Based on Multi-Activity Integrated Strategy to Screening, Characterization and Quantification of Bioactive Compounds from Red Wine
Abstract
:1. Introduction
2. Result and Discussion
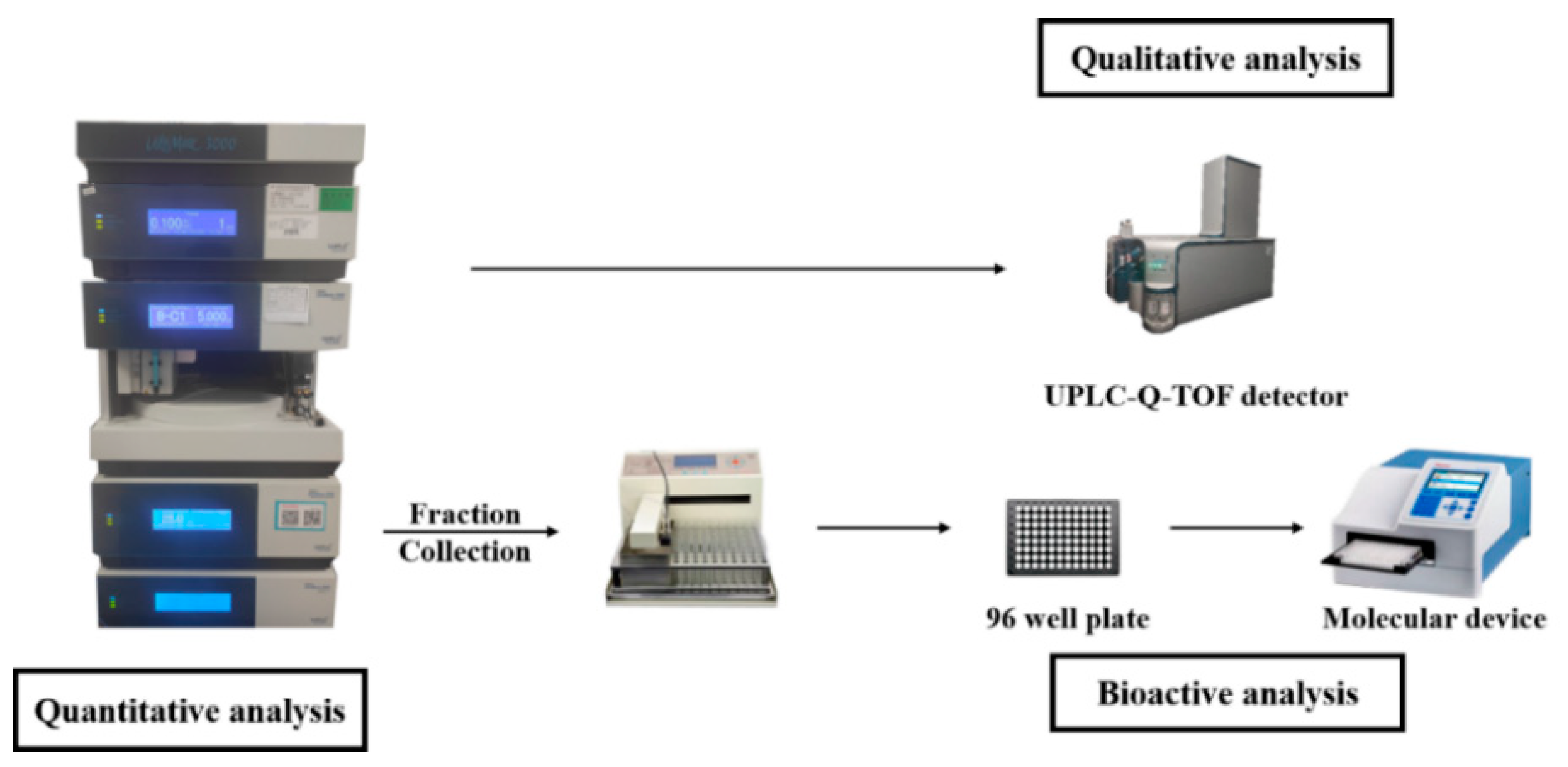
2.1. Establishment of Multi-Activity Integrated Strategy
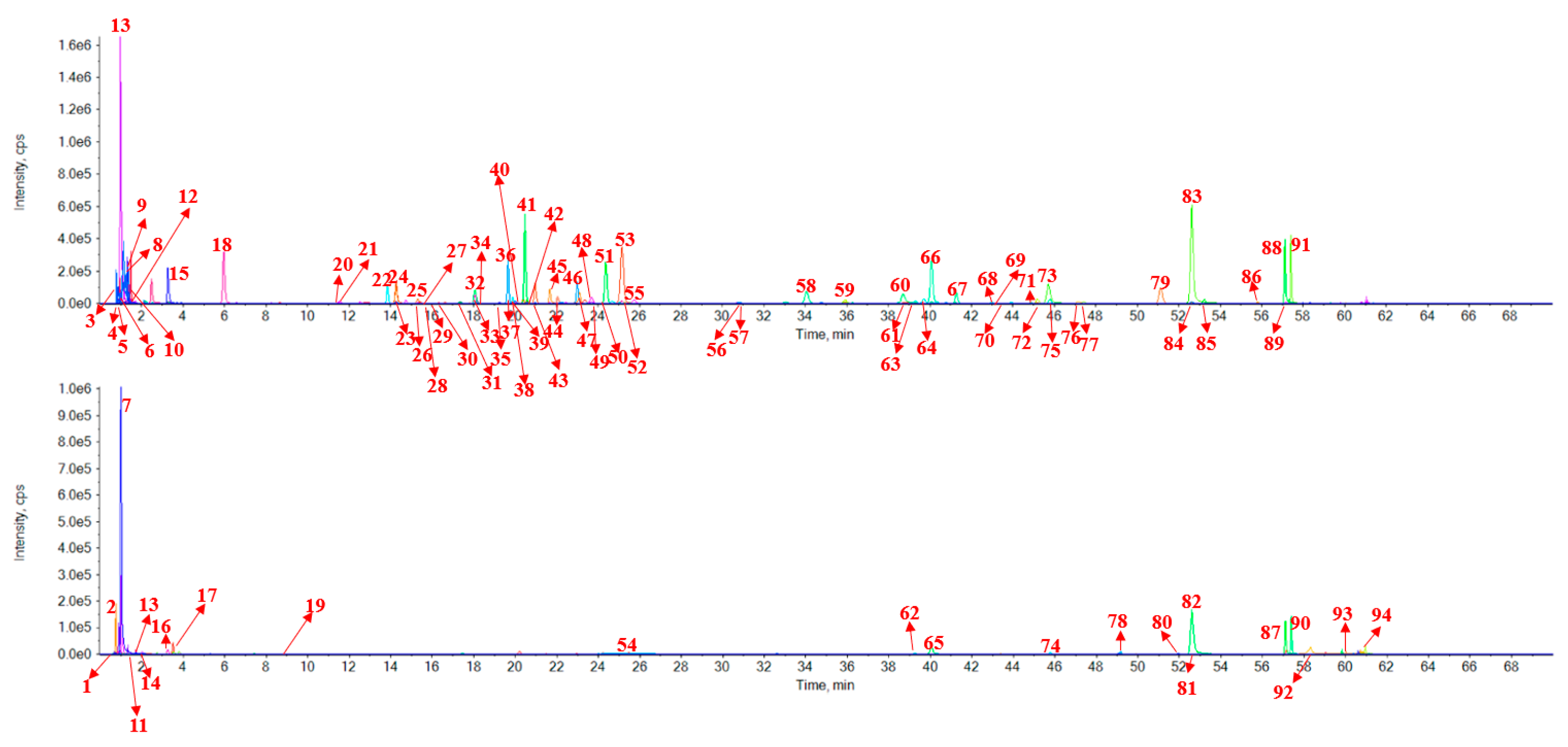
2.2. Identification of 94 Components from Red Wine
2.3. Active Ingredients of Red Wine
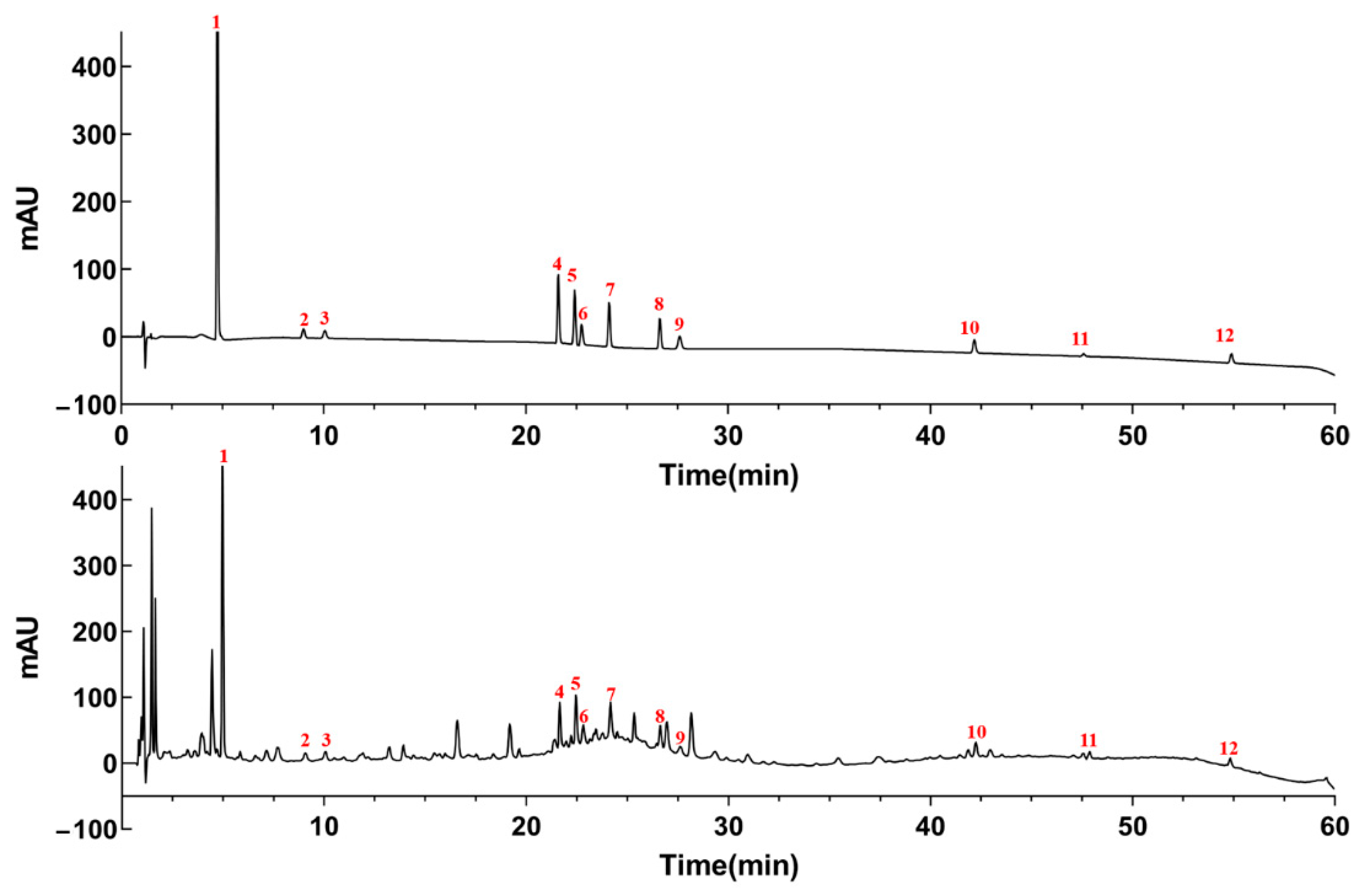
2.4. UHPLC Analysis
2.5. Method Validation
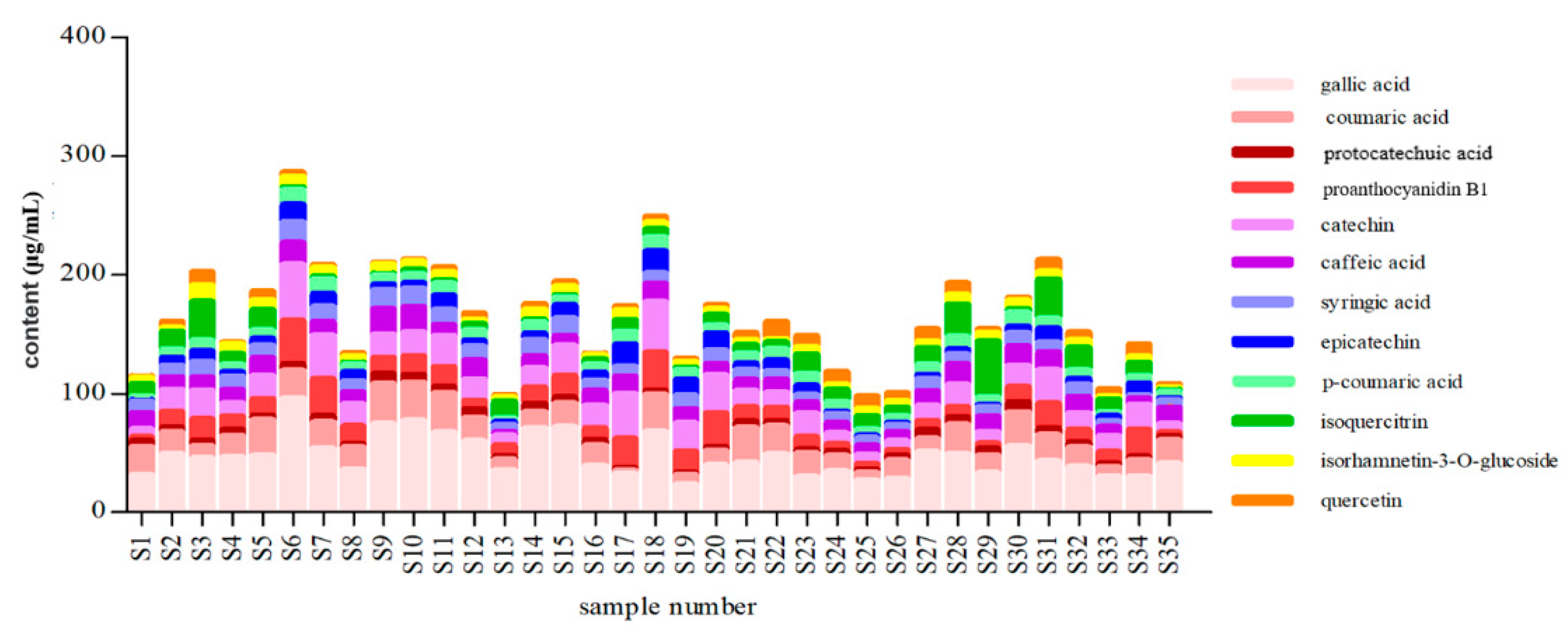
2.6. Contents of the Bioactive Components from Red Wine
2.7. Confirmation the Activity of the 12 Compounds
2.8. The Application of Multi-Activity Integrated Strategy
3. Materials and Methods
3.1. Reagents and Chemical
3.2. Sample and Standard Solution Preparation
3.3. Preparation of the Red Wine Fractions
3.4. UHPLC Analysis
3.5. UFLC/Q-TOF-MS Analysis
3.6. Antioxidant Activity Assay
3.7. Thrombin Inhibitory Activity Assay
3.8. Lipase Inhibitory Activity Assay
3.9. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rodrigo, R.; Miranda, A.; Vergara, L. Modulation of endogenous antioxidant system by wine polyphenols in human disease. Clin. Chim. Acta 2011, 412, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, L.; Narváez, A.; Izzo, L.; Graziani, G.; Gaspari, A.; Di Minno, G.; Ritieni, A. Red wine consumption and cardiovascular health. Molecules 2019, 24, 3626. [Google Scholar] [CrossRef] [Green Version]
- Renaud, S.D.; de Lorgeril, M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet 1992, 339, 1523–1526. [Google Scholar] [CrossRef]
- Snopek, L.; Mlcek, J.; Sochorova, L.; Baron, M.; Hlavacova, I.; Jurikova, T.; Sochor, J. Contribution of red wine consumption to human health protection. Molecules 2018, 23, 1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Statistics 2019: Monitoring Health for the SDGs, Sustainable Development Goals. Available online: https://apps.who.int/iris/handle/10665/324835 (accessed on 21 May 2019).
