CD123-Targeted Nano-Curcumin Molecule Enhances Cytotoxic Efficacy in Leukemic Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Reagents
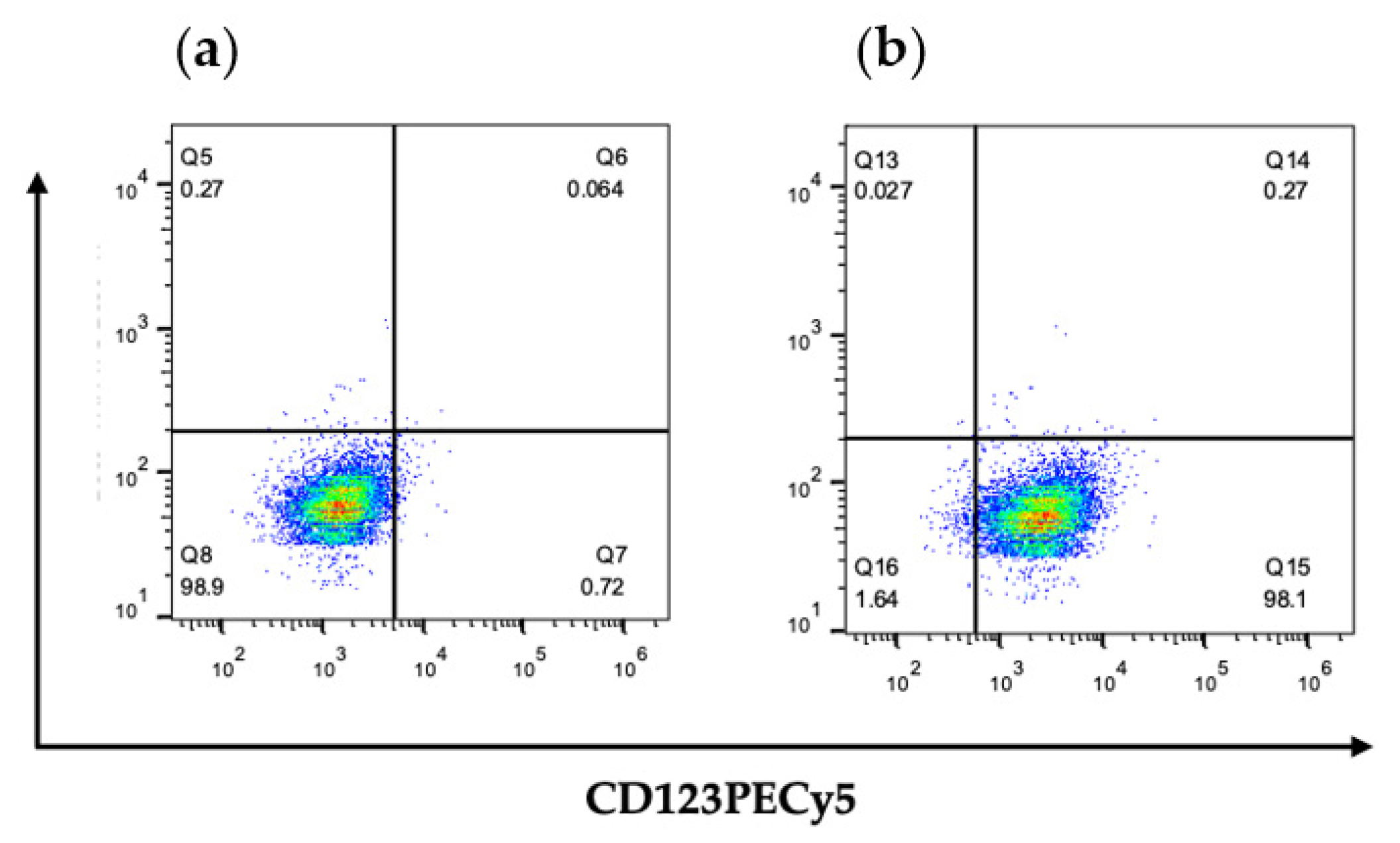
2.2. Analysis of Surface CD123 Expression through Flow Cytometry
2.3. Preparation of Curcumin-Loaded PLGA/Poloxamer Nanoparticles (Cur-NPs) with or without the Targeting Ab
2.3.1. Preparation of Curcumin-Loaded PLGA/Poloxamer Nanoparticles (Cur-NPs)
2.3.2. Conjugation of CD123 Antibody to Cur-NPs (Anti-CD123-Cur-NPs)
2.4. NP Characterization
2.5. Cellular Uptake of Anti-CD123-Cur-NPs into the CD123-Expressing KG-1a Cell Line
2.6. In Vitro Cytotoxicity Assay
2.7. Cytotoxicity of Peripheral Blood Mononuclear Cells (PBMCs)
2.8. Apoptosis Assay
2.9. Statistical Analysis
3. Results
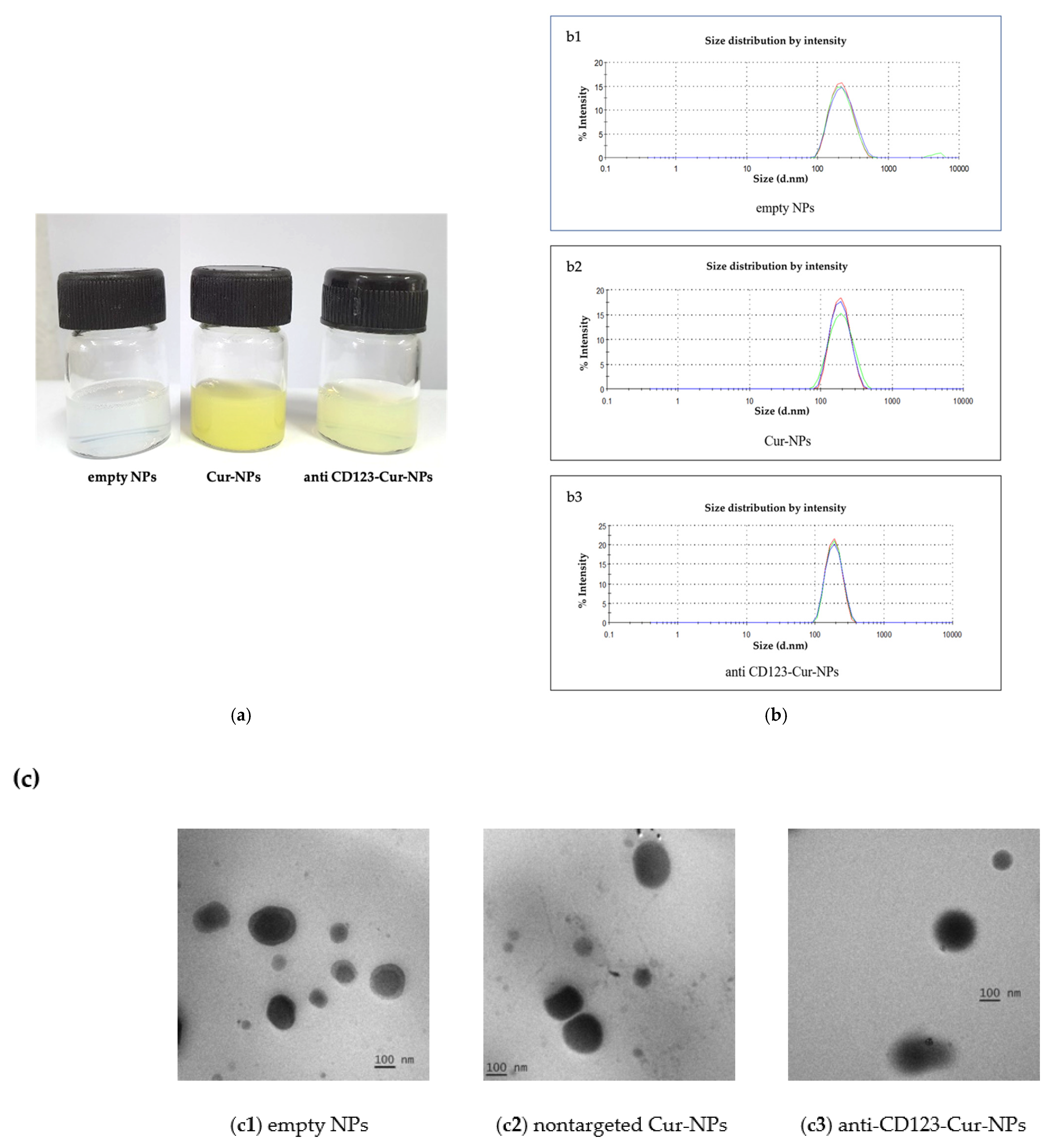
3.1. NP Formulation
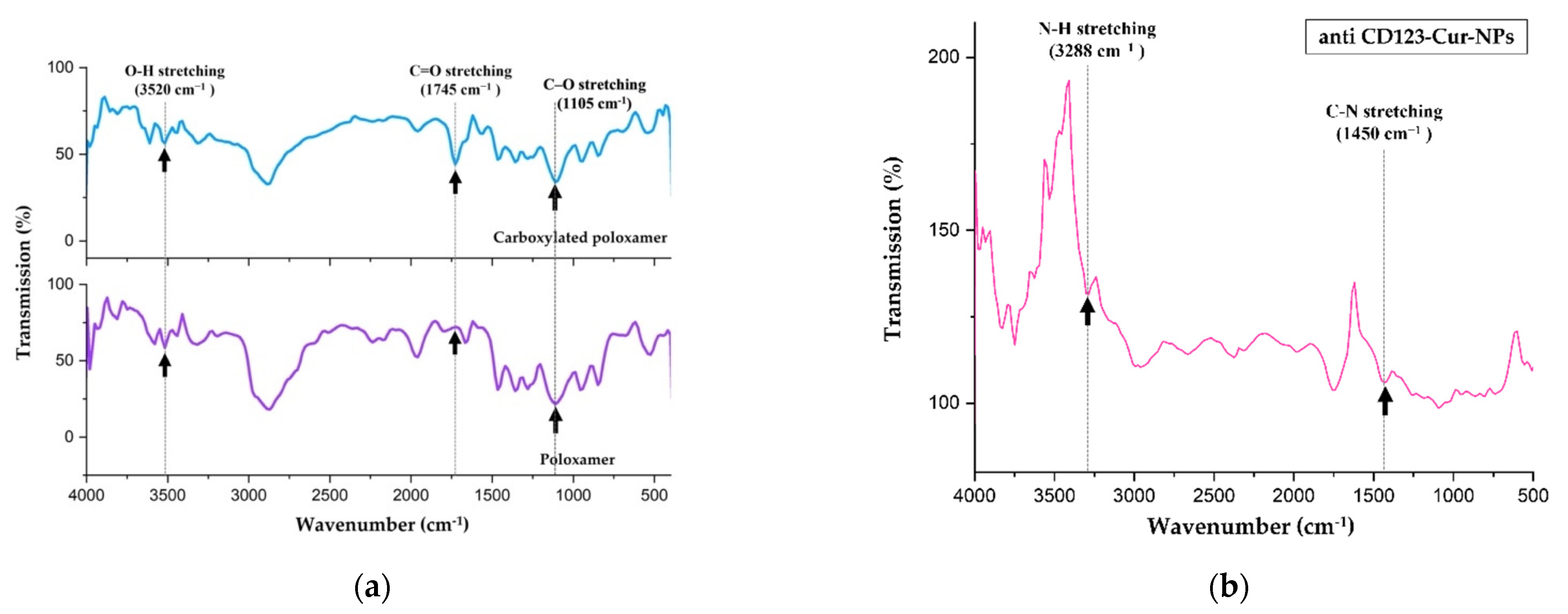
3.2. Characterization of Nanoparticles
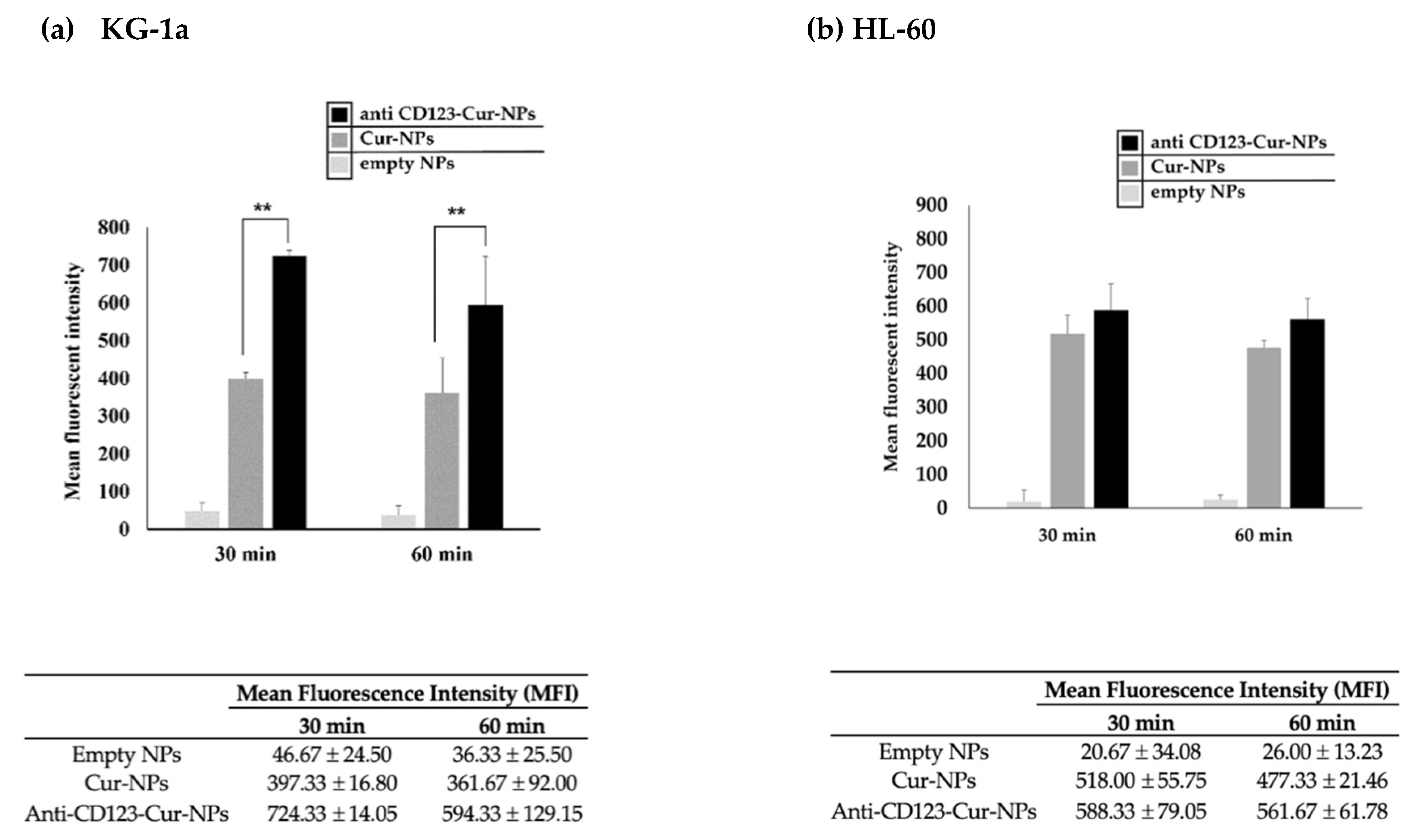
3.3. Cellular Uptake of Anti-CD123-Cur-NPs
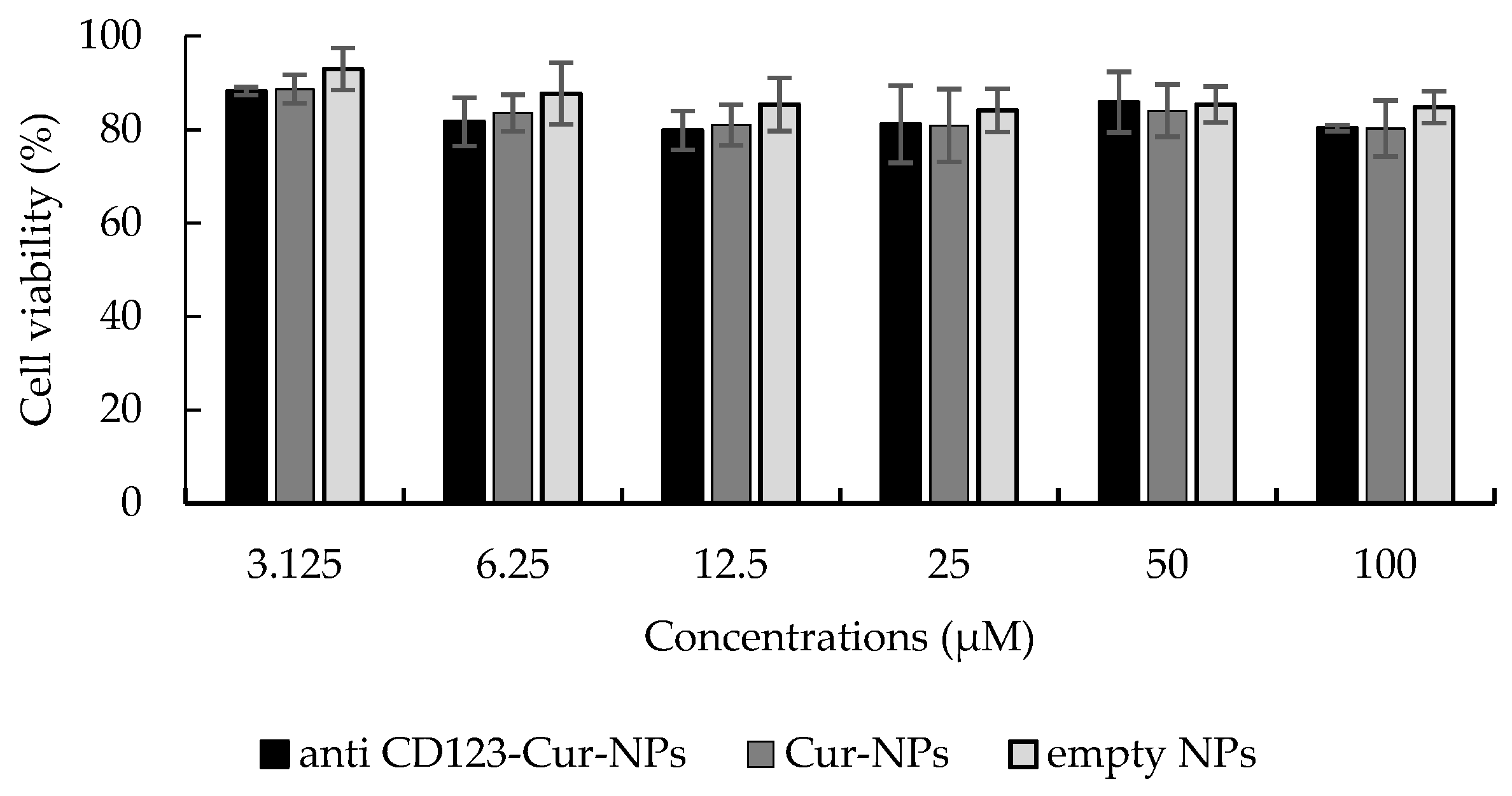
3.4. Cytotoxicity Assay
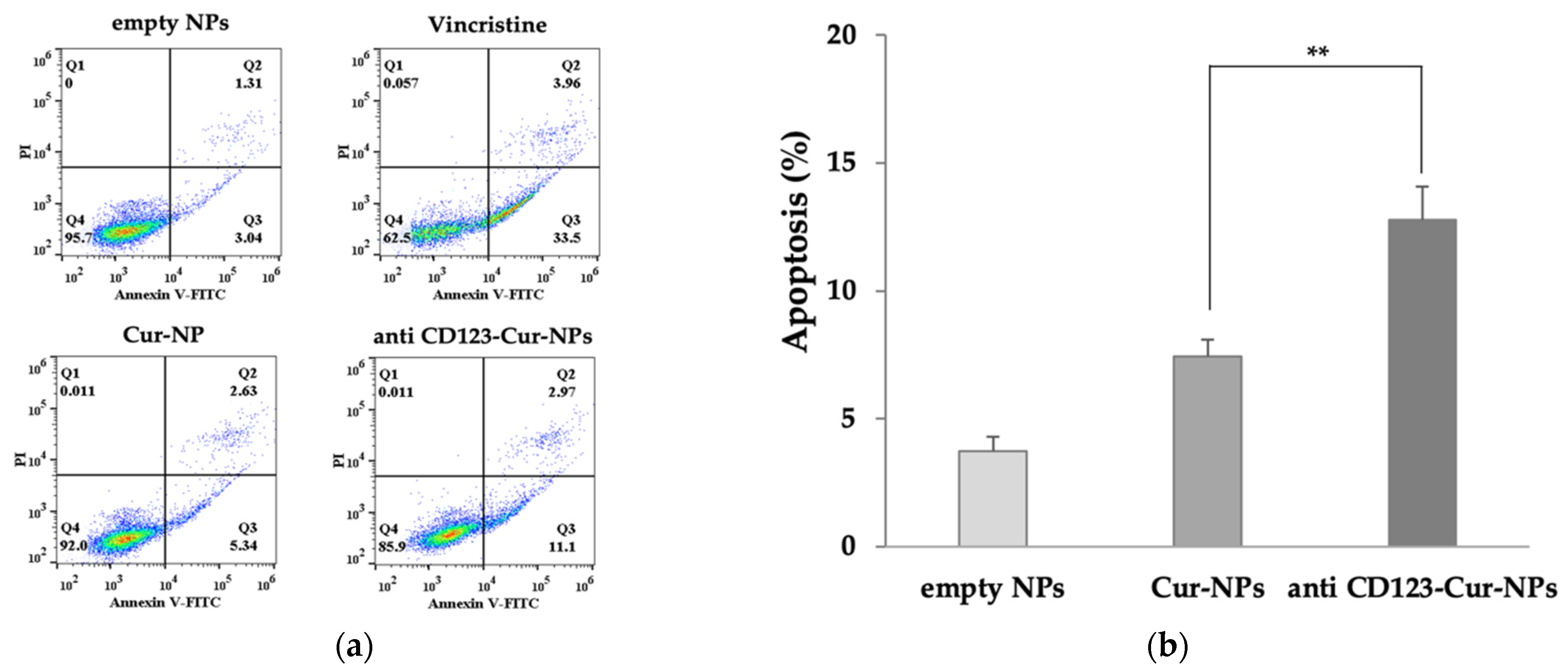
3.5. Anti-CD123-Cur-NPs Induce Apoptosis of KG-1a Cell Line
4. Discussion and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ladikou, E.E.; Sivaloganathan, H.; Pepper, A.; Chevassut, T. Acute myeloid leukaemia in its niche: The bone marrow microenvironment in acute myeloid leukaemia. Curr. Oncol. Rep. 2020, 22, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, M.; Wang, F.; Loghavi, S.; Bueso-Ramos, C.; Gumbs, C.; Little, L.; Song, X.; Zhang, J.; Kadia, T.; Borthakur, G.; et al. Late relapse in acute myeloid leukemia (AML): Clonal evolution or therapy-related leukemia. Blood Cancer J. 2019, 9, 7. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the european leukemia. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef]
