Combined Mass Spectrometry-Based Metabolite Profiling of Different Pigmented Rice (Oryza sativa L.) Seeds and Correlation with Antioxidant Activities
Abstract
:1. Introduction
2. Results and Discussion
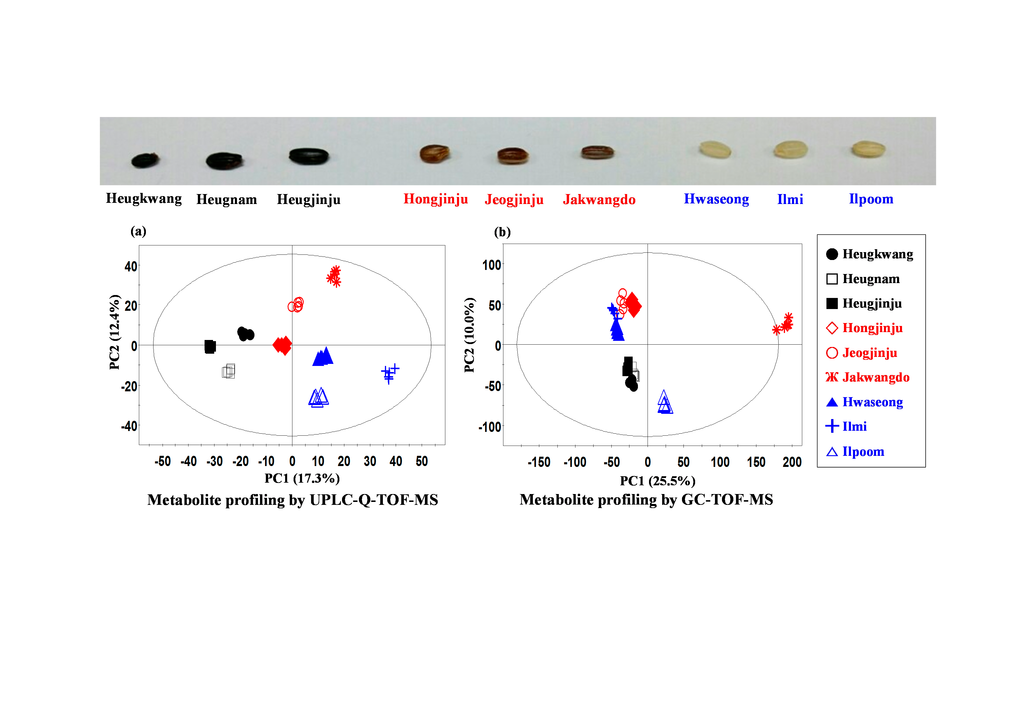
2.1. Metabolite Profiling of Different Pigmented Rice Seeds by UPLC-Q-TOF-MS
 HoJJ (Hongjinju),
HoJJ (Hongjinju),  JJJ (Jeogjinju),
JJJ (Jeogjinju),  JKD (Jakwangdo); White rice seed:
JKD (Jakwangdo); White rice seed:  HS (Hwaseong),
HS (Hwaseong), 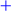 IM (Ilmi),
IM (Ilmi),  IP (Ilpoom). * Target identified.
IP (Ilpoom). * Target identified.
 HoJJ (Hongjinju),
HoJJ (Hongjinju),  JJJ (Jeogjinju),
JJJ (Jeogjinju),  JKD (Jakwangdo); White rice seed:
JKD (Jakwangdo); White rice seed:  HS (Hwaseong),
HS (Hwaseong),  IM (Ilmi),
IM (Ilmi),  IP (Ilpoom). * Target identified.
IP (Ilpoom). * Target identified.
2.2. Metabolite Profiling of Different Pigmented Rice Seeds by GC-TOF-MS
 HoJJ (Hongjinju),
HoJJ (Hongjinju),  JJJ (Jeogjinju),
JJJ (Jeogjinju),  JKD (Jakwangdo); White rice seed:
JKD (Jakwangdo); White rice seed:  HS (Hwaseong),
HS (Hwaseong), 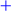 IM (Ilmi),
IM (Ilmi),  IP (Ilpoom).
IP (Ilpoom).
 HoJJ (Hongjinju),
HoJJ (Hongjinju),  JJJ (Jeogjinju),
JJJ (Jeogjinju),  JKD (Jakwangdo); White rice seed:
JKD (Jakwangdo); White rice seed:  HS (Hwaseong),
HS (Hwaseong), 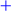 IM (Ilmi),
IM (Ilmi),  IP (Ilpoom).