- Liu, P.; Zhou, B.; Gu, D.; Zhang, L.; Han, Z. Endothelial progenitor cell therapy in atherosclerosis: A double-edged sword. Ageing Res. Rev. 2009, 8, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Ceriello, A.; Assaloni, R.; Da Ros, R.; Maier, A.; Piconi, L.; Quagliaro, L.; Esposito, K.; Giugliano, D. Effect of atorvastatin and irbesartan, alone and in combination, on postprandial endothelial dysfunction, oxidative stress, and inflammation in type 2 diabetic patients. Circulation 2005, 111, 2518–2524. [Google Scholar] [CrossRef]
- Pothineni, N.; Subramany, S.; Kuriakose, K.; Shirazi, L.F.; Romeo, F.; Shah, P.K.; Mehta, J.L. Infections, atherosclerosis, and coronary heart disease. Eur. Heart J. 2017, 38, 3195–3201. [Google Scholar] [CrossRef]
- Yan, Y.L.; Qiu, B.; Hu, L.J.; Jing, X.D.; Liu, Y.J.; Deng, S.B.; Du, J.L.; She, Q. Efficacy and safety evaluation of intensive statin therapy in older patients with coronary heart disease: A systematic review and meta-analysis. Eur. J. Clin. Pharmacol. 2013, 69, 2001–2009. [Google Scholar] [CrossRef]
- Divakaran, S.; Loscalzo, J. The role of nitroglycerin and other nitrogen oxides in cardiovascular therapeutics. J. Am. Coll. Cardiol. 2017, 70, 2393–2410. [Google Scholar] [CrossRef]
- Demartini, C.; Greco, R.; Zanaboni, A.M.; Sances, G.; De Icco, R.; Borsook, D.; Tassorelli, C. Nitroglycerin as a comparative experimental model of migraine pain: From animal to human and back. Prog. Neurobiol. 2019, 177, 15–32. [Google Scholar] [CrossRef]
- Chang, Y.X.; Ge, A.H.; Donnapee, S.; Li, J.; Bai, Y.; Liu, J.; Gao, X.M. The multi-targets integrated fingerprinting for screening anti-diabetic compounds from a Chinese medicine Jinqi Jiangtang Tablet. J. Ethnopharmacol. 2015, 164, 210–222. [Google Scholar] [CrossRef]
- Huang, X.; Kong, L.; Li, X.; Chen, X.; Guo, M.; Zou, H. Strategy for analysis and screening of bioactive compounds in traditional Chinese medicines. J. Chromatogr. B 2004, 812, 71–84. [Google Scholar] [CrossRef]
- Kang, H.J.; Yang, H.J.; Kim, M.J.; Han, E.S.; Kim, H.J.; Kwon, D.Y. Metabolomic analysis of meju during fermentation by ultra performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS). Food Chem. 2011, 127, 1056–1064. [Google Scholar] [CrossRef]
- He, Q.; Huang, S.; Wu, Y.; Zhang, W.; Wang, F.; Cao, J.; Ou, W.B. Comparative study on the composition of free amino acids and derivatives in the two botanical origins of an edible Chinese herb “Xiebai”, i.e., Allium chinense G. Don and Allium macrostemon Bunge species. Food Res. Int. 2018, 106, 446–457. [Google Scholar] [CrossRef]
- Konya, Y.; Taniguchi, M.; Fukusaki, E. Novel high-throughput and widely-targeted liquid chromatography–time of flight mass spectrometry method for d-amino acids in foods. J. Biosci. Bioeng. 2017, 123, 126–133. [Google Scholar] [CrossRef]
- Cellar, N.A.; McClure, S.C.; Salvati, L.M.; Reddy, T.M. A new sample preparation and separation combination for precise, accurate, rapid, and simultaneous determination of vitamins B1, B2, B3, B5, B6, B7, and B9 in infant formula and related nutritionals by LC-MS/MS. Anal. Chim. Acta. 2016, 934, 180–185. [Google Scholar] [CrossRef]
- Hu, H.; Xiao, L.; Zheng, B.; Wei, X.; Ellis, A.; Liu, Y.M. Identification of chemical markers in Cordyceps sinensis by HPLC-MS/MS. Anal. Bioanal Chem. 2015, 407, 8059–8066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Wang, Y.; Ouyang, H.; Lu, Y.; Qiu, Y.; Feng, Y.; Yang, S. A novel dereplication strategy for the identification of two new trace compounds in the extract of Gastrodia elata using UHPLC/Q-TOF-MS/MS. J. Chromatogr. B 2015, 988, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Ramabulana, A.T.; Steenkamp, P.; Madala, N.; Dubery, I.A. Profiling of chlorogenic acids from bidens pilosa and differentiation of closely related positional isomers with the aid of UHPLC-QTOF-MS/MS-based in-source collision-induced dissociation. Metabolites 2020, 10, 178. [Google Scholar] [CrossRef]
- Fattahi, A.; Shakeri, A.; Tayarani-Najaran, Z.; Kharbach, M.; Segers, K.; Heyden, Y.V.; Asili, J. UPLC-PDA-ESI-QTOF-MS/MS and GC-MS analysis of Iranian Dracocephalum moldavica L. Food Sci. Nutr. 2021. [Google Scholar] [CrossRef] [PubMed]
- Dhananjeyan, M.R.; Bykowski, C.; Trendel, J.A.; Sarver, J.G.; Ando, H.; Erhardt, P.W. Simultaneous determination of procaine and para-aminobenzoic acid by LC-MS/MS method. J. Chromatogr. B 2007, 847, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Kıvrak, İ. Analytical methods applied to assess chemical composition, nutritional value and in vitro bioactivities of Terfezia olbiensis and Terfezia claveryi from Turkey. Food Anal. Methods 2015, 8, 1279–1293. [Google Scholar] [CrossRef]
- Boğa, M.; Ertaş, A.; Yılmaz, M.A.; Kızıl, M.; Çeken, B.; Haşimi, N.; Deveci, Ö. UHPLC-ESI-MS/MS and GC-MS analyses on phenolic, fatty acid and essential oil of Verbascum pinetorum with antioxidant, anticholinesterase, antimicrobial and DNA damage protection effects. Iran. J. Pharm. Res. 2016, 15, 393–405. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5149026/ (accessed on 11 July 2021).