- Jonas, B.A. On the origin of relapse in AML. Sci. Transl. Med. 2017, 9, eaan8205. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Lanza, F.; Dabusti, M.; Tieghi, A.; Campioni, D.; Dominici, M.; Castoldi, G. CD123 (interleukin 3 receptor alpha chain). J. Biol. Regul. Homeost. Agents 2001, 15, 98–100. [Google Scholar]
- Rahmani, A.H.; Alsahli, M.A.; Aly, S.M.; Khan, M.A.; Aldebasi, Y.H. Role of curcumin in disease prevention and treatment. Adv. Biomed. Res. 2018, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Sahebkar, A.; Serban, M.C.; Ursoniu, S.; Banach, M. Effect of curcuminoids on oxidative stress: A systematic review and meta-analysis of randomized controlled trials. J. Funct. Foods 2015, 18, 898–909. [Google Scholar] [CrossRef]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. A review of curcumin and its derivatives as anticancer agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar]
- Adamczak, A.; Ożarowski, M.; Karpiński, T.M. Curcumin, a natural antimicrobial agent with strain-specific activity. Pharmaceuticals 2020, 13, 153. [Google Scholar] [CrossRef]
- Anuchapreeda, S.; Thanarattanakorn, P.; Sittipreechacharn, S.; Chanarat, P.; Limtrakul, P. Curcumin inhibits WT1 gene expression in human leukemic K562 cells. Acta Pharmacol. Sin. 2006, 27, 360–366. [Google Scholar] [CrossRef]
- Anuchapreeda, S.; Thanarattanakorn, P.; Sittipreechacharn, S.; Tima, S.; Chanarat, P.; Limtrakul, P. Inhibitory effect of curcumin on MDR1 gene expression in patient leukemic cells. Arch. Pharm. Res. 2006, 29, 866–873. [Google Scholar]
- Panyajai, P.; Tima, S.; Chiampanichayakul, S.; Anuchapreeda, S. Dietary turmeric bisdemethoxycurcumin suppresses Wilms’ tumor 1 and CD34 protein expressions in KG-1a leukemic stem cells. Nutr. Cancer 2019, 71, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.L.; Hsu, C.H.; Lin, J.K.; Hsu, M.M.; Ho, Y.F.; Shen, T.S.; Ko, J.Y.; Lin, J.T.; Lin, B.R.; Shiang, W.M.; et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001, 21, 2895–2900. [Google Scholar]