IP (Ilpoom).
2.3. Correlation between Rice Seed Metabolites and Antioxidant Activities

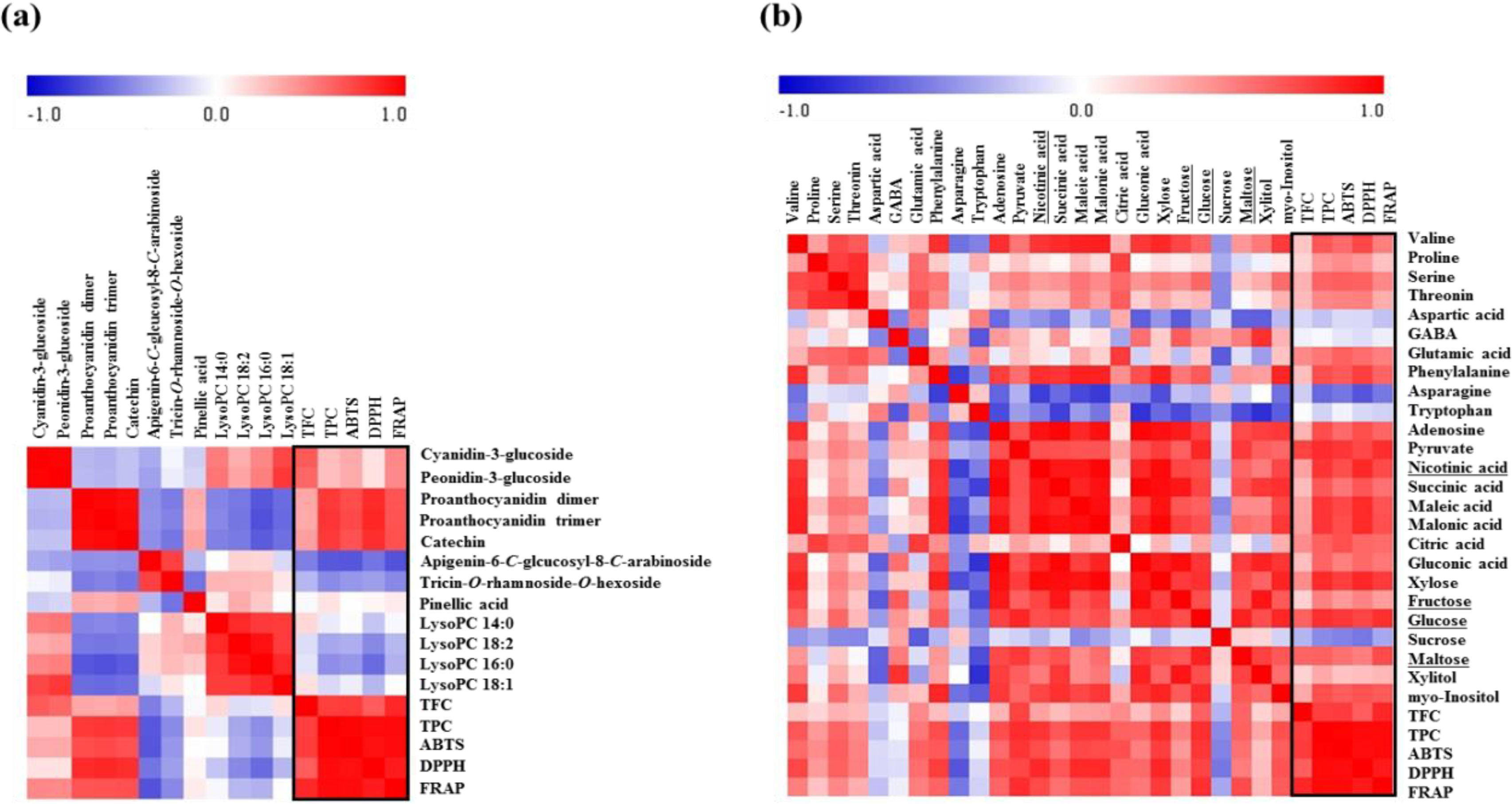
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Plant Materials
3.3. Sample Preparation
3.4. GC-TOF-MS Analysis
3.5. UPLC-Q-TOF-MS and LC-IT-MS/MS Analysis
3.6. Determination of Antioxidant Activities Using ABTS, DPPH, and FRAP Assays
3.7. Determination of Total Phenolic Contents (TPC) and Total Flavonoid Contents (TFC)
3.8. Data Processing and Multivariate Analysis
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bhattacharjee, P.; Singhal, R.S.; Kulkarni, P.R. Basmati rice: A review. Int. J. Food Sci. Technol. 2002, 37, 1–12. [Google Scholar] [CrossRef]
- Deng, G.F.; Xu, X.R.; Zhang, Y.; Li, D.; Gan, R.Y.; Li, H.B. Phenolic compounds and bioactivities of pigmented rice. Crit. Rev. Food Sci. 2013, 53, 296–306. [Google Scholar] [CrossRef]
- Yoshida, H.; Tomiyama, Y.; Mizushina, Y. Lipid components, fatty acids and triacylglycerol molecular species of black and red rices. Food Chem. 2010, 123, 210–215. [Google Scholar] [CrossRef]
- Abdel-Aal, E.S.M.; Young, J.C.; Rabalski, I. Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 2006, 54, 4696–4704. [Google Scholar] [CrossRef] [PubMed]
- Vuchapong, J.; Sookserm, M.; Srijesdaruk, V.; Swatsitang, P.; Srijaranai, S. High performance liquid chromatigraphic analysis of phenolic compounds and their antioxidant activities in rice varieties. LWT-Food Sci. Technol. 2010, 43, 1325–1330. [Google Scholar] [CrossRef]
- Min, B.; Gu, L.; McClung, A.M.; Bergman, C.J.; Chen, M.H. Free and bound total phenolic concentrations, antioxidant capacities, and profiles of proanthocyanidins and anthocyanins in whole grain rice (Oryza sativa L.) of different bran colours. Food Chem. 2012, 133, 715–722. [Google Scholar] [CrossRef]
- Nam, S.H.; Choi, S.P.; Kang, M.Y.; Koh, H.J.; Kozukue, N.; Friedman, M. Antioxidative activities of bran extracts from twenty one pigmented rice cultivars. Food Chem. 2006, 94, 613–620. [Google Scholar] [CrossRef]
- Tsuda, T.; Horio, F.; Osawa, T. Cyanidin 3-O-beta-d-glucoside suppresses nitric oxide production during a zymosan treatment in rats. J. Nutr. Sci. Vitaminol. 2002, 48, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Mi, M.T.; Ling, W.H. Effects of anthocyanin-rich extract from black rice alone or combined with chemotherapeutic agents on proliferation of different cacner cells. Acta Acad. Med. Mlitaris. 2007, 29, 1943–1946. [Google Scholar]
- Guo, H.H.; Ling, W.H.; Wang, Q.; Liu, C.; Hu, Y.; Xia, M.; Feng, X.; Xia, X.D. Effect of Anthocyanin-Rich Extract from Black Rice (Oryza sativa L. indica) on Hyperlipidemia and Insulin Resistance in Fructose-Fed Rats. Plant Food. Hum. Nutr. 2007, 62, 1–6. [Google Scholar] [CrossRef]