- Jeszka-Skowron, M.; Zgoła-Grześkowiak, A.; Frankowski, R. Cistus incanus a promising herbal tea rich in bioactive compounds: LC-MS/MS determination of catechins, flavonols, phenolic acids and alkaloids—A comparison with Camellia sinensis, Rooibos and Hoan Ngoc herbal tea. J. Food Compost Anal. 2018, 74, 71–81. [Google Scholar] [CrossRef]
- Gajula, R.; Pilli, N.R.; Ravi, V.B.; Maddela, R.; Inamadugu, J.K.; Polagani, S.R.; Busa, S. Simultaneous determination of atorvastatin and acetylsalicylic acid in human plasma by LC-MS/MS: Its pharmacokinetic application. Sci. Pharm. 2012, 80, 923–940. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.B.; Rai, D.K.; Brunton, N.P.; Martin-Diana, A.B.; Barry-Ryan, C. Characterization of phenolic composition in Lamiaceae spices by LC-ESI-MS/MS. J. Agric. Food Chem. 2010, 58, 10576–10581. [Google Scholar] [CrossRef]
- Kammerer, D.; Claus, A.; Carle, R.; Schieber, A. Polyphenol screening of pomace from red and white grape varieties (Vitis vinifera L.) by HPLC-DAD-MS/MS. J. Agric. Food Chem. 2004, 52, 4360–4367. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Guo, K.; Xia, J.; Zhou, J.; Yang, J.; Westaway, D.; Li, L. Development of Isotope labeling liquid chromatography mass spectrometry for mouse urine metabolomics: Quantitative metabolomic study of transgenic mice related to Alzheimer’s disease. J. Proteome Res. 2014, 13, 4457–4469. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Fernández, M.; Yang, P.Y.T.; Ludwig, I.A.; Clifford, M.N.; Cid, C.; Rodriguez-Mateos, A. In vivo study of the bioavailability and metabolic profile of (poly) phenols after sous-vide artichoke consumption. Food Chem. 2022, 367, 130620. [Google Scholar] [CrossRef]
- Zhang, H.; Shao, Y.; Bao, J.; Beta, T. Phenolic compounds and antioxidant properties of breeding lines between the white and black rice. Food Chem. 2015, 172, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yin, P.; Ma, C.; Liu, Y. Method development and validation for pharmacokinetic and tissue distributions of ellagic acid using ultrahigh performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Molecules 2014, 19, 18923–18935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuengchamnong, N.; Krittasilp, K.; Ingkaninan, K. Characterisation of phenolic antioxidants in aqueous extract of Orthosiphon grandiflorus tea by LC-ESI-MS/MS coupled to DPPH assay. Food Chem. 2011, 127, 1287–1293. [Google Scholar] [CrossRef]
- Dall’Asta, M.; Calani, L.; Tedeschi, M.; Jechiu, L.; Brighenti, F.; Del Rio, D. Identification of microbial metabolites derived from in vitro fecal fermentation of different polyphenolic food sources. Nutrition 2012, 28, 197–203. [Google Scholar] [CrossRef]
- Todoroki, K.; Ishii, Y.; Miyauchi, C.; Kitagawa, S.; Min, J.Z.; Inoue, K.; Toyo’oka, T. Simple and sensitive analysis of histamine and Tyramine in Japanese soy sauces and their intermediates using the stable isotope dilution HILIC-MS/MS method. J. Agric. Food Chem. 2014, 62, 6206–6211. [Google Scholar] [CrossRef] [PubMed]
- Mazzotti, F.; Benabdelkamel, H.; Di Donna, L.; Maiuolo, L.; Napoli, A.; Sindona, G. Assay of tyrosol and hydroxytyrosol in olive oil by tandem mass spectrometry and isotope dilution method. Food Chem. 2012, 135, 1006–1010. [Google Scholar] [CrossRef]
- Liu, Z.; Ezernieks, V.; Reddy, P.; Elkins, A.; Krill, C.; Murphy, K.; Spangenberg, G. A simple GC-MS/MS method for determination of smoke taint-related volatile phenols in grapes. Metabolites 2020, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.; Heck, N.D.V.; Ferreira, I.; Göethel, G.; Somacal, S.; Emanuelli, T.; Augusti, P.R. Ochratoxin A presence in Cabernet Sauvignon wine changes antioxidant activity in vitro and oxidative stress markers in vivo. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess 2020, 37, 1755–1764. [Google Scholar] [CrossRef]
- Arimboor, R.; Arumughan, C. HPLC-DAD-MS/MS profiling of antioxidant flavonoid glycosides in sea buckthorn (Hippophae rhamnoides L.) seeds. Int. J. Food Sci. Nutr. 2012, 63, 730–738. [Google Scholar] [CrossRef]
- Hong, Y.; Liao, X.; Chen, Z. Determination of bioactive components in the fruits of Cercis chinensis Bunge by HPLC-MS/MS and quality evaluation by principal components and hierarchical cluster analysis. J. Pharm. Anal. 2020. [Google Scholar] [CrossRef]
- Hubner, A.; Sobreira, F.; Vetore Neto, A.; Pinto, C.A.S.D.O.; Dario, M.F.; Díaz, I.E.C.; Bacchi, E.M. The synergistic behavior of antioxidant phenolic compounds obtained from winemaking waste’s valorization, increased the efficacy of a sunscreen system. Antioxidants 2019, 8, 530. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.F.; Guo, Y.X.; Zheng, G.; Wang, W.J. Chemical constituents comparison between Rhizoma Smilacis Glabrae and Rhizoma Smilacis Chinae by HPLC-DAD-MS/MS. Nat. Prod. Res. 2013, 27, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Tetik, M.A.; Sevindik, O.; Kelebek, H.; Selli, S. Screening of key odorants and anthocyanin compounds of cv. Okuzgozu (Vitis vinifera L.) red wines with a free run and pressed pomace using GC-MS-Olfactometry and LC-MS-MS. J. Mass Spectrom. 2018, 53, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.J.; Park, J.E.; Lee, K.S.; Seo, W.D.; Song, S.B.; Lee, M.H.; Lee, J.H. Identification of anthocyanin compositions in black seed coated Korean adzuki bean (Vigna angularis) by NMR and UPLC-Q-Orbitrap-MS/MS and screening for their antioxidant properties using different solvent systems. Food Chem. 2021, 346, 128882. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liang, F.; Bin, Y.; Li, P.; Duan, C. Screening non-colored phenolics in red wines using liquid chromatography/ultraviolet and mass spectrometry/mass spectrometry libraries. Molecules 2007, 12, 679–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Mu, Y.; Xiong, S.; Sun, P.; Deng, Z. A UPLC-MS/MS method for comparative pharmacokinetics study of morusin and morin in normal and diabetic rats. Biomed. Chromatogr. 2019, 33, e4516. [Google Scholar] [CrossRef]