- Anand, P.; Kunnumakkara, A.; Newman, R.; Aggarwal, B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar]
- Heger, M.; van Golen, R.F.; Broekgaarden, M.; Michel, M.C. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol. Rev. 2014, 66, 222–307. [Google Scholar] [CrossRef]
- Hewlings, S.J.; Kalman, D.S. Curcumin: A Review of Its Effects on Human Health. Foods. 2017, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Khudhayer Oglah, M.; Fakri Mustafa, Y. Curcumin analogs: Synthesis and biological activities. Med. Chem. Res. 2020, 29, 479–486. [Google Scholar] [CrossRef]
- Youssef, K.M.; El-Sherbeny, M.A.; El-Shafie, F.S.; Farag, H.A.; Al-Deeb, O.A.; Awadalla, S.A.A. Synthesis of curcumin analogues as potential antioxidant, cancer chemopreventive agents. Arch. Pharm. 2004, 337, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zheng, Y.; Liu, X.; Fang, W.; Chen, X.; Liao, W.; Jing, X.; Lei, M.; Tao, E.; Ma, Q.; et al. Curcumin-loaded PLGA-PEG nanoparticles conjugated with B6 peptide for potential use in Alzheimer’s disease. Drug Deliv. 2018, 25, 1091–1102. [Google Scholar]
- Orunoğlu, M.; Kaffashi, A.; Pehlivan, S.B.; Şahin, S.; Söylemezoğlu, F.; Oğuz, K.K.; Mut, M. Effects of curcumin-loaded PLGA nanoparticles on the RG2 rat glioma model. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 78, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Mirakabad, F.S.T.; Nejati-Koshki, K.; Akbarzadeh, A.; Yamchi, M.R.; Milani, M.; Zarghami, N. PLGA-Based Nanoparticles as Cancer Drug Delivery Systems. Asian Pac. J. Cancer Prev. 2014, 15, 517–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukerjee, A.; Vishwanatha, J.K. Formulation, characterization and evaluation of curcumin-loaded PLGA nanosphere for cancer therapy. Anticancer Res. 2009, 29, 3867–3875. [Google Scholar] [PubMed]
