Isolation and Characterization of an Antibacterial Peptide Fraction from the Pepsin Hydrolysate of Half-Fin Anchovy (Setipinna taty)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Identification of Antibacterial Fraction from HAHp
2.1.1. Gel Permeation Chromatography
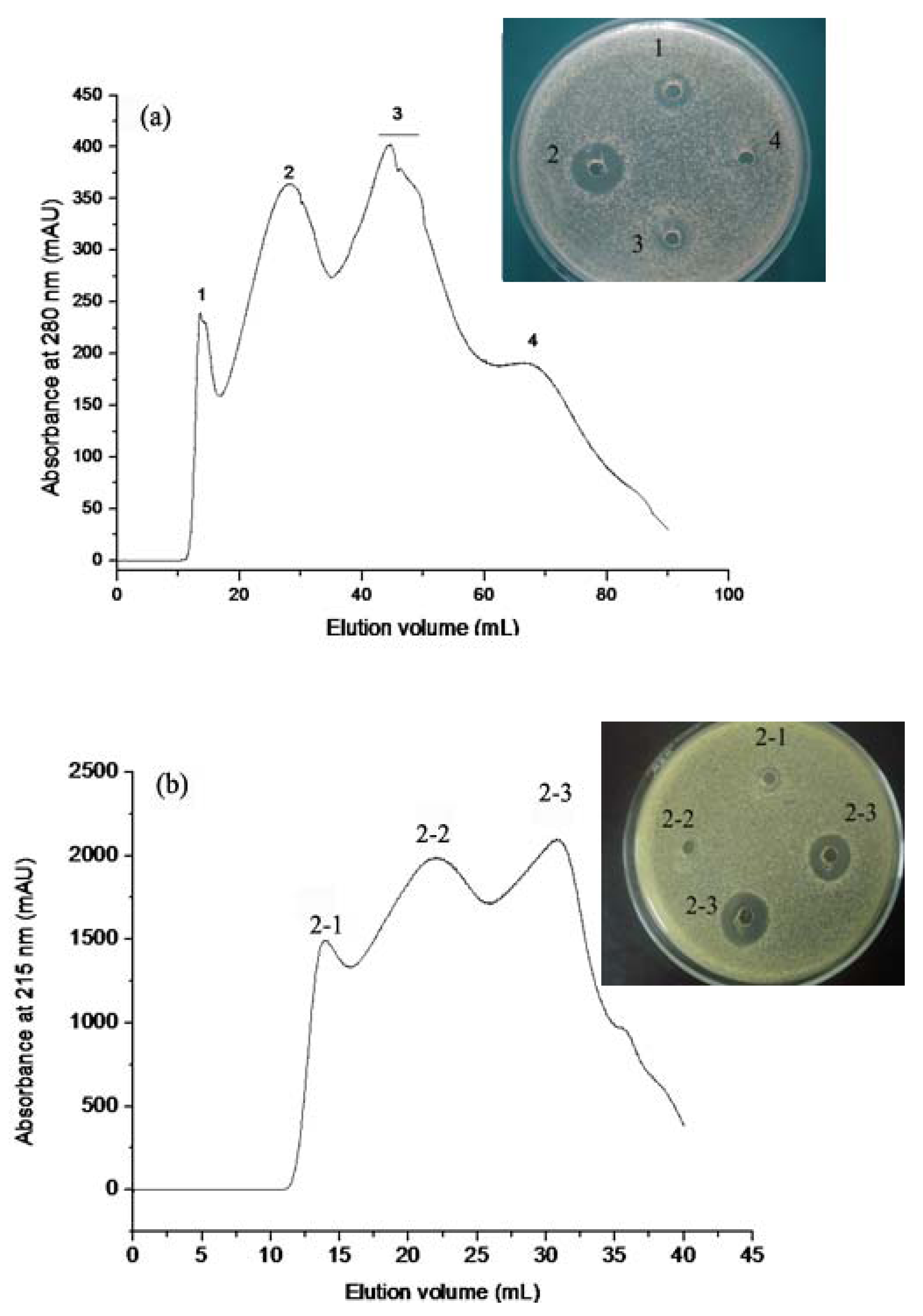
2.1.2. RP-HPLC and Source 5RPC ST purification
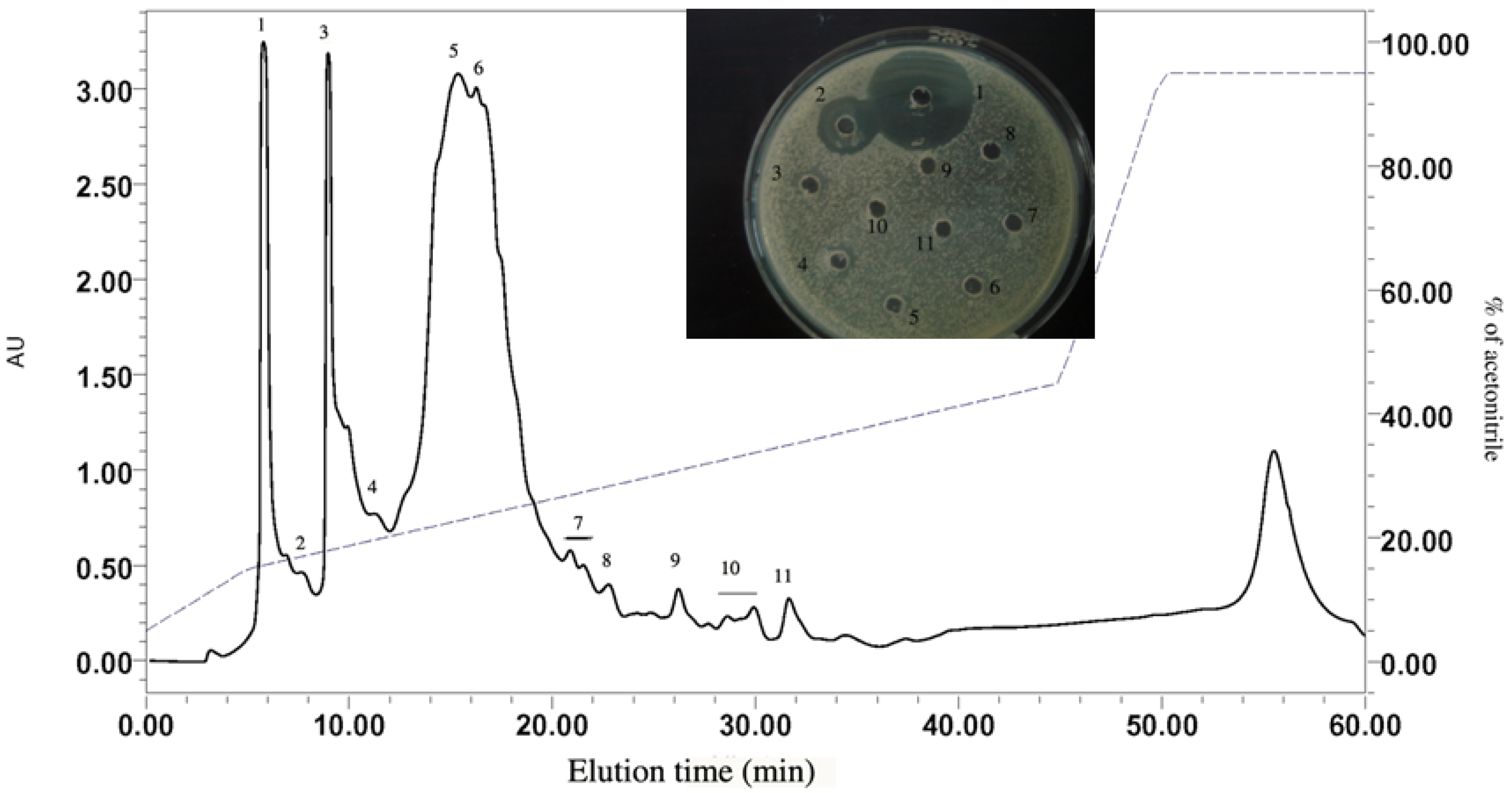
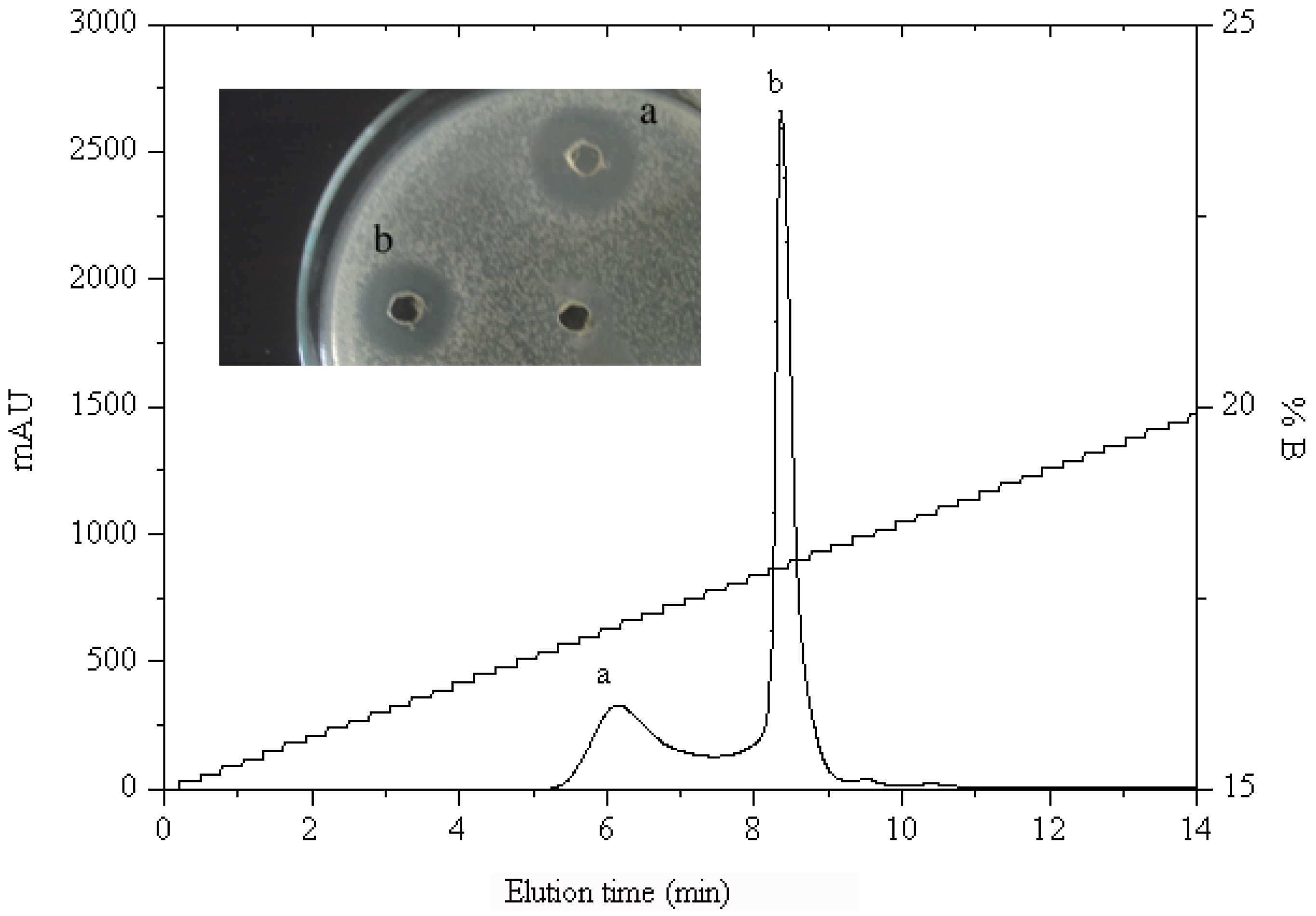
2.2. Peptide Sequence Analysis and Physicochemical Properties
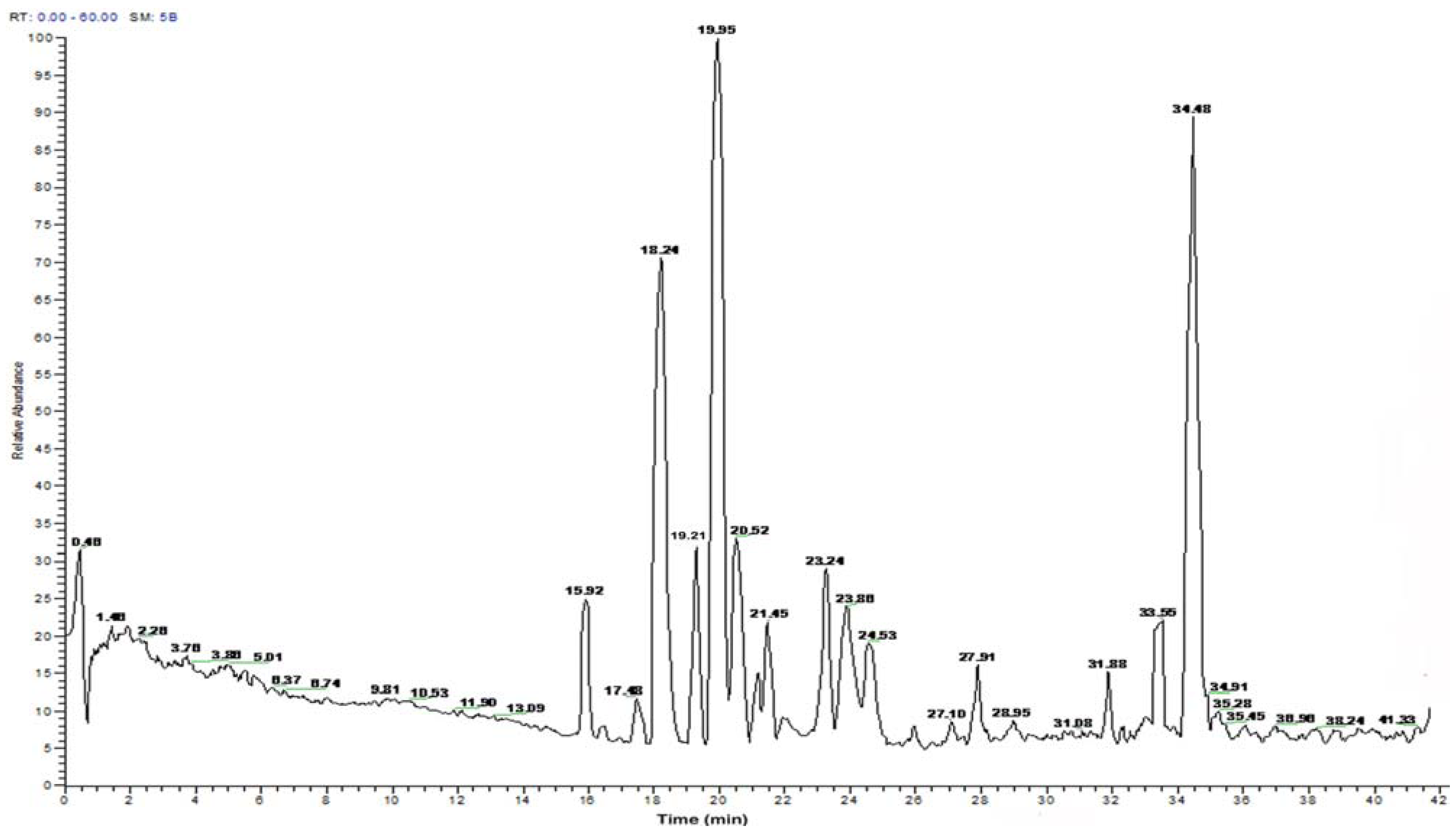
| Time | Amino acid sequence | Calculated mass | Net | Hydrophobic ratio b (%) |
|---|---|---|---|---|
| (min) | /observed mass | charge a | ||
| 15.92 | MLTTPPHAKYVLQW | 1685.0/1665.92 | +2 | 42 |
| 18.24 | LRSKAAAPAEQYE | 1433.5/1414.78 | 0 | 38 |
| 19.21 | TPGALLEHPTL | 1148.3/1129.60 | 0 | 36 |
| 19.95 | SHAATKAPPKNGNY | 1455.5/1436.91 | +3 | 21 |
| 20.52 | LATVSVGAVELCY | 1324.5/1305.73 | −1 | 61 |
| 21.45 | PTAGVANALQHA | 1149.2/1130.67 | +1 | 50 |
| 23.24 | QLGTHSAQPVPF | 1281.4/1262.62 | +1 | 33 |
| 23.86 | VNVDERWRKL | 1314.5/1295.65 | +1 | 40 |
| 33.55 | NPEFLASGDHLDNLQ | 1669.7/1650.89 | −2 | 33 |
| 34.48 | PEVVYECLHW | 1274.4/1255.72 | −1 | 50 |
2.3. Prediction of Peptide Secondary Structure
