AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Independently of PGRMC1 and Related MAPR Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell lines and Cell Culture
2.2. Chemical Compounds
2.3. Cell Viability Assay
2.4. Inhibition Techniques (siRNA Transfection or AG-205 Addition)
2.5. RNA Extraction
2.6. Quantitative Real-Time PCR
2.7. RNA Sequencing
2.8. Bioinformatics Analysis Workflow
2.9. Immunolabeling
2.10. Cell Fractionation and Western Blot Analysis
2.11. Statistical Analysis
3. Results
3.1. Setup of AG-205 Use in Endometrial Cell Culture
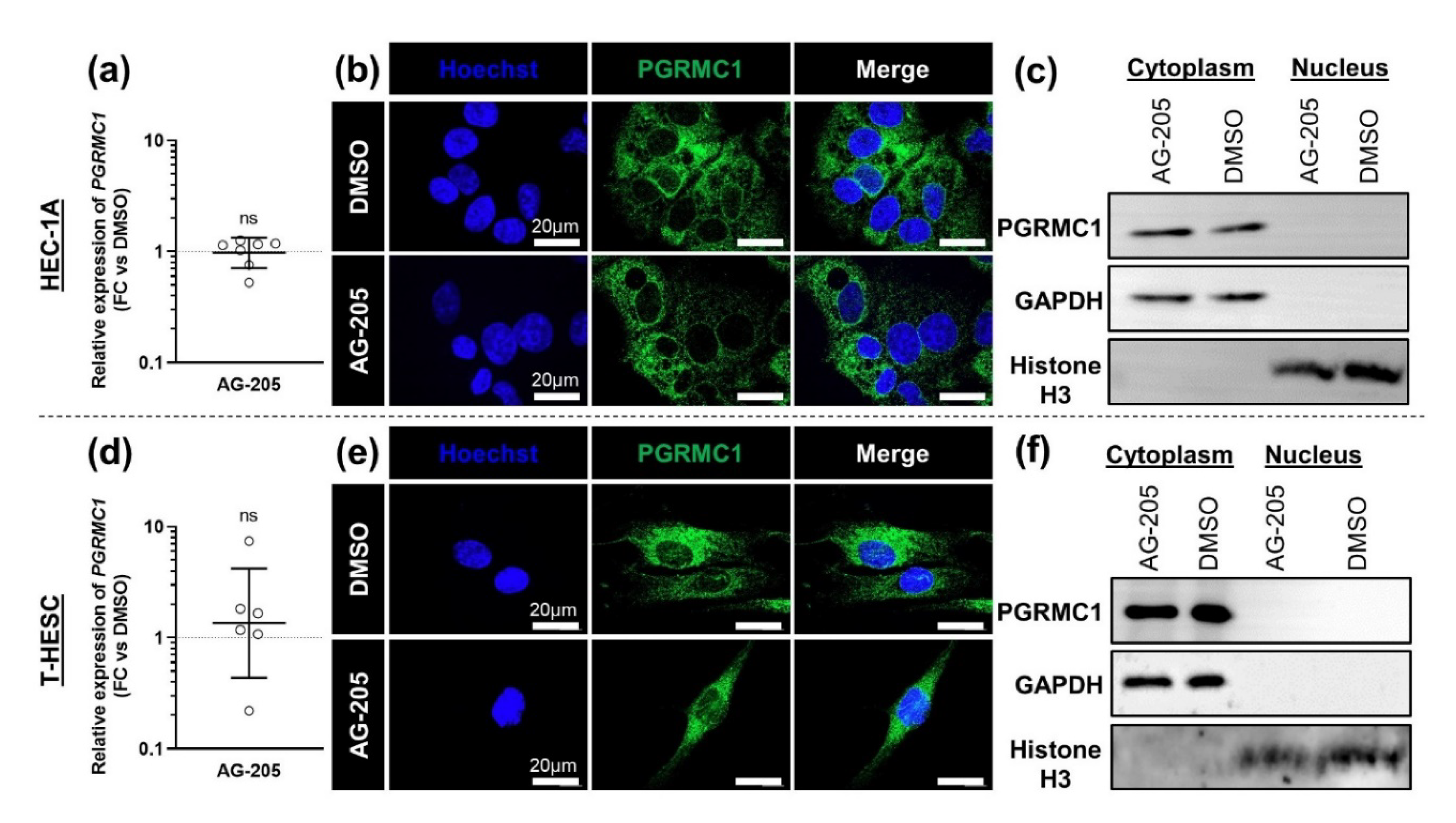