- Guerrouache, M.; Carole, K.; Gaillet, C.; Canva, M.; Millot, M. Immobilization of a functionalized poly(ethylene glycol) onto β-cyclodextrin-coated surfaces by formation of inclusion complexes: Application to the coupling of proteins. J. Appl. Polym. Sci. 2006, 100, 2362–2370. [Google Scholar] [CrossRef]
- Chittasupho, C.; Xie, S.-X.; Baoum, A.; Yakovleva, T.; Siahaan, T.J.; Berkland, C.J. ICAM-1 targeting of doxorubicin-loaded PLGA nanoparticles to lung epithelial cells. Eur. J. Pharm. Sci. 2009, 37, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Cirstoiu-Hapca, A.; Bossy-Nobs, L.; Buchegger, F.; Gurny, R.; Delie, F. Differential tumor cell targeting of anti-HER2 (Herceptin) and anti-CD20 (Mabthera) coupled nanoparticles. Int. J. Pharm. 2007, 331, 190–196. [Google Scholar]
- Punfa, W.; Suzuki, S.; Pitchakarn, P.; Yodkeeree, S.; Naiki, T.; Takahashi, S.; Limtrakul, P. Curcumin-loaded PLGA nanoparticles conjugated with anti-P-glycoprotein antibody to overcome multidrug resistance. Asian Pac. J. Cancer Prev. 2014, 15, 9249–9258. [Google Scholar] [CrossRef] [Green Version]
- Shaikh, J.; Ankola, D.D.; Beniwal, V.; Singh, D.; Kumar, M.N.V.R. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur. J. Pharm. Sci. 2009, 37, 223–230. [Google Scholar] [CrossRef]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamay, Y.; Golan, M.; Tyomkin, D.; David, A. Assessing the therapeutic efficacy of VEGFR-1-targeted polymer drug conjugates in mouse tumor models. J. Control. Release 2016, 229, 192–199. [Google Scholar]
- Zhou, J. Identification and targeting leukemia stem cells: The path to the cure for acute myeloid leukemia. World J. Stem Cells 2014, 6, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, R. Targeting leukemia stem cell resistance in chronic myelogenous leukemia. Trans. Am. Clin. Climatol. Assoc. 2019, 130, 246–254. [Google Scholar]