| Peptides | Amino acid sequence a | Secondary structure b |
|---|---|---|
| HAHp-cationic1 | MLTTPPHAKYVLQW | |
| ccccccccceeecc | Extended strand (21.43%), random coil (78.57%) | |
| HAHp-cationic2 | SHAATKAPPKNGNY | |
| cccccccccccccc | Random coil (100%) | |
| HAHp-cationic3 | PTAGVANALQHA | |
| cccchhhhhccc | Alpha helix (41.67%), random coil (58.33%) | |
| HAHp-cationic4 | QLGTHSAQPVPF | |
| cccccccccccc | Random coil (100%) | |
| HAHp-cationic5 | VNVDERWRKL | |
| cccchhhccc | Alpha helix (30.00%), random coil (70.00%) | |
| HAHp-anionic1 | LATVSVGAVELCY | |
| ceeeeeeeeeecc | Extended strand (76.92%), random coil (23.08%) | |
| HAHp-anionic2 | NPEFLASGDHLDNLQ | |
| ccceccccccccccc | Extended strand (6.67%), random coil (93.33%) | |
| HAHp-anionic3 | PEVVYECLHW | |
| cceeeeeecc | Extended strand (60.00%), random coil (40.00%) |
2.4. Scanning Electron Microscopy (SEM)
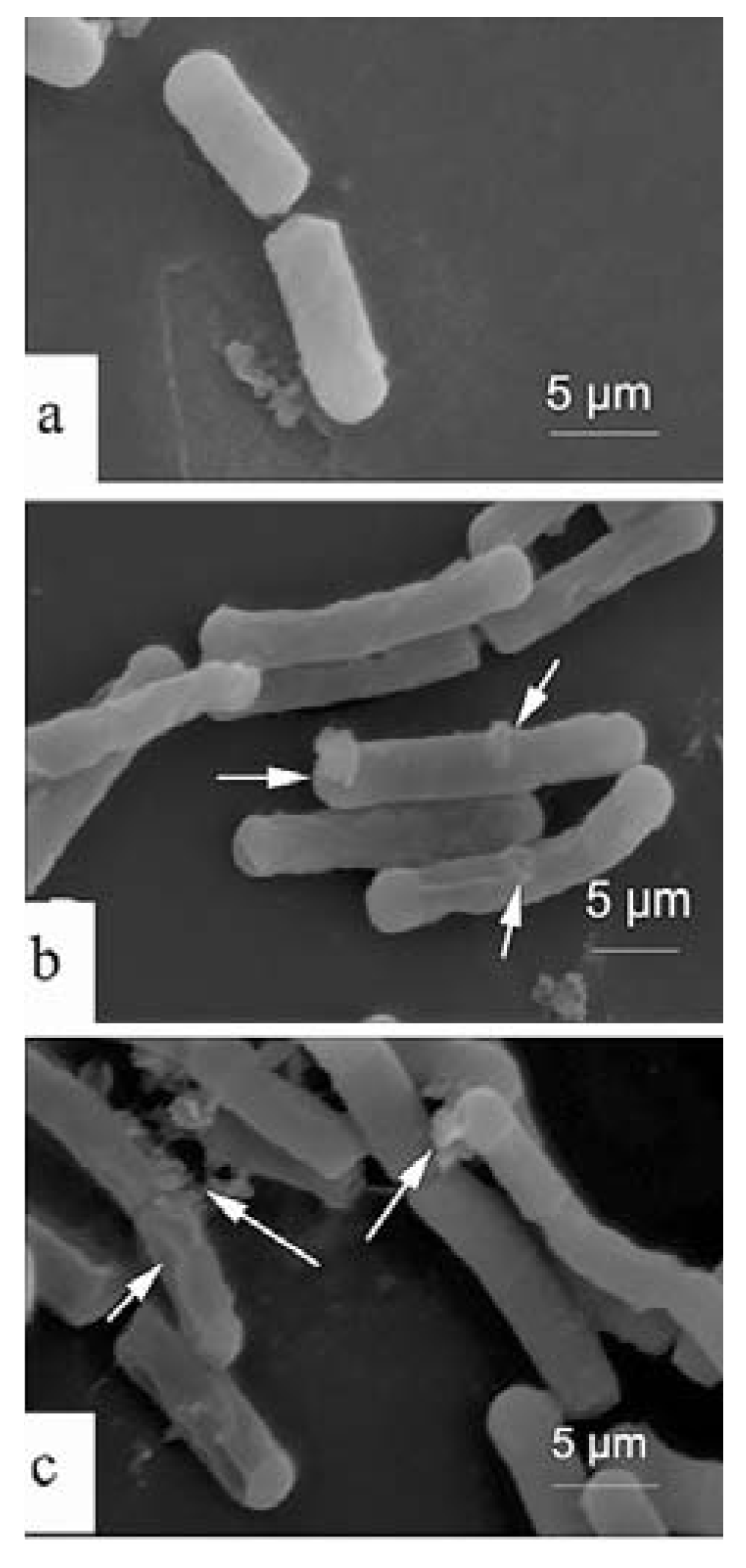
3. Experimental
3.1. Materials
3.2. Preparation of the Pepsin Hydrolysate of Half-Fin Anchovy (HAHp)
3.3. Antibacterial Activity against Escherichia coli
3.4. Isolation of Antibacterial Fraction from HAHp
3.5. Peptide Sequence Analysis and Physicochemical Properties
3.6. Prediction of the Secondary Structure of Peptide
3.7. Scanning Electron Microscopy (SEM)
4. Conclusions