- De Rosso, M.; Panighel, A.; Vedova, A.D.; Gardiman, M.; Flamini, R. Characterization of non-anthocyanic flavonoids in some hybrid red grape extracts potentially interesting for industrial uses. Molecules 2015, 20, 18095–18106. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, V.; López, R.; Cacho, J.F. Quantitative determination of the odorants of young red wines from different grape varieties. J. Agric. Sci. 2000, 80, 1659–1667. [Google Scholar] [CrossRef]
- Yang, L.; Jiang, H.; Xing, X.; Yan, M.; Guo, X.; Man, W.; Yang, L. A biosensor-based quantitative analysis system of major active ingredients in Lonicera japonica thunb. Using UPLC-QDa and chemometric analysis. Molecules 2019, 24, 1787. [Google Scholar] [CrossRef] [Green Version]
- Abass, K.; Reponen, P.; Mattila, S.; Pelkonen, O. Metabolism of α-thujone in human hepatic preparations in vitro. Xenobiotica 2011, 41, 101–111. [Google Scholar] [CrossRef]
- Thaipong, K.; Boonprakob, U.; Crosby, K.; Cisneros-Zevallos, L.; Byrne, D.H. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compost. Anal. 2006, 19, 669–675. [Google Scholar] [CrossRef]
- Jasprica, I.; Bojic, M.; Mornar, A.; Besic, E.; Bucan, K.; Medic-Saric, M. Evaluation of antioxidative activity of croatian propolis samples using DPPH· and ABTS·+ stable free radical assays. Molecules 2007, 12, 1006–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baburajeev, C.P.; Mohan, C.D.; Pandey, V.; Rangappa, S.; Shivalingegowda, N.; Kalash, L.; Rangappa, K.S. Synthesis of CC, CN coupled novel substituted dibutyl benzothiazepinone derivatives and evaluation of their thrombin inhibitory activity. Bioorg. Chem. 2019, 87, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.K.; Weng, Z.M.; Ge, G.B.; Li, H.L.; Ding, L.L.; Dai, Z.R.; Hou, J. Biflavones from Ginkgo biloba as novel pancreatic lipase inhibitors: Inhibition potentials and mechanism. Int. J. Biol. Macromol. 2018, 118, 2216–2223. [Google Scholar] [CrossRef] [PubMed]


| Antioxidant Activity Inhibition (Trolox (%)) | Thrombin Inhibitory Activity Inhibition (%) | Lipase Inhibitory Activity Inhibition (%) | |
|---|---|---|---|
| F1 | 31.07 | 37.82 | 69.67 |
| F2 | no | 44.42 | 87.30 |
| F3 | 45.35 | 24.65 | no |
| F4 | 55.64 | 26.68 | no |
| F5 | 29.63 | 29.77 | no |
| F6 | 42.56 | 26.61 | no |
| F7 | no | no | no |
| F8 | no | no | no |
| F9 | 24.08 | 21.21 | no |
| F10 | 23.10 | no | no |
| F11 | no | no | 87.35 |
| F12 | no | no | no |
| F13 | 25.70 | no | no |
| F14 | no | 22.44 | no |
| F15 | no | no | no |
| F16 | 36.79 | no | 100.20 |
| F17 | 55.64 | no | 66.44 |
| F18 | 29.63 | no | no |
| F19 | 42.56 | no | 124.96 |
| F20 | no | no | 70.68 |
| F21 | no | no | 113.43 |
| F22 | 19.34 | 19.22 | 136.53 |
| F23 | 30.56 | no | 94.14 |
| F24 | 23.59 | no | 103.15 |
| F25 | 42.87 | 18.56 | 66.12 |
| F26 | 54.32 | 35.06 | 97.89 |
| F27 | 48.31 | 36.21 | 67.39 |
| F28 | 43.88 | 48.47 | 68.79 |
| F29 | 39.03 | 29.44 | 127.97 |
| F30 | 40.83 | 18.07 | 90.74 |
| F31 | 32.31 | 24.49 | no |
| F32 | 30.82 | no | no |
| F33 | no | no | no |
| F34 | no | no | no |
| F35 | no | no | no |
| F36 | 30.36 | 35.12 | no |
| F37 | 24.07 | no | no |
| F38 | no | no | no |
| F39 | 19.44 | no | no |
| F40 | 19.48 | no | no |
| F41 | no | no | no |
| F42 | 22.28 | no | 71.92 |
| F43 | 22.64 | no | 77.88 |
| S44 | 25.36 | no | 84.26 |
| F45 | no | 27.81 | no |
| F46 | 25.08 | no | no |
| F47 | no | no | no |
| F48 | no | no | 79.38 |
| F49 | no | no | 68.49 |
| F50 | no | no | no |
| F51 | no | 19.33 | no |
| F52 | no | no | no |
| F53 | no | no | no |
| F54 | 27.09 | no | no |
| F55 | 25.94 | no | 73.85 |
| F56 | 25.27 | 27.66 | 72.88 |
| F57 | 36.68 | 20.91 | 66.70 |
| F58 | no | 20.65 | 76.42 |
| F59 | 22.80 | no | 69.65 |
| F60 | 23.21 | no | 67.84 |
| Peak No. | RT (min) | Ion | Found at Mass | MS/MS Fragmentation Ions | Formula | Identification | ppm |
|---|---|---|---|---|---|---|---|
| 1 | 0.70 | [M+H]+ | 116.0702 | 70.0647 | C5H9NO2 | D-proline | 0.8 |
| 2 | 0.76 | [M+H]+ | 175.1189 | 70.0652 | C6H14N4O2 | L-arginine | −0.3 |
| 3 | 0.79 | [M−H]− | 179.0562 | 99.0087, 71.0137, 59.0139 | C6H12O6 | D-glucose | 0.3 |
| 4 | 0.88 | [M-H]− | 181.0720 | 83.0143, 71.0140, 59.0140 | C6H14O6 | D-mannitol | 1.5 |
| 5 | 0.96 | [M−H]− | 191.0199 | 85.0296, 59.0134 | C6H8O7 | citric acid | 1.0 |
| 6 | 0.99 | [M−H]− | 149.0091 | 105.0217, 87.0085, 72.9928 | C4H6O6 | tartaric acid | −0.7 |
| 7 | 1.04 | [M+H]+ | 138.0548 | 120.0433, 92.0495, 78.0339 | C7H7NO2 | 4-aminobenzoic acid | −1.5 |