- Van Gils, N.; Denkers, F.; Smit, L. Escape from treatment; the different faces of leukemic stem cells and therapy resistance in acute myeloid leukemia. Front. Oncol. 2021, 11, 1454. [Google Scholar] [CrossRef]
- Felipe Rico, J.; Hassane, D.C.; Guzman, M.L. Acute myelogenous leukemia stem cells: From bench to bedside. Cancer Lett. 2013, 338, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Hope, K.; Jin, L.; Dick, J. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 2004, 5, 738–743. [Google Scholar] [CrossRef]
- Horton, S.J.; Huntly, B.J.P. Recent advances in acute myeloid leukemia stem cell biology. Haematologica 2012, 97, 966–974. [Google Scholar] [PubMed] [Green Version]
- Kumari, M.; Sharma, N.; Manchanda, R.; Gupta, N.; Syed, A.; Bahkali, A.H.; Nimesh, S. PGMD/curcumin nanoparticles for the treatment of breast cancer. Sci. Rep. 2021, 11, 3824. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Tao, Q.; Zou, Y.; Zhang, F.; Guo, M.; Wang, Y.; Wang, H.; Zhou, Q.; Yu, S. PLGA nanoparticles improve the oral bioavailability of curcumin in rats: Characterizations and mechanisms. J. Agric. Food Chem. 2011, 59, 9280–9289. [Google Scholar] [CrossRef]
- Yallapu, M.M.; Gupta, B.K.; Jaggi, M.; Chauhan, S.C. Fabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cells. J. Colloid Interface Sci. 2010, 351, 19–29. [Google Scholar]
- Tabatabaei Mirakabad, F.S.; Akbarzadeh, A.; Milani, M.; Zarghami, N.; Taheri-Anganeh, M.; Zeighamian, V.; Badrzadeh, F.; Rahmati-Yamchi, M. A Comparison between the cytotoxic effects of pure curcumin and curcumin-loaded PLGA-PEG nanoparticles on the MCF-7 human breast cancer cell line. Artif. Cells Nanomed. Biotechnol. 2016, 44, 423–430. [Google Scholar] [CrossRef]