Acknowledgments
References and Notes
- Steiner, H.; Hultmark, D.; Engström, Å.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef]
- Broekaert, W.F.; Terras, F.R.; Cammue, B.P.; Osborn, R.W. Plant defensins: Novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995, 108, 1353–1358. [Google Scholar]
- Mkrtchyan, H.; Gibbons, S.; Heidelberger, S.; Zloh, M.; Limaki, H.K. Purification, characterisation and identification of acidocin LCHV, an antimicrobial peptide produced by Lactobacillus acidophilus n.v. Er 317/402 strain Narine. Int. J. Antimicrob. Agents 2010, 35, 255–260. [Google Scholar]
- Rajanbabu, V.; Chen, J.Y. Applications of antimicrobial peptides from fish and perspectives for the future. Peptides 2011, 32, 415–420. [Google Scholar] [CrossRef]
- Hill, R.D.; Lahav, E.; Givol, D. A rennin-sensitive bond in alpha-s1 B-casein. J. Dairy Res. 1974, 41, 147–153. [Google Scholar] [CrossRef]
- Jones, F.S.; Simms, H.S. The bacterial growth inhibitor (lactenin) of milk. J. Exp. Med. 1930, 51, 327–339. [Google Scholar] [CrossRef]
- Zucht, H.D.; Raida, M.; Adermann, K.; Mägert, H.J.; Forssmann, W.G. Casocidin-I: A casein-αs2 derived peptide exhibits antibacterial activity. FEBS Lett. 1995, 372, 185–188. [Google Scholar] [CrossRef]