| 8 | 1.12 | [M−H]− | 341.1088 | 161.0459, 71.0137, 59.0138 | C12H22O11 | trehalose | −0.3 |
| 9 | 1.15 | [M−H]− | 149.0455 | 75.0221, 59.0133 | C5H10O5 | D-ribose | −0.2 |
| 10 | 1.31 | [M−H]− | 133.0142 | 115.0018, 71.0136 | C4H6O5 | L-malic acid | −0.5 |
| 11 | 1.33 | [M+H]+ | 118.0862 | 72.0809 | C5H11NO2 | L-valine | −0.4 |
| 12 | 1.48 | [M−H]− | 173.0454 | 137.0225, 93.0343 | C7H10O5 | shikimic acid | −1.0 |
| 13 | 1.71 | [M+H]+ | 150.0586 | 74.0243, 61.0108 | C5H11NO2S | L-methionine | 1.8 |
| 14 | 2.03 | [M+H]+ | 124.0393 | 80.0506, 55.9346 | C6H5NO2 | niacin | 0.2 |
| 15 | 3.27 | [M−H]− | 117.0192 | 99.9254, 73.0291 | C4H6O4 | succinic acid | −1.2 |
| 16 | 3.51 | [M+H]+ | 165.0547 | 119.0479, 95.0503, 77.0388, 59.9302 | C9H8O3 | trans-4-hydroxycinnamic acid | 0.7 |
| 17 | 3.85 | [M+H]+ | 137.0913 | 121.0616, 103.0616, 95.0561, 91.0556, 77.0391 | C8H11NO | tyramine | −0.5 |
| 18 | 5.97 | [M−H]− | 169.0141 | 125.0244, 107.0156, 79.0186 | C7H6O5 | gallic acid | −1.0 |
| 19 | 8.77 | [M+H]+ | 141.0182 | 123.0080, 95.0127, 67.0187, 55.9453 | C6H4O4 | coumalic acid | −0.1 |
| 20 | 11.49 | [M−H]− | 153.0558 | 123.0085, 108.0221, 95.0139, 78.9590 | C8H10O3 | hydroxytyrosol | 0.2 |
| 21 | 11.57 | [M−H]− | 153.0194 | 109.0298, 91.0298, 65.0029 | C7H6O4 | protocatechuic acid | 0.3 |
| 22 | 13.85 | [M−H]− | 305.0664 | 179.0360, 165.0194, 137.0243, 125.0246 | C15H14O7 | (+)-gallocatechin | −1.0 |
| 23 | 14.27 | [M−H]− | 179.0349 | 135.0560, 89.0394, 79.0550 | C9H8O4 | acetylsalicylic acid | −0.7 |
| 24 | 14.27 | [M−H]− | 179.0348 | 161.0263, 135.0454, 117.0351 | C13H12O9 | caftaric acid | −1.0 |
| 25 | 15.05 | [M−H]− | 153.0194 | 123.0061, 109.0298, 91.0298 | C7H6O4 | gentisic acid | 0.4 |
| 26 | 15.21 | [M−H]− | 137.0255 | 108.0213 | C7H6O3 | protocatechualdehyde | −2.7 |
| 27 | 15.31 | [M−H]− | 181.0506 | 135.0453 | C9H10O4 | homovanillic acid | 0.0 |
| 28 | 15.43 | [M−H]− | 183.0299 | 124.0234, 95.0136 | C8H8O5 | methyl gallate | 0.0 |
| 29 | 15.90 | [M−H]− | 137.0244 | 93.0340 | C7H6O3 | 4-hydroxybenzoic acid | 0.4 |
| 30 | 16.32 | [M−H]− | 353.0877 | 191.0616, 179.0341, 135.0439 | C16H18O9 | chlorogenic acid | −0.2 |
| 31 | 17.36 | [M−H]− | 295.0458 | 163.0400, 119.0503, 87.0094 | C13H12O8 | p-coutaric acid | 0.1 |
| 32 | 18.08 | [M−H]− | 163.0401 | 119.0498, 93.0347 | C9H8O3 | m-coumaric acid | 0.2 |
| 33 | 18.08 | [M−H]− | 295.0458 | 163.0400, 119.0498 | C13H12O8 | coutaric acid | −0.4 |
| 34 | 18.15 | [M−H]− | 145.0507 | 73.0273, 55.0191 | C6H10O4 | dimethyl succinate | 0.6 |
| 35 | 19.24 | [M−H]− | 121.0296 | 108.0212, 92.0265 | C7H6O2 | benzoic acid | 0.8 |
| 36 | 19.66 | [M−H]− | 577.1346 | 425.0864, 407.0775, 289.0714, 161.0238, 125.0243 | C30H26O12 | procyanidin B1 | −0.9 |
| 37 | 19.89 | [M−H]− | 305.0664 | 245.0462, 219.0652, 139.0401, 125.0244 | C15H14O7 | (−)-epigallocatechin | 0.2 |
| 38 | 19.98 | [M−H]− | 577.1346 | 407.0768, 289.0709 | C30H26O12 | procyanidin B3 | 0.0 |
| 39 | 20.01 | [M−H]− | 167.0350 | 108.0212 | C8H8O4 | vanillic acid | 0.2 |
| 40 | 20.21 | [M−H]− | 163.0400 | 119.0502, 93.0343 | C9H8O3 | 2-Hydroxycinnamic acid | −0.1 |
| 41 | 20.47 | [M−H]− | 289.0713 | 245.0843, 203.0713, 151.0401, 109.0294 | C15H14O6 | catechin | −1.5 |
| 42 | 20.58 | [M−H]− | 325.0563 | 193.0490, 117.0341 | C14H14O9 | fertaric acid | −0.5 |
| 43 | 20.98 | [M−H]− | 179.0349 | 135.0449, 89.0393 | C9H8O4 | caffeic acid | −1.0 |
| 44 | 21.90 | [M−H]− | 441.0186 | 289.0735, 217.0167, 191.0337, 151.0386 | C22H18O10 | epicatechin gallate | 2.2 |
| 45 | 22.06 | [M−H]− | 197.0454 | 182.0256, 167.0014, 138.0317, 123.0090, 95.0142 | C9H10O5 | syringic acid | −0.3 |
| 46 | 23.01 | [M−H]− | 577.1346 | 425.0893, 407.0782, 289.0721 | C30H26O12 | procyanidin B2 | −0.7 |
| 47 | 23.11 | [M−H]− | 325.0926 | 163.0394, 145.0294, 117.0343 | C15H18O8 | p-coumaric acid glucoside | −0.5 |
| 48 | 23.37 | [M−H]− | 189.0769 | 71.0503 | C8H14O5 | diethyl malate | 0.3 |
| 49 | 23.69 | [M−H]− | 165.0556 | 119.0527, 103.0565 | C9H10O3 | desaminotyrosine | −0.5 |
| 50 | 24.37 | [M−H]− | 137.0244 | 93.0470, 65.0473 | C7H6O3 | 2-hydroxybenzoic acid | 2.2 |
| 51 | 24.38 | [M−H]− | 289.0713 | 203.0710, 187.0396, 151.0401, 123.0449, 109.0290 | C15H14O6 | epicatechin | −0.7 |
| 52 | 24.68 | [M−H]− | 167.0350 | 123.0436, 108.0212 | C8H8O4 | 3-hydroxy-4-methoxybenzoic acid | 2.5 |
| 53 | 25.16 | [M−H]− | 197.0454 | 169.0142, 125.0239, 79.0184 | C9H10O5 | ethyl gallate | −0.9 |
| 54 | 25.32 | [M]+ | 493.1341 | 331.0822, 316.0553 | C23H25O12 | malvidin-3-O-glucoside | 0.1 |
| 55 | 25.74 | [M−H]− | 163.0401 | 119.0503, 93.0347 | C9H8O3 | p-coumaric acid | −0.6 |
| 56 | 30.75 | [M−H]− | 167.0350 | 123.0450, 65.0024 | C8H8O4 | homogentisic acid | 0.8 |
| 57 | 30.80 | [M−H]− | 577.1346 | 425.0886, 407.0789, 289.0734 | C30H26O12 | procyanidin B7 | 0.6 |
| 58 | 34.06 | [M−H]− | 479.0826 | 316.0225, 271.0249, 151.0034 | C21H20O13 | myricetin-3-O-galactoside | −1.0 |
| 59 | 35.93 | [M−H]− | 389.1242 | 227.0716, 185.0606, 143.0505 | C20H22O8 | trans-piceid | −0.2 |
| 60 | 38.70 | [M−H]− | 300.9987 | 257.011, 229.0126, 145.0295 | C14H6O8 | ellagic acid | −0.9 |
| 61 | 38.84 | [M−H]− | 181.0506 | 109.0286, 65.0032 | C9H10O4 | dihydrocaffeic acid | 0.5 |
| 62 | 39.28 | [M]+ | 465.1036 | 303.051, 229.0488 | C21H21O12 | delphinidin-3-O-glucoside | 1.9 |