- Gunaratne, A.; Wu, K.; Li, D.; Bentota, A.; Corke, H.; Cai, Y.Z. Antioxidant activity and nutritional quality of traditional red-grained rice varieties containing proanthocyanidins. Food Chem. 2013, 138, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.V.; Shepherd, L.V.T.; Stewart, D.; Frank, T.; Röhling, R.M.; Engel, K.H. Metabolome variability in crop plant species—when, where, how much and so what? Regul. Toxicol. Pharm. 2010, 58, S54–S61. [Google Scholar]
- Frank, T.; Meuleye, B.S.; Miller, A.; Shu, Q.Y.; Engel, K.H. Metabolite profiling of two low phytic acid (lpa) rice mutants. J. Agric. Food Chem. 2007, 55, 11011–11019. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Do, S.G.; Kim, S.Y.; Kim, J.; Jin, Y.; Lee, C.H. Mass spectrometry-based metabolite profiling and antioxidant activity of aloe vera (Aloe barbadensis Miller) in different growth stages. J. Agric. Food Chem. 2012, 60, 11222–11228. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.S.; Lee, S.; Lim, S.H.; Ha, S.H.; Liu, K.H.; Lee, C.H. Metabolite profiling of the short-term responses of rice leaves (Oryza sativa cv. Ilmi) cultivated under different LED lights and its correlations with antioxidant activities. Plant Sci. 2013, 210, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Shyur, L.F.; Yang, N.S. Metabolomics for phytomedicine research and drug development. Curr. Opin. Chem. Biol. 2008, 12, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Schauer, N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 2009, 25, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Luo, G.A.; Liang, Q.L.; Hu, P.; Wang, Y.M. Rapid qualitative and quantitative analyses of Asian ginseng in adulterated American ginseng preparations by UPLC/Q-TOF-MS. J. Pharmaceut. Biomed. 2010, 52, 66–72. [Google Scholar] [CrossRef]
- Frank, T.; Reichardt, B.; Shu, Q.; Engel, K.H. Metabolite profiling of colored rice (Oryza sativa L.) grains. J. Cereal Sci. 2012, 55, 112–119. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, S.Y.; Chu, S.M.; Lim, S.H.; Suh, S.C.; Lee, Y.T.; Cho, H.S.; Ha, S.H. Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J. Agric. Food Chem. 2010, 58, 12804–12809. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Park, S.Y.; Lim, S.H.; Yeo, Y.; Cho, H.S.; Ha, S.H. Comparative metabolic profiling of pigmented rice (Oryza sativa L.) cultivars reveals primary. J. Cereal Sci. 2013, 57, 14–20. [Google Scholar] [CrossRef]
- Shu, X.L.; Frank, T.; Shu, Q.Y.; Engel, K.H. Metabolite profiling of germinating rice seeds. J. Agric. Food Chem. 2008, 56, 11612–11620. [Google Scholar] [CrossRef] [PubMed]