- Liu, Z.Y.; Dong, S.Y.; Xu, J.; Zeng, M.Y.; Song, H.X.; Zhao, Y.H. Production of cysteine-rich antimicrobial peptide by digestion of oyster (Crassostrea gigas) with alcalase and bromelin. Food Control 2008, 19, 231–235. [Google Scholar] [CrossRef]
- Mine, Y.; Ma, F.P.; Lauriau, S. Antimicrobial peptides released by enzymatic hydrolysis of hen egg white lysozyme. J. Agric. Food Chem. 2004, 52, 1088–1094. [Google Scholar] [CrossRef]
- Pellegrini, A.; Hülsmeier, A.J.; Hunziker, P.; Thomas, U. Proteolytic fragments of ovalbumin display antimicrobial activity. Biochim. Biophys. Acta 2004, 1672, 76–85. [Google Scholar] [CrossRef]
- Bolscher, J.G.M.; van der Kraan, M.I.A.; Nazmi, K.; Kalay, H.; Grün, C.H.; van’t Hof, W.; Veerman, E.C.I.; Nieuw Amerongen, A.V. A one-enzyme strategy to release an antimicrobial peptide from the LFampin-domain of bovine lactoferrin. Peptides 2006, 27, 1–9. [Google Scholar] [CrossRef]
- Subramanian, S.; Ross, N.W.; Mackinnon, S.L. Comparison of the biochemical composition of normale pidermal mucus and extruded slime of hagfish (Myxine glutinosa L.). Fish Shellfish Immunol. 2008, 25, 625–632. [Google Scholar]
- Noga, E.J.; Ullal, A.J.; Corrales, J.; Fernandes, J.M. Application of antimicrobial polypeptide host defenses to aquaculture: Exploitation of downregulation and upregulation responses. Comp. Biochem. Physiol. D 2011, 6, 44–45. [Google Scholar]