| 63 | 39.31 | [M−H]− | 463.0884 | 301.0379, 271.0245, 243.0291, 151.0031 | C21H20O12 | hyperoside | 0.4 |
| 64 | 39.71 | [M−H]− | 449.1090 | 285.0413, 151.0041 | C21H22O11 | quercitrin | 0.2 |
| 65 | 39.83 | [M]+ | 627.1462 | 303.0351 | C27H31O17 | delphinidin-3,5-O-diglucoside | 1.1 |
| 66 | 40.08 | [M−H]− | 477.0670 | 301.0352, 255.0296, 151.0034 | C21H18O13 | quercetin-3-O-glucuronide | −0.9 |
| 67 | 41.28 | [M−H]− | 493.0985 | 330.0381, 315.0155, 271.0249, 151.0034 | C22H22O13 | laricitrin-3-O-glucoside | −0.6 |
| 68 | 42.86 | [M−H]− | 447.0936 | 301.0372, 271.0256, 255.0295, 151.0020 | C21H20O11 | luteolin -3’-glucoside | 1.2 |
| 69 | 42.98 | [M−H]− | 449.1090 | 285.0413, 151.0041 | C21H22O11 | astilbin | 0.9 |
| 70 | 43.45 | [M−H]− | 463.0884 | 301.0379, 255.0305, 151.0039 | C21H20O12 | isoquercitrin | 0.8 |
| 71 | 44.78 | [M−H]− | 447.0936 | 301.0372, 255.0293, 151.0016 | C21H20O11 | astragalin | 0.8 |
| 72 | 45.19 | [M−H]− | 389.1242 | 227.0994, 185.0606 | C20H22O8 | cis-piceid | 0.0 |
| 73 | 45.72 | [M−H]− | 317.0300 | 245.0504, 137.0241, 109.0292, 151.0035 | C15H10O8 | myricetin | −0.9 |
| 74 | 45.77 | [M]+ | 479.1193 | 317.0648, 302.0429, 285.0362, 153.0184 | C22H23O12 | petunidin-3-O-glucoside | 1.9 |
| 75 | 45.80 | [M−H]− | 477.1041 | 315.0530, 300.0263, 271.0255, 243.0306, 151.0036 | C22H22O12 | isorhamnetin-3-O-glucoside | 0.6 |
| 76 | 47.17 | [M−H]− | 227.0715 | 185.0587, 143.0501, 115.0556 | C14H12O3 | trans-resveratrol | 0.6 |
| 77 | 47.42 | [M−H]− | 435.1300 | 273.0769, 179.0349, 167.0352 | C21H24O10 | phloridzin | 0.7 |
| 78 | 49.16 | [M]+ | 507.1141 | 303.0767, 187.0626, 113.0234 | C23H23O13 | delphindin-3-O-(6-O-acetylglucoside) | 1.6 |
| 79 | 51.13 | [M−H]− | 207.0662 | 179.0433, 135.0446 | C11H12O4 | ethyl caffeate | −0.6 |
| 80 | 51.94 | [M]+ | 639.1719 | 477.1176, 419.0853, 331.0874 | C32H31O14 | malvidin-3-O-(6-O-coumaroylglucoside) | 1.7 |
| 81 | 52.60 | [M+H]+ | 303.0497 | 285.0398, 257.0456, 229.0492, 153.0188 | C15H10O7 | morin | −0.8 |
| 82 | 52.60 | [M]+ | 303.0497 | 229.0492, 153.0188 | C15H11O7 | delphinidin | −0.8 |
| 83 | 52.63 | [M−H]− | 301.0351 | 245.0447, 229.0513, 151.0034, 121.0292 | C15H10O7 | quercetin | −1.0 |
| 84 | 52.67 | [M−H]− | 227.0715 | 143.0504 | C14H12O3 | resveratrol | −0.9 |
| 85 | 53.23 | [M−H]− | 331.0459 | 316.0231, 271.0251, 178.9991, 151.0039 | C16H12O8 | laricitrin | 0.0 |
| 86 | 55.82 | [M−H]− | 271.0613 | 229.0500, 177.0190, 151.0048, 119.0618 | C15H12O5 | naringenin | 0.4 |
| 87 | 57.10 | [M]+ | 287.0549 | 213.0559, 153.0180, 137.0233 | C15H11O6 | cyanidin | −0.3 |
| 88 | 57.12 | [M−H]− | 285.0403 | 239.0356, 187.0403, 151.004 | C15H10O6 | kamepferol | −0.6 |
| 89 | 57.32 | [M−H]− | 345.0618 | 330.0385, 315.0153, 259.0252, 187.0394 | C17H14O8 | syringetin | 0.6 |
| 90 | 57.39 | [M]+ | 317.0656 | 229.0498, 153.0188 | C16H13O7 | petunidin | 0.0 |
| 91 | 57.41 | [M−H]− | 315.0508 | 300.0270, 271.0241, 255.0299, 227.0241, 151.0036 | C16H12O7 | isorhamnetin | −0.6 |
| 92 | 58.32 | [M+H]+ | 193.1588 | 56.9419 | C13H20O | ionone | 0.6 |
| 93 | 60.04 | [M+H]+ | 173.1536 | 76.0221 | C10H20O2 | ethyl caprylate | 0.2 |
| 94 | 60.96 | [M+H]+ | 195.1748 | 177.1634, 107.0855, 77.0387 | C13H22O | theaspirane | 2.1 |
| Fraction Number | Peak Number | Identification | Antioxidant Activity | Thrombin Inhibitory Activity | Lipase Inhibitory Activity |
|---|---|---|---|---|---|
| F1 | 5, 6, 10, 12 | citric acid, tartaric acid, L-malic acid, shikimic acid | yes | yes | yes |
| F2 | 12 | shikimic acid | yes | yes | |
| F3 | 15, 16 | succinic acid, trans-4-hydroxycinnamic acid | yes | yes | |
| F4 | 17 | tyramine | yes | yes | |
| F5 | 18 | gallic acid | yes | yes | |
| F6 | 18 | gallic acid | yes | yes | |
| F9 | 19 | coumalic acid | yes | yes | |
| F10 | 21 | protocatechuic acid | yes | ||
| F11 | 21 | protocatechuic acid | yes | ||
| F13 | 22 | (+)-gallocatechin | yes | ||
| F14 | 23, 24 | acetylsalicylic acid, caftaric acid | yes | ||
| F16 | 25, 26, 27, 28 | gentisic acid, protocatechualdehyde, homovanillic acid, methyl gallate | yes | yes | |
| F17 | 29 | 4-hydroxybenzoic acid | yes | yes | |
| F18 | 30 | chlorogenic acid | yes | ||
| F19 | 31 | p-coutaric acid | yes | yes | |
| F20 | 32, 33, 34 | m-coumaric acid, coutaric acid, dimethyl succinate | yes | ||
| F21 | 35 | benzoic acid | yes | ||
| F22 | 36, 37, 38 | procyanidin B1, (−)-epigallocatechin, procyanidin B3 | yes | yes | yes |
| F23 | 39, 40, 41, 42, 43 | vanillic acid, 2-Hydroxycinnamic acid, catechin, fertaric acid, caffeic acid | yes | yes | |
| F24 | 44 | epicatechin gallate | yes | yes | |
| F25 | 45 | syringic acid | yes | yes | yes |
| F26 | 46, 47, 48 | procyanidin B2, p-coumaric acid glucoside, diethyl malate | yes | yes | yes |
| F27 | 49, 50, 51 | desaminotyrosine, 2-hydroxybenzoic acid, epicatechin | yes | yes | yes |
| F28 | 53, 54 | ethyl gallate, malvidin-3-O-glucoside | yes | yes | yes |
| F29 | 55 | p-coumaric acid | yes | yes | yes |
| F30 | 55 | p-coumaric acid | yes | yes | yes |
| F31 | 56 | homogentisic acid | yes | yes | |
| F32 | 57 | procyanidin B7 | yes | ||
| F36 | 58 | myricetin-3-O-galactoside | yes | yes | |
| F37 | 59 | trans-piceid | yes | ||
| F39 | 60, 61 | ellagic acid, dihydrocaffeic acid | yes | ||
| F40 | 62, 63 | delphinidin-3-O-glucoside, hyperoside | yes | ||
