Morano, K.A. Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryotic Cell 2008, 7, 938–948. [Google Scholar] [CrossRef]
- Mosley, A.L.; Pattenden, S.G.; Carey, M.; Venkatesh, S.; Gilmore, J.M.; Florens, L.; Workman, J.L.; Washburn, M.P. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 2009, 34, 168–178. [Google Scholar] [CrossRef]
- Xiang, K.; Manley, J.L.; Tong, L. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat. Commun. 2012, 3, 946. [Google Scholar] [CrossRef]
- Egloff, S.; Zaborowska, J.; Laitem, C.; Kiss, T.; Murphy, S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell 2012, 45, 111–122. [Google Scholar] [CrossRef]
- Jeronimo, C.; Forget, D.; Bouchard, A.; Li, Q.; Chua, G.; Poitras, C.; Therien, C.; Bergeron, D.; Bourassa, S.; Greenblatt, J.; et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 2007, 27, 262–274. [Google Scholar] [CrossRef]
- Nestel, F.P.; Colwill, K.; Harper, S.; Pawson, T.; Anderson, S.K. RS cyclophilins: Identification of an NK-TR1-related cyclophilin. Gene 1996, 180, 151–155. [Google Scholar] [CrossRef]
- Fichtinger, G.; Deguet, A.; Masamune, K.; Balogh, E.; Fischer, G.S.; Mathieu, H.; Taylor, R.H.; Zinreich, S.J.; Fayad, L.M. Image overlay guidance for needle insertion in CT scanner. IEEE Trans. Biomed. Eng. 2005, 52, 1415–1424. [Google Scholar] [CrossRef]
- Bourquin, J.P.; Stagljar, I.; Meier, P.; Moosmann, P.; Silke, J.; Baechi, T.; Georgiev, O.; Schaffner, W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997, 25, 2055–2061. [Google Scholar] [CrossRef]
- Yuryev, A.; Patturajan, M.; Litingtung, Y.; Joshi, R.V.; Gentile, C.; Gebara, M.; Corden, J.L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 6975–6980. [Google Scholar]
- Brown, N.R.; Noble, M.E.; Endicott, J.A.; Johnson, L.N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999, 1, 438–443. [Google Scholar] [CrossRef]
- Weiwad, M.; Kullertz, G.; Schutkowski, M.; Fischer, G. Evidence that the substrate backbone conformation is critical to phosphorylation by p42 MAP kinase. FEBS Lett. 2000, 478, 39–42. [Google Scholar] [CrossRef]
- Zhou, X.Z.; Kops, O.; Werner, A.; Lu, P.J.; Shen, M.; Stoller, G.; Kullertz, G.; Stark, M.; Fischer, G.; Lu, K.P. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell 2000, 6, 873–883. [Google Scholar] [CrossRef]
- Jeffrey, P.D.; Russo, A.A.; Polyak, K.; Gibbs, E.; Hurwitz, J.; Massague, J.; Pavletich, N.P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 1995, 376, 313–320. [Google Scholar] [CrossRef]
- Schutkowski, M.; Bernhardt, A.; Zhou, X.Z.; Shen, M.; Reimer, U.; Rahfeld, J.U.; Lu, K.P.; Fischer, G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry 1998, 37, 5566–5575. [Google Scholar] [CrossRef]
- Hsin, J.P.; Manley, J.L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012, 26, 2119–2137. [Google Scholar] [CrossRef]
- Compe, E.; Egly, J.M. TFIIH: When transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012, 13, 343–354. [Google Scholar] [CrossRef]
- Lu, H.; Zawel, L.; Fisher, L.; Egly, J.M.; Reinberg, D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 1992, 358, 641–645. [Google Scholar] [CrossRef]
- Yankulov, K.Y.; Bentley, D.L. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997, 16, 1638–1646. [Google Scholar] [CrossRef]
- Ganuza, M.; Santamaria, D. Cdk7: Open questions beyond the prevailing model. Cell Cycle 2012, 11, 3519–3520. [Google Scholar] [CrossRef]
- Morgan, D.O. Principles of CDK regulation. Nature 1995, 374, 131–134. [Google Scholar] [CrossRef]
- Roy, R.; Adamczewski, J.P.; Seroz, T.; Vermeulen, W.; Tassan, J.P.; Schaeffer, L.; Nigg, E.A.; Hoeijmakers, J.H.; Egly, J.M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 1994, 79, 1093–1101. [Google Scholar] [CrossRef]