- Chen, S.L.; Xu, M.Y.; Ji, X.S.; Yu, G.C.; Liu, Y. Cloning, characterization, and expression analysis of hepcidin gene from red sea bream (Chrysophrys major). Antimicrob. Agents Chemother. 2005, 49, 1608–1612. [Google Scholar] [CrossRef]
- Dong, X.Z.; Xu, H.B.; Huang, K.X.; Liou, Q.; Zhou, J. The preparation and characterization of an antimicrobial polypeptide from the loach, Misgurnus anguillicaudatus. Protein Expr. Purif. 2002, 26, 235–242. [Google Scholar] [CrossRef]
- Hirono, I.; Hwang, J.Y.; Ono, Y.; Kurobe, T.; Ohira, T.; Nozaki, R.; Aoki, T. Two different types of hepcidins from the Japanese flounder Paralichthys olivaceus. FEBS J. 2005, 272, 5257–5264. [Google Scholar] [CrossRef]
- Song, R.; Wei, R.B.; Zhang, B.; Wang, D.F. Optimization of the antibacterial activity of half-fin anchovy (Setipinna taty) hydrolysates. Food Bioprocess Technol. 2011. [Google Scholar] [CrossRef]
- Friedrich, C.L.; Moyles, D.; Beveridge, T.J.; Hancock, R.E.W. Antibacterial action of structurally diverse cationic peptides on Gram-positive bacteria. Antimicrob. Agents Chemother. 2000, 44, 2086–2092. [Google Scholar] [CrossRef]