- Sriseadka, T.; Wongpornchai, S.; Rayanakorn, M. Quantification of flavonoid in black rice by liquid chromatography-negative electrospray ionization tandem mass spectrometry. J. Agric. Food Chem. 2012, 60, 11723–11732. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Ha, S.H.; Lim, S.H.; Jung, J.Y.; Lee, S.M.; Yeo, Y.; Kim, J.K. Determination of phenolic acids in Korean rice (Oryza sativa L.) cultivars using gas chromatography-time-of-flight mass spectrometry. Food Sci. Biotechnol. 2012, 21, 1141–1148. [Google Scholar] [CrossRef]
- Finocciaro, F.; Ferrari, B.; Gianinetti, A. A study of biodiversity of flavonoid content in the rice caryopsis evidencing simultaneous accumulation of anthocyanins and proanthocyanidins in a black-grained genotype. J. Cereal Sci. 2010, 10, 28–34. [Google Scholar] [CrossRef]
- Lim, S.H.; Ha, S.H. Marker development for the identification of rice seeds color. Plant Biotechnol. Rep. 2013, 7, 391–398. [Google Scholar] [CrossRef]
- Ferreres, F.; Andrade, P.B.; Valentão, P.; Gil-Izquierdo, A. Further knowledge on barley (Hordeum vulgare L.) leaves O-glycosyl-C-glycosyl flavones by liquid chromatography-UV diode-array detection-electrospray ionisation mass spectrometry. J. Chromatogr. A. 2008, 1182, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Markham, K.R.; Tanner, G.J.; Caasi-Lit, M.; Whitecross, M.I.; Nayudu, M.; Mitchell, K.A. Possible protective role for 3',4'-dihydrowyflavones induced by enhanced UV-B in a UV-tolerant rice cultivar. Phytochemistry 1998, 49, 1913–1919. [Google Scholar] [CrossRef]
- Moheb, A.; Ibrahim, R.K.; Roy, R.; Sarhan, F. Changes in wheat leaf phenolome in response to cold acclimation. Phytochemistry 2011, 72, 2294–2307. [Google Scholar] [CrossRef] [PubMed]
- Suemune, H.; Harabe, T.; Sakai, K. Syntheses of unsaturated trihydroxy C-18 fatty acids isolated from rice plants suffering from rice blast disease. Chem. Pharm. Bull. 1988, 36, 3632–3637. [Google Scholar] [CrossRef]
- Kasai, M.; Ohishi, K.; Shimada, A.; Hatae, K. Taste properties of cooked rice based on an analysis of the cooked rice extracts. J. Cookery Sci. Jap. 2001, 34, 373–379. [Google Scholar]
- Saikusa, T.; Horino, T.; Mori, Y. Distribution of free amino acids in the rice kernel and kernel fractions and the effect of water soaking on the distribution. J. Agric. Food Chem. 1994, 42, 1122–1125. [Google Scholar] [CrossRef]
- Okada, T.; Sugishita, T.; Murakami, T.; Murai, H.; Saikusa, T.; Horino, T.; Onoda, A.; Kajimoto, O.; Takahashi, R.; Takahashi, T. Effect of the defatted rice germ enriched with GABA for sleeplessness, depression, autonomic disorder by oral administration. J. Jpn. Soc. Food Sci. 2000, 47, 596–603. [Google Scholar] [CrossRef]
- Ganji, S.H.; Zhang, L.H.; Kamanna, V.S.; Kashyap, M.L. Effect of niacin on lipoproteins and atherosclerosis. Future Med. 2006, 1, 549–557. [Google Scholar]
- Swain, T.; Williams, C.A. The role of phenylalanine in flavonoid biosynthesis. Phytochemistry 1970, 9, 2115–2122. [Google Scholar] [CrossRef]