- Serizawa, H.; Makela, T.P.; Conaway, J.W.; Conaway, R.C.; Weinberg, R.A.; Young, R.A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 1995, 374, 280–282. [Google Scholar] [CrossRef]
- Shiekhattar, R.; Mermelstein, F.; Fisher, R.P.; Drapkin, R.; Dynlacht, B.; Wessling, H.C.; Morgan, D.O.; Reinberg, D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 1995, 374, 283–287. [Google Scholar] [CrossRef]
- Adamczewski, J.P.; Rossignol, M.; Tassan, J.P.; Nigg, E.A.; Moncollin, V.; Egly, J.M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996, 15, 1877–1884. [Google Scholar]
- Zhovmer, A.; Oksenych, V.; Coin, F. Two sides of the same coin: TFIIH complexes in transcription and DNA repair. TheScientificWorldJOURNAL 2010, 10, 633–643. [Google Scholar] [CrossRef]
- Svejstrup, J.Q.; Feaver, W.J.; Kornberg, R.D. Subunits of yeast RNA polymerase II transcription factor TFIIH encoded by the CCL1 gene. J. Biol. Chem. 1996, 271, 643–645. [Google Scholar] [CrossRef]
- Keogh, M.C.; Cho, E.J.; Podolny, V.; Buratowski, S. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell Biol. 2002, 22, 1288–1297. [Google Scholar] [CrossRef]
- Espinoza, F.H.; Farrell, A.; Erdjument-Bromage, H.; Tempst, P.; Morgan, D.O. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 1996, 273, 1714–1717. [Google Scholar] [CrossRef]
- Fisher, R.P. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J. Cell Sci. 2005, 118, 5171–5180. [Google Scholar] [CrossRef]
- Woychik, N.A.; Hampsey, M. The RNA polymerase II machinery: Structure illuminates function. Cell 2002, 108, 453–463. [Google Scholar] [CrossRef]
- Yudkovsky, N.; Ranish, J.A.; Hahn, S. A transcription reinitiation intermediate that is stabilized by activator. Nature 2000, 408, 225–229. [Google Scholar] [CrossRef]
- Holstege, F.C.; van der Vliet, P.C.; Timmers, H.T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996, 15, 1666–1677. [Google Scholar]
- Dvir, A.; Conaway, J.W.; Conaway, R.C. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. 2001, 11, 209–214. [Google Scholar] [CrossRef]
- Feaver, W.J.; Svejstrup, J.Q.; Henry, N.L.; Kornberg, R.D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 1994, 79, 1103–1109. [Google Scholar] [CrossRef]
- Buratowski, S. Progression through the RNA polymerase II CTD cycle. Mol. Cell 2009, 36, 541–546. [Google Scholar] [CrossRef]
- Akoulitchev, S.; Makela, T.P.; Weinberg, R.A.; Reinberg, D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 1995, 377, 557–560. [Google Scholar] [CrossRef]
- Liu, Y.; Kung, C.; Fishburn, J.; Ansari, A.Z.; Shokat, K.M.; Hahn, S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell Biol. 2004, 24, 1721–1735. [Google Scholar] [CrossRef]
- Sogaard, T.M.; Svejstrup, J.Q. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 2007, 282, 14113–14120. [Google Scholar] [CrossRef]
- Lu, H.; Flores, O.; Weinmann, R.; Reinberg, D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. USA 1991, 88, 10004–10008. [Google Scholar] [CrossRef]
- Rodriguez, C.R.; Cho, E.J.; Keogh, M.C.; Moore, C.L.; Greenleaf, A.L.; Buratowski, S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell Biol. 2000, 20, 104–112. [Google Scholar] [CrossRef]
- Cho, E.J.; Takagi, T.; Moore, C.R.; Buratowski, S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997, 11, 3319–3326. [Google Scholar] [CrossRef]
- Komarnitsky, P.; Cho, E.J.; Buratowski, S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000, 14, 2452–2460. [Google Scholar] [CrossRef]
- Schwer, B.; Shuman, S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol. Cell 2011, 43, 311–318. [Google Scholar] [CrossRef]
- Suh, M.H.; Meyer, P.A.; Gu, M.; Ye, P.; Zhang, M.; Kaplan, C.D.; Lima, C.D.; Fu, J. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J. Biol. Chem. 2010, 285, 34027–34038. [Google Scholar]