- Mandal, S.M.; Dey, S.; Mandal, M.; Sarkar, S.; Maria-Neto, S.; Franco, O.L. Identification and structural insights of three novel antimicrobial peptidesisolated from green coconut water. Peptides 2009, 30, 633–637. [Google Scholar] [CrossRef]
- Pouny, Y.; Rapaport, D.; Mor, A.; Nicolas, P.; Shai, Y. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry 1992, 31, 12416–12423. [Google Scholar] [CrossRef]
- Kustanovich, I.; Shalev, D.E.; Mikhlin, M.; Gaidukov, L.; Mor, A. Structural requirements for potent versus selective cytotoxicity for antimicrobial dermaseptin S4 derivatives. J. Biol. Chem. 2002, 277, 16941–16945. [Google Scholar]
- Zelezetsky, I.; Pag, U.; Sahl, H.G.; Tossi, A. Tuning the biological properties of amphipathic alpha-helical antimicrobial peptides: rational use of minimal amino acid substitutions. Peptides 2005, 26, 2368–2376. [Google Scholar] [CrossRef]
- Bello, J.; Bello, H.R.; Granados, E. Conformation and aggregation of melittin: Dependence onpH and concentration. Biochemistry 1982, 21, 461–465. [Google Scholar] [CrossRef]
- Falla, T.J.; Karunaratne, D.N.; Hancock, R.E.W. Mode of action of the antimicrobial peptide indolicidin. J. Biol. Chem. 1996, 271, 19298–19303. [Google Scholar]
- Gennaro, R.; Zanetti, M. Structural features and biological activities of thecathelicidin-derivedantimicrobial peptides. Biopolymers 2000, 55, 31–49. [Google Scholar] [CrossRef]