- Chang, P.-Y.; Peng, S.-F.; Lee, C.-Y.; Lu, C.-C.; Tsai, S.-C.; Shieh, T.-M.; Wu, T.-S.; Tu, M.-G.; Chen, M.Y.; Yang, J.-S. Curcumin-loaded nanoparticles induce apoptotic cell death through regulation of the function of MDR1 and reactive oxygen species in cisplatin-resistant CAR human oral cancer cells. Int. J. Oncol. 2013, 43, 1141–1150. [Google Scholar] [CrossRef] [Green Version]
- Azandeh, S.S.; Abbaspour, M.; Khodadadi, A.; Khorsandi, L.; Orazizadeh, M.; Heidari-Moghadam, A. Anticancer activity of curcumin-loaded PLGA nanoparticles on PC3 prostate cancer cells. Iran. J. Pharm. Res. IJPR 2017, 16, 868–879. [Google Scholar] [PubMed]
- Punfa, W.; Yodkeeree, S.; Pitchakarn, P.; Ampasavate, C.; Limtrakul, P. Enhancement of cellular uptake and cytotoxicity of curcumin-loaded PLGA nanoparticles by conjugation with anti-P-glycoprotein in drug resistance cancer cells. Acta Pharmacol. Sin. 2012, 33, 823–831. [Google Scholar] [CrossRef]
- Khalil, N.M.; do Nascimento, T.C.F.; Casa, D.M.; Dalmolin, L.F.; de Mattos, A.C.; Hoss, I.; Romano, M.A.; Mainardes, R.M. Pharmacokinetics of curcumin-loaded PLGA and PLGA–PEG blend nanoparticles after oral administration in rats. Colloids Surf. B Biointerfaces 2013, 101, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Mezghrani, O.; Zhang, L.; Chen, Y.; Ke, X.; Ci, T. GE11 peptide modified and reduction-responsive hyaluronic acid-based nanoparticles induced higher efficacy of doxorubicin for breast carcinoma therapy. Int. J. Nanomed. 2016, 11, 5125–5147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, C.T.; Upchurch, D.; Szilvassy, S.J.; Guzman, M.L.; Howard, D.S.; Pettigrew, A.L.; Meyerrose, T.; Rossi, R.; Grimes, B.; Rizzieri, D.A.; et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 2000, 14, 