- Baumli, S.; Hole, A.J.; Wang, L.Z.; Noble, M.E.; Endicott, J.A. The CDK9 tail determines the reaction pathway of positive transcription elongation factor b. Structure 2012, 20, 1788–1795. [Google Scholar] [CrossRef]
- Guo, Z.; Stiller, J.W. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics 2004, 5, 69. [Google Scholar] [CrossRef]
- Pei, Y.; Shuman, S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J. Biol. Chem. 2003, 278, 43346–43356. [Google Scholar] [CrossRef]
- Bartkowiak, B.; Mackellar, A.L.; Greenleaf, A.L. Updating the CTD Story: From Tail to Epic. Genet. Res. Int. 2011, 2011, 623–718. [Google Scholar]
- Bartkowiak, B.; Greenleaf, A.L. Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription 2011, 2, 115–119. [Google Scholar] [CrossRef]
- Qiu, H.; Hu, C.; Hinnebusch, A.G. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 2009, 33, 752–762. [Google Scholar] [CrossRef]
- Burger, K.; Muhl, B.; Rohrmoser, M.; Coordes, B.; Heidemann, M.; Kellner, M.; Gruber-Eber, A.; Heissmeyer, V.; Strasser, K.; Eick, D. Cyclin-dependent kinase 9 Links RNA polymerase II transcription to processing of ribosomal RNA. J. Biol. Chem. 2013, 288, 21173–21183. [Google Scholar] [CrossRef]
- Bres, V.; Yoh, S.M.; Jones, K.A. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008, 20, 334–340. [Google Scholar] [CrossRef]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Mayer, A.; Lidschreiber, M.; Siebert, M.; Leike, K.; Soding, J.; Cramer, P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010, 17, 1272–1278. [Google Scholar] [CrossRef]
- Zhou, K.; Kuo, W.H.; Fillingham, J.; Greenblatt, J.F. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. USA 2009, 106, 6956–6961. [Google Scholar] [CrossRef]
- Svejstrup, J.Q. Transcription: Another mark in the tail. EMBO J. 2012, 31, 2753–2754. [Google Scholar] [CrossRef]
- Lee, J.M.; Greenleaf, A.L. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc. Natl. Acad. Sci. USA 1989, 86, 3624–3628. [Google Scholar] [CrossRef]
- Wood, A.; Shilatifard, A. Bur1/Bur2 and the Ctk complex in yeast: The split personality of mammalian P-TEFb. Cell Cycle 2006, 5, 1066–1068. [Google Scholar] [CrossRef]
- Liu, J.; Kipreos, E.T. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): Differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 2000, 17, 1061–1074. [Google Scholar] [CrossRef]
- Chen, H.H.; Wang, Y.C.; Fann, M.J. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol. Cell Biol. 2006, 26, 2736–2745. [Google Scholar] [CrossRef]
- Chen, H.H.; Wong, Y.H.; Geneviere, A.M.; Fann, M.J. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem. Biophys. Res. Commun. 2007, 354, 735–740. [Google Scholar] [CrossRef]
- Blazek, D.; Kohoutek, J.; Bartholomeeusen, K.; Johansen, E.; Hulinkova, P.; Luo, Z.; Cimermancic, P.; Ule, J.; Peterlin, B.M. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011, 25, 2158–2172. [Google Scholar] [CrossRef]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010, 24, 2303–2316. [Google Scholar] [CrossRef]
- Ahn, S.H.; Kim, M.; Buratowski, S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol. Cell 2004, 13, 67–76. [Google Scholar] [CrossRef]
- Cho, E.J.; Kobor, M.S.; Kim, M.; Greenblatt, J.; Buratowski, S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001, 15, 3319–3329. [Google Scholar] [CrossRef]
- Ni, Z.; Schwartz, B.E.; Werner, J.; Suarez, J.R.; Lis, J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 2004, 13, 55–65. [Google Scholar] [CrossRef]
- Rahl, P.B.; Lin, C.Y.; Seila, A.C.; Flynn, R.A.; McCuine, S.; Burge, C.B.; Sharp, P.A.; Young, R.A. c-Myc regulates transcriptional pause release. Cell 2010, 141, 432–445. [Google Scholar] [CrossRef]
- Nechaev, S.; Adelman, K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 2011, 1809, 34–45. [Google Scholar]