- Park, C.B.; Yi, K.S.; Matsuzaki, K.; Kim, M.S.; Kim, S.C. Structure-activity analysis of buforin II, a histone H2A derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA 2000, 97, 8245–8250. [Google Scholar]
- Zhang, L.; Rozek, A.; Hancock, R.E.W. Interaction of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 2001, 276, 35714–35722. [Google Scholar] [CrossRef]
- Zasloff, M.; Martin, B.; Chen, H.C. Antimicrobial activity of synthetic magainin peptides and several analogues. Proc. Natl. Acad. Sci. USA 1988, 85, 910–913. [Google Scholar] [CrossRef]
- Ladokhin, A.S.; White, S.H. Detergent-like’ permeabilization of anionic lipid vesicles by melittin. Biochim. Biophys. Acta 2001, 1514, 253–260. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alphahelical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Song, R.; Wei, R.B.; Zhang, B.; Yang, Z.S.; Wang, D.F. Antioxidant and antiproliferative activities of heated sterilized pepsin hydrolysatederived from half-fin anchovy (Setipinna taty). Mar. Drugs 2011, 9, 1142–1156. [Google Scholar] [CrossRef]
- Murthy, P.S.; Manonmani, H.K. Physico-chemical, antioxidant and antimicrobial properties of Indian monsooned coffee. Eur. Food Res. Technol. 2009, 229, 645–650. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic. Acids Res. 2003, 31, 3784–3788. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Sample Availability: Contact the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Song, R.; Wei, R.-B.; Luo, H.-Y.; Wang, D.-F. Isolation and Characterization of an Antibacterial Peptide Fraction from the Pepsin Hydrolysate of Half-Fin Anchovy (Setipinna taty). Molecules 2012, 17, 2980-2991. https://doi.org/10.3390/molecules17032980
Song R, Wei R-B, Luo H-Y, Wang D-F. Isolation and Characterization of an Antibacterial Peptide Fraction from the Pepsin Hydrolysate of Half-Fin Anchovy (Setipinna taty). Molecules. 2012; 17(3):2980-2991. https://doi.org/10.3390/molecules17032980
Chicago/Turabian StyleSong, Ru, Rong-Bian Wei, Hong-Yu Luo, and Dong-Feng Wang. 2012. "Isolation and Characterization of an Antibacterial Peptide Fraction from the Pepsin Hydrolysate of Half-Fin Anchovy (Setipinna taty)" Molecules 17, no. 3: 2980-2991. https://doi.org/10.3390/molecules17032980
APA StyleSong, R., Wei, R.-B., Luo, H.-Y., & Wang, D.-F. (2012). Isolation and Characterization of an Antibacterial Peptide Fraction from the Pepsin Hydrolysate of Half-Fin Anchovy (Setipinna taty). Molecules, 17(3), 2980-2991. https://doi.org/10.3390/molecules17032980