- Sompong, R.; Siebenhandl-Ehn, S.; Linsberger-Martin, G.; Berghofer, E. Physicochemical and antioxidative properties of red and black rice varieties from Thailand, China and Sri Lanka. Food Chem. 2011, 124, 132–140. [Google Scholar] [CrossRef]
- Morelli, R.; Russo-Volpe, S.; Bruno, N.; Scalzo, R.L. Fenton-dependent damage to carbohydrates: Free radical scavengin activity of some simple sugars. J. Agric. Food Chem. 2003, 51, 7418–7425. [Google Scholar] [CrossRef] [PubMed]
- Ogata, S.; Takeuchi, M.; Teradaira, S.; Yamamoto, N.; Iwata, K.; Okumura, K.; Taguchi, H. Radical scavenging activities of niacin-related compounds. Biosci. Biotechnol. Biochem. 2002, 66, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidanat activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Dietz, B.M.; Kang, Y.H.; Liu, G.; Eggler, A.L.; Yao, P.; Chadwick, L.R.; Pauli, G.F.; Farnsworth, N.R.; Mesecar, A.D.; van Breemen, R.B.; et al. Xanthohumol isolated from Humulus lupulus inhibits menadione-induced DNA damage through induction of quinone reductase. Chem. Res. Toxicol. 2005, 18, 1296–1305. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability if plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Wageningen UR for Quality of Life. Available online: http://www.metalign.nl (accessed on 25 September 2014).
- TM4: Microarray Software Suite. Available online: http://www.tm4.org/ (accessed on 25 September 2014).
- Sample Availability: No available.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, G.R.; Jung, E.S.; Lee, S.; Lim, S.-H.; Ha, S.-H.; Lee, C.H. Combined Mass Spectrometry-Based Metabolite Profiling of Different Pigmented Rice (Oryza sativa L.) Seeds and Correlation with Antioxidant Activities. Molecules 2014, 19, 15673-15686. https://doi.org/10.3390/molecules191015673
Kim GR, Jung ES, Lee S, Lim S-H, Ha S-H, Lee CH. Combined Mass Spectrometry-Based Metabolite Profiling of Different Pigmented Rice (Oryza sativa L.) Seeds and Correlation with Antioxidant Activities. Molecules. 2014; 19(10):15673-15686. https://doi.org/10.3390/molecules191015673
Chicago/Turabian StyleKim, Ga Ryun, Eun Sung Jung, Sarah Lee, Sun-Hyung Lim, Sun-Hwa Ha, and Choong Hwan Lee. 2014. "Combined Mass Spectrometry-Based Metabolite Profiling of Different Pigmented Rice (Oryza sativa L.) Seeds and Correlation with Antioxidant Activities" Molecules 19, no. 10: 15673-15686. https://doi.org/10.3390/molecules191015673
APA StyleKim, G. R., Jung, E. S., Lee, S., Lim, S.-H., Ha, S.-H., & Lee, C. H. (2014). Combined Mass Spectrometry-Based Metabolite Profiling of Different Pigmented Rice (Oryza sativa L.) Seeds and Correlation with Antioxidant Activities. Molecules, 19(10), 15673-15686. https://doi.org/10.3390/molecules191015673