1777–1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busfield, S.J.; Biondo, M.; Wong, M.; Ramshaw, H.; Lee, E.M.; Ghosh, S.; Braley, H.; Panousis, C.; Roberts, A.; He, S.Z.; et al. Targeting of acute myeloid leukemia in vitro and in vivo with an anti-CD123 mAB engineered for optimal ADCC. Leukemia 2014, 28, 2213–2221. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G. CD123 as a therapeutic target in the treatment of hematological malignancies. Cancers 2019, 11, 1358. [Google Scholar]
- Haubner, S.; Perna, F.; Köhnke, T.; Schmidt, C.; Berman, S.; Augsberger, C.; Schnorfeil, F.M.; Krupka, C.; Lichtenegger, F.S.; Liu, X.; et al. Coexpression profile of leukemic stem cell markers for combinatorial targeted therapy in AML. Leukemia 2019, 33, 64–74. [Google Scholar]
- Lei, T.; Srinivasan, S.; Tang, Y.; Manchanda, R.; Nagesetti, A.; Fernandez, A.; McGoron, A.J. Comparing cellular uptake and cytotoxicity of targeted drug carriers in cancer cell lines with different drug resistance mechanisms. Nanomedicine 2011, 7, 324–332. [Google Scholar] [CrossRef] [Green Version]
- Attia, M.; Anton, N.; Wallyn, J.; Omran, Z.; Vandamme, T. An overview of active and passive targeting strategies to improve the nanocarriers efficiency to tumour sites. J. Pharm. Pharm. 2019, 71, 1185–1198. [Google Scholar] [CrossRef] [Green Version]
- Duvoix, A.; Morceau, F.; Schnekenburger, M.; Delhalle, S.; Galteau, M.M.; Dicato, M.; Diederich, M. Curcumin-induced cell death in two leukemia cell lines: K562 and Jurkat. Ann. N. Y. Acad. Sci. 2003, 1010, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Ghosh, U.; Bhattacharyya, N.P.; Bhattacharya, R.K.; Roy, M. Inhibition of telomerase activity and induction of apoptosis by curcumin in K-562 cells. Mutat. Res. 2006, 596, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.L.; Li, J.; Qin, Z.H.; Liang, Z.Q. Autophagic and apoptotic mechanisms of curcumin-induced death in K562 cells. J. Asian Nat. Prod. Res. 2009, 11, 918–928. [Google Scholar] [PubMed]