- Ahn, S.H.; Keogh, M.C.; Buratowski, S. Ctk1 promotes dissociation of basal transcription factors from elongating RNA polymerase II. EMBO J. 2009, 28, 205–212. [Google Scholar] [CrossRef]
- Tassan, J.P.; Jaquenoud, M.; Leopold, P.; Schultz, S.J.; Nigg, E.A. Identification of human cyclin-dependent kinase 8, a putative protein kinase partner for cyclin C. Proc. Natl. Acad. Sci. USA 1995, 92, 8871–8875. [Google Scholar] [CrossRef]
- Maldonado, E.; Shiekhattar, R.; Sheldon, M.; Cho, H.; Drapkin, R.; Rickert, P.; Lees, E.; Anderson, C.W.; Linn, S.; Reinberg, D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature 1996, 381, 86–89. [Google Scholar] [CrossRef]
- Pan, G.; Aso, T.; Greenblatt, J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 1997, 272, 24563–24571. [Google Scholar] [CrossRef]
- Liao, S.M.; Zhang, J.; Jeffery, D.A.; Koleske, A.J.; Thompson, C.M.; Chao, D.M.; Viljoen, M.; van Vuuren, H.J.; Young, R.A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 1995, 374, 193–196. [Google Scholar] [CrossRef]
- Hengartner, C.J.; Myer, V.E.; Liao, S.M.; Wilson, C.J.; Koh, S.S.; Young, R.A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 1998, 2, 43–53. [Google Scholar] [CrossRef]
- Rickert, P.; Corden, J.L.; Lees, E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 1999, 18, 1093–1102. [Google Scholar] [CrossRef]
- Borggrefe, T.; Davis, R.; Erdjument-Bromage, H.; Tempst, P.; Kornberg, R.D. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 2002, 277, 44202–44207. [Google Scholar]
- Tsai, K.L.; Sato, S.; Tomomori-Sato, C.; Conaway, R.C.; Conaway, J.W.; Asturias, F.J. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013, 20, 611–619. [Google Scholar] [CrossRef]
- Knuesel, M.T.; Meyer, K.D.; Bernecky, C.; Taatjes, D.J. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009, 23, 439–451. [Google Scholar] [CrossRef]
- Akoulitchev, S.; Chuikov, S.; Reinberg, D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 2000, 407, 102–106. [Google Scholar] [CrossRef]
- Galbraith, M.D.; Donner, A.J.; Espinosa, J.M. CDK8: A positive regulator of transcription. Transcription 2010, 1, 4–12. [Google Scholar] [CrossRef]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef]
- Donner, A.J.; Ebmeier, C.C.; Taatjes, D.J.; Espinosa, J.M. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010, 17, 194–201. [Google Scholar] [CrossRef]
- Alarcon, C.; Zaromytidou, A.I.; Xi, Q.; Gao, S.; Yu, J.; Fujisawa, S.; Barlas, A.; Miller, A.N.; Manova-Todorova, K.; Macias, M.J.; et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 2009, 139, 757–769. [Google Scholar] [CrossRef]
- Galbraith, M.D.; Allen, M.A.; Bensard, C.L.; Wang, X.; Schwinn, M.K.; Qin, B.; Long, H.W.; Daniels, D.L.; Hahn, W.C.; Dowell, R.D.; et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 2013, 153, 1327–1339. [Google Scholar] [CrossRef]
- Hirst, M.; Kobor, M.S.; Kuriakose, N.; Greenblatt, J.; Sadowski, I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 1999, 3, 673–678. [Google Scholar] [CrossRef]
- Rohde, J.R.; Trinh, J.; Sadowski, I. Multiple signals regulate GAL transcription in yeast. Mol. Cell Biol. 2000, 20, 3880–3886. [Google Scholar] [CrossRef]
- Vincent, O.; Kuchin, S.; Hong, S.P.; Townley, R.; Vyas, V.K.; Carlson, M. Interaction of the Srb10kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 5790–5796. [Google Scholar] [CrossRef]
- Mo, X.; Kowenz-Leutz, E.; Xu, H.; Leutz, A. Ras induces mediator complex exchange on C/EBP beta. Mol. Cell 2004, 13, 241–250. [Google Scholar] [CrossRef]
- Phatnani, H.P.; Greenleaf, A.L. Identifying phosphoCTD-associating proteins. Meth. Mol. Biol. 2004, 257, 17–28. [Google Scholar]
- Phatnani, H.P.; Jones, J.C.; Greenleaf, A.L. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: Novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 2004, 43, 15702–15719. [Google Scholar] [CrossRef]