| F42 | 65, 66 | delphinidin-3,5-O-diglucoside, quercetin-3-O-glucuronide | yes | yes | |
| F43 | 67, 68, 69, 70 | laricitrin-3-O-glucoside, luteolin -3-O-glucoside, astilbin, isoquercitrin | yes | yes | |
| F44 | 70 | isoquercitrin | yes | yes | |
| F45 | 71 | astragalin | yes | ||
| F46 | 72 | cis-piceid | yes | ||
| F48 | 73, 74, 75 | myricetin, petunidin-3-O-glucoside, isorhamnetin-3-O-glucoside | yes | ||
| F49 | 76, 77 | trans-resveratrol, phloridzin | yes | ||
| F51 | 778 | delphindin-3-O-(6-O-acetylglucoside) | yes | ||
| F54 | 79, 80 | ethyl caffeate, malvidin-3-O-(6-O-coumaroylglucoside) | yes | ||
| F55 | 81, 82, 83, 84 | morin, delphinidin, quercetin, resveratrol | yes | yes | |
| F56 | 85 | laricitrin | yes | yes | yes |
| F57 | 85 | laricitrin | yes | yes | yes |
| F58 | 86 | naringenin | yes | yes | |
| F59 | 87 | cyanidin | yes | yes | |
| F60 | 88, 89, 90 | kamepferol, syringetin, petunidin | yes | yes |
| Analytes | Precision | Stability | Linearity | LOQ (μg/mL) | LOD (μg/mL) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intra-Day, RSD (%), (n = 3) | Inter-Day RSD (%) (n = 9) | RSD (%) (n = 6) | Range (μg/mL) | Equation | r2 | |||||
| Low | Middle | High | ||||||||
| gallic acid | 2.2 | 2.2 | 1.3 | 3.4 | 2.5 | 20.00~320.0 | y = 0.3627x + 0.0831 | 0.9998 | 0.12 | 0.040 |
| coumalic acid | 3.2 | 1.5 | 1.9 | 4.4 | 1.8 | 6.030~96.48 | y = 0.2339x − 0.0078 | 0.9994 | 2.0 | 0.75 |
| protocatechuic acid | 4.9 | 2.2 | 1.7 | 4.2 | 1.1 | 1.005~16.08 | y = 0.1498x + 0.0290 | 0.9993 | 0.12 | 0.040 |
| proanthocyanidin B1 | 3.6 | 2.1 | 1.8 | 4.6 | 2.2 | 6.000~96.0 | y = 0.1156x + 0.0121 | 0.9996 | 0.50 | 0.12 |
| catechin | 5.0 | 1.3 | 2.0 | 3.3 | 2.5 | 6.030~96.5 | y = 0.2149x + 0.0288 | 0.9997 | 0.12 | 0.040 |
| caffeic acid | 2.5 | 2.3 | 1.3 | 4.7 | 2.2 | 3.000~48.00 | y = 0.2645x + 0.0428 | 0.9992 | 0.12 | 0.040 |
| syringic acid | 4.8 | 2.4 | 2.1 | 4.8 | 1.8 | 2.973~47.56 | y = 0.5234x + 0.0300 | 0.9997 | 0.25 | 0.08 |
| epicatechin | 4.2 | 2.0 | 1.2 | 4.4 | 2.9 | 3.030~48.48 | y = 0.3627x + 0.0831 | 0.9998 | 0.50 | 0.12 |
| p-coumaric acid | 4.4 | 1.7 | 1.4 | 4.1 | 2.1 | 3.015~48.24 | y = 0.2339x + 0.0078 | 0.9994 | 0.25 | 0.08 |
| isoquercitrin | 4.6 | 1.9 | 1.3 | 4.6 | 1.8 | 0.2500~64.64 | y = 0.1495x + 0.0431 | 0.9994 | 0.25 | 0.08 |
| isorhamnetin-3-O-glucoside | 5.0 | 2.1 | 1.7 | 3.3 | 2.3 | 0.990~15.84 | y = 0.1156x + 0.0121 | 0.9996 | 0.50 | 0.12 |
| quercetin | 4.3 | 2.6 | 2.2 | 4.6 | 2.5 | 0.7500~31.46 | y = 0.2166x − 0.0094 | 0.9995 | 0.75 | 0.25 |
| Analytes | Original (μg) | Spiked (μg) | Found (μg) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| gallic acid | 29.34 | 20.00 | 45.81 | 99.8 | 1.8 |
| 40.00 | 68.85 | 97.0 | 1.4 | ||
| 60.00 | 89.14 | 99.8 | 1.0 | ||
| coumalic acid | 9.724 | 6.030 | 14.65 | 87.8 | 4.1 |
| 12.06 | 21.19 | 95.1 | 2.2 | ||
| 18.09 | 26.77 | 86.7 | 3.1 | ||
| protocatechuic acid | 3.298 | 1.005 | 3.720 | 83.4 | 4.3 |
| 2.010 | 4.864 | 90.6 | 2.5 | ||
| 3.015 | 6.338 | 98.4 | 1.4 | ||
| proanthocyanidin B1 | 9.027 | 6.000 | 14.01 | 89.7 | 4.0 |
| 12.00 | 21.58 | 103.6 | 2.3 | ||
| 18.00 | 26.27 | 94.1 | 2.1 | ||
| catechin | 11.43 | 3.000 | 14.15 | 97.7 | 3.6 |
| 6.000 | 17.50 | 100.1 | 0.7 | ||
| 9.000 | 20.61 | 100.0 | 0.3 | ||
| caffeic acid | 6.539 | 2.972 | 9.33 | 96.3 | 3.4 |
| 5.945 | 13.14 | 105.0 | 2.1 | ||
| 8.918 | 15.26 | 98.9 | 1.0 | ||
| syringic acid | 6.204 | 2.973 | 8.97 | 97.7 | 3.4 |
| 5.945 | 12.08 | 97.5 | 3.2 | ||
| 8.918 | 14.95 | 101.7 | 1.8 | ||
| epicatechin | 3.922 | 3030 | 6.932 | 101.2 | 1.6 |
| 6.060 | 10.04 | 101.9 | 0.7 | ||
| 9.090 | 13.18 | 105.9 | 1.4 | ||
| p-coumaric acid | 4.473 | 3.015 | 7.590 | 105.6 | 3.6 |
| 6.030 | 10.15 | 92.5 | 1.1 | ||
| 9.045 | 13.78 | 100.6 | 0.6 | ||
| isoquercitrin | 8.802 | 4.040 | 12.75 | 98.1 | 3.2 |
| 8.080 | 16.75 | 98.8 | 2.1 | ||
| 12.12 | 20.64 | 98.8 | 1.9 | ||
| isorhamnetin-3-O-glucoside | 3.144 | 0.9900 | 4.053 | 98.1 | 1.1 |
| 1.980 | 5.123 | 97.4 | 2.6 | ||
| 2.970 | 6.019 | 102.1 | 2.4 | ||
| quercetin | 3.146 | 1.966 | 4.928 | 95.5 | 1.6 |
| 3.933 | 6.950 | 96.7 | 1.3 | ||
| 5.899 | 8.82 | 92.2 | 4.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Yu, X.-a.; Wang, B.; Yin, G.; Wang, J.; Wang, T.; Bi, K. Based on Multi-Activity Integrated Strategy to Screening, Characterization and Quantification of Bioactive Compounds from Red Wine. Molecules 2021, 26, 6750. https://doi.org/10.3390/molecules26216750
Gao Y, Yu X-a, Wang B, Yin G, Wang J, Wang T, Bi K. Based on Multi-Activity Integrated Strategy to Screening, Characterization and Quantification of Bioactive Compounds from Red Wine. Molecules. 2021; 26(21):6750. https://doi.org/10.3390/molecules26216750
Chicago/Turabian StyleGao, Yuye, Xie-an Yu, Bing Wang, Guo Yin, Jue Wang, Tiejie Wang, and Kaishun Bi. 2021. "Based on Multi-Activity Integrated Strategy to Screening, Characterization and Quantification of Bioactive Compounds from Red Wine" Molecules 26, no. 21: 6750. https://doi.org/10.3390/molecules26216750
APA StyleGao, Y., Yu, X.-a., Wang, B., Yin, G., Wang, J., Wang, T., & Bi, K. (2021). Based on Multi-Activity Integrated Strategy to Screening, Characterization and Quantification of Bioactive Compounds from Red Wine. Molecules, 26(21), 6750. https://doi.org/10.3390/molecules26216750