| Nanoparticles | Size (nm) | PdI | ZP (mV) | EE (%) | LC (%w/w) |
|---|---|---|---|---|---|
| Empty NPs | 214.37 ± 3.23 | 0.09 ± 0.05 | −30.32 ± 3.10 | - | - |
| Cur-NPs | 183.78 ± 4.38 | 0.06 ± 0.03 | −32.87 ± 1.63 | 88.64 | 0.42 |
| Anti-CD123-Cur-NPs | 181.27 ± 0.07 | 0.07 ± 0.03 | −46.21 ± 0.07 | 77.56 | 0.25 |
| Sample | IC50 (μM) |
|---|---|
| Anti-CD123-Cur-NPs | 41.45 ± 5.49 ** |
| Cur-NPs | 74.20 ± 6.71 |
| Empty NPs | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nirachonkul, W.; Ogonoki, S.; Thumvijit, T.; Chiampanichayakul, S.; Panyajai, P.; Anuchapreeda, S.; Tima, S.; Chiampanichayakul, S. CD123-Targeted Nano-Curcumin Molecule Enhances Cytotoxic Efficacy in Leukemic Stem Cells. Nanomaterials 2021, 11, 2974. https://doi.org/10.3390/nano11112974
Nirachonkul W, Ogonoki S, Thumvijit T, Chiampanichayakul S, Panyajai P, Anuchapreeda S, Tima S, Chiampanichayakul S. CD123-Targeted Nano-Curcumin Molecule Enhances Cytotoxic Efficacy in Leukemic Stem Cells. Nanomaterials. 2021; 11(11):2974. https://doi.org/10.3390/nano11112974
Chicago/Turabian StyleNirachonkul, Wariya, Siriporn Ogonoki, Tarika Thumvijit, Supanimit Chiampanichayakul, Pawaret Panyajai, Songyot Anuchapreeda, Singkome Tima, and Sawitree Chiampanichayakul. 2021. "CD123-Targeted Nano-Curcumin Molecule Enhances Cytotoxic Efficacy in Leukemic Stem Cells" Nanomaterials 11, no. 11: 2974. https://doi.org/10.3390/nano11112974
APA StyleNirachonkul, W., Ogonoki, S., Thumvijit, T., Chiampanichayakul, S., Panyajai, P., Anuchapreeda, S., Tima, S., & Chiampanichayakul, S. (2021). CD123-Targeted Nano-Curcumin Molecule Enhances Cytotoxic Efficacy in Leukemic Stem Cells. Nanomaterials, 11(11), 2974. https://doi.org/10.3390/nano11112974