- Jeronimo, C.; Bataille, A.R.; Robert, F. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem. Rev. 2013, 113, 8491–8522. [Google Scholar] [CrossRef]
- Zhang, D.W.; Rodriguez-Molina, J.B.; Tietjen, J.R.; Nemec, C.M.; Ansari, A.Z. Emerging views on the CTD code. Genet. Res. Int. 2012, 2012, 347214. [Google Scholar]
- Kubicek, K.; Cerna, H.; Holub, P.; Pasulka, J.; Hrossova, D.; Loehr, F.; Hofr, C.; Vanacova, S.; Stefl, R. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012, 26, 1891–1896. [Google Scholar] [CrossRef]
- Arigo, J.T.; Eyler, D.E.; Carroll, K.L.; Corden, J.L. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 2006, 23, 841–851. [Google Scholar] [CrossRef]
- Kim, M.; Vasiljeva, L.; Rando, O.J.; Zhelkovsky, A.; Moore, C.; Buratowski, S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell 2006, 24, 723–734. [Google Scholar] [CrossRef]
- Carroll, K.L.; Pradhan, D.A.; Granek, J.A.; Clarke, N.D.; Corden, J.L. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell Biol. 2004, 24, 6241–6252. [Google Scholar] [CrossRef]
- Gudipati, R.K.; Villa, T.; Boulay, J.; Libri, D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008, 15, 786–794. [Google Scholar] [CrossRef]
- Steinmetz, E.J.; Conrad, N.K.; Brow, D.A.; Corden, J.L. RNA-binding protein Nrd1 directs poly(A)-independent 3'-end formation of RNA polymerase II transcripts. Nature 2001, 413, 327–331. [Google Scholar] [CrossRef]
- Vasiljeva, L.; Kim, M.; Mutschler, H.; Buratowski, S.; Meinhart, A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008, 15, 795–804. [Google Scholar] [CrossRef]
- Steinmetz, E.J.; Ng, S.B.; Cloute, J.P.; Brow, D.A. cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell Biol. 2006, 26, 2688–2696. [Google Scholar] [CrossRef]
- Lykke-Andersen, S.; Jensen, T.H. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie 2007, 89, 1177–1182. [Google Scholar] [CrossRef]
- Samaranayake, D.; Atencio, D.; Morse, R.; Wade, J.T.; Chaturvedi, V.; Hanes, S.D. Role of Ess1 in growth, morphogenetic switching, and RNA polymerase II transcription in Candida albicans. PloS One 2013, 8, e59094. [Google Scholar]
- Barilla, D.; Lee, B.A.; Proudfoot, N.J. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2001, 98, 445–450. [Google Scholar]
- Licatalosi, D.D.; Geiger, G.; Minet, M.; Schroeder, S.; Cilli, K.; McNeil, J.B.; Bentley, D.L. Functional interaction of yeast pre-mRNA 3'-end processing factors with RNA polymerase II. Mol. Cell 2002, 9, 1101–1111. [Google Scholar] [CrossRef]
- Meinhart, A.; Cramer, P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature 2004, 430, 223–226. [Google Scholar] [CrossRef]
- Noble, C.G.; Hollingworth, D.; Martin, S.R.; Ennis-Adeniran, V.; Smerdon, S.J.; Kelly, G.; Taylor, I.A.; Ramos, A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 2005, 12, 144–151. [Google Scholar] [CrossRef]
- Lunde, B.M.; Reichow, S.L.; Kim, M.; Suh, H.; Leeper, T.C.; Yang, F.; Mutschler, H.; Buratowski, S.; Meinhart, A.; Varani, G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2010, 17, 1195–1201. [Google Scholar] [CrossRef]
- Furuichi, Y.; Shatkin, A.J. Viral and cellular mRNA capping: Past and prospects. Adv. Virus Res. 2000, 55, 135–184. [Google Scholar] [CrossRef]
- Issur, M.; Picard-Jean, F.; Bisaillon, M. The RNA capping machinery as an anti-infective target. Wiley Interdiscip. Rev. RNA 2011, 2, 184–192. [Google Scholar] [CrossRef]
- Burley, S.K.; Sonenberg, N. Gimme phospho-serine five! Capping enzyme guanylyltransferase recognition of the RNA polymerase II CTD. Mol. Cell 2011, 43, 163–165. [Google Scholar] [CrossRef]
- Ho, C.K.; Shuman, S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 1999, 3, 405–411. [Google Scholar] [CrossRef]
- Schroeder, S.C.; Schwer, B.; Shuman, S.; Bentley, D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000, 14, 2435–2440. [Google Scholar] [CrossRef]
- West, M.L.; Corden, J.L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 1995, 140, 1223–1233. [Google Scholar]
- Chu, C.; Das, K.; Tyminski, J.R.; Bauman, J.D.; Guan, R.; Qiu, W.; Montelione, G.T.; Arnold, E.; Shatkin, A.J. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. USA 2011, 108, 10104–10108. [Google Scholar] [CrossRef]
- Fabrega, C.; Shen, V.; Shuman, S.; Lima, C.D. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell 2003, 11, 1549–1561. [Google Scholar] [CrossRef]
- Ghosh, A.; Shuman, S.; Lima, C.D. Structural insights to how mammalian capping enzyme reads the CTD code. Mol. Cell 2011, 43, 299–310. [Google Scholar] [CrossRef]
- Gu, M.; Rajashankar, K.R.; Lima, C.D. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 2010, 18, 216–227. [Google Scholar] [CrossRef]
- Kim, M.; Krogan, N.J.; Vasiljeva, L.; Rando, O.J.; Nedea, E.; Greenblatt, J.F.; Buratowski, S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 2004, 432, 517–522. [Google Scholar] [CrossRef]
- West, S.; Gromak, N.; Proudfoot, N.J. Human 5' → 3' exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 2004, 432, 522–525. [Google Scholar] [CrossRef]
- Tollervey, D. Molecular biology: Termination by torpedo. Nature 2004, 432, 456–457. [Google Scholar] [CrossRef]
- Pearson, E.L.; Moore, C.L. Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1. J. Biol. Chem. 2013, 288, 19750–19759. [Google Scholar] [CrossRef]
- Becker, R.; Loll, B.; Meinhart, A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2008, 283, 22659–22669. [Google Scholar] [CrossRef]
- Patturajan, M.; Wei, X.; Berezney, R.; Corden, J.L. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell Biol. 1998, 18, 2406–2415. [Google Scholar]
- Yaffe, M.B.; Schutkowski, M.; Shen, M.; Zhou, X.Z.; Stukenberg, P.T.; Rahfeld, J.U.; Xu, J.; Kuang, J.; Kirschner, M.W.; Fischer, G.; et al. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science 1997, 278, 1957–1960. [Google Scholar] [CrossRef]
- Shen, M.; Stukenberg, P.T.; Kirschner, M.W.; Lu, K.P. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998, 12, 706–720. [Google Scholar] [CrossRef]
- Korzhnev, D.M.; Religa, T.L.; Banachewicz, W.; Fersht, A.R.; Kay, L.E. A transient and low-populated protein-folding intermediate at atomic resolution. Science 2010, 329, 1312–1316. [Google Scholar] [CrossRef]
- Evans, P.A.; Dobson, C.M.; Kautz, R.A.; Hatfull, G.; Fox, R.O. Proline isomerism in staphylococcal nuclease characterized by NMR and site-directed mutagenesis. Nature 1987, 329, 266–268. [Google Scholar] [CrossRef]
- Wang, X.J.; Xu, B.; Mullins, A.B.; Neiler, F.K.; Etzkorn, F.A. Conformationally locked isostere of phosphoSer-cis-Pro inhibits Pin1 23-fold better than phosphoSer-trans-Pro isostere. J. Am. Chem. Soc. 2004, 126, 15533–15542. [Google Scholar] [CrossRef]
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yogesha, S.D.; Mayfield, J.E.; Zhang, Y. Cross-Talk of Phosphorylation and Prolyl Isomerization of the C-terminal Domain of RNA Polymerase II. Molecules 2014, 19, 1481-1511. https://doi.org/10.3390/molecules19021481
Yogesha SD, Mayfield JE, Zhang Y. Cross-Talk of Phosphorylation and Prolyl Isomerization of the C-terminal Domain of RNA Polymerase II. Molecules. 2014; 19(2):1481-1511. https://doi.org/10.3390/molecules19021481
Chicago/Turabian StyleYogesha, S. D., Joshua E. Mayfield, and Yan Zhang. 2014. "Cross-Talk of Phosphorylation and Prolyl Isomerization of the C-terminal Domain of RNA Polymerase II" Molecules 19, no. 2: 1481-1511. https://doi.org/10.3390/molecules19021481
APA StyleYogesha, S. D., Mayfield, J. E., & Zhang, Y. (2014). Cross-Talk of Phosphorylation and Prolyl Isomerization of the C-terminal Domain of RNA Polymerase II. Molecules, 19(2), 1481-1511. https://doi.org/10.3390/molecules19021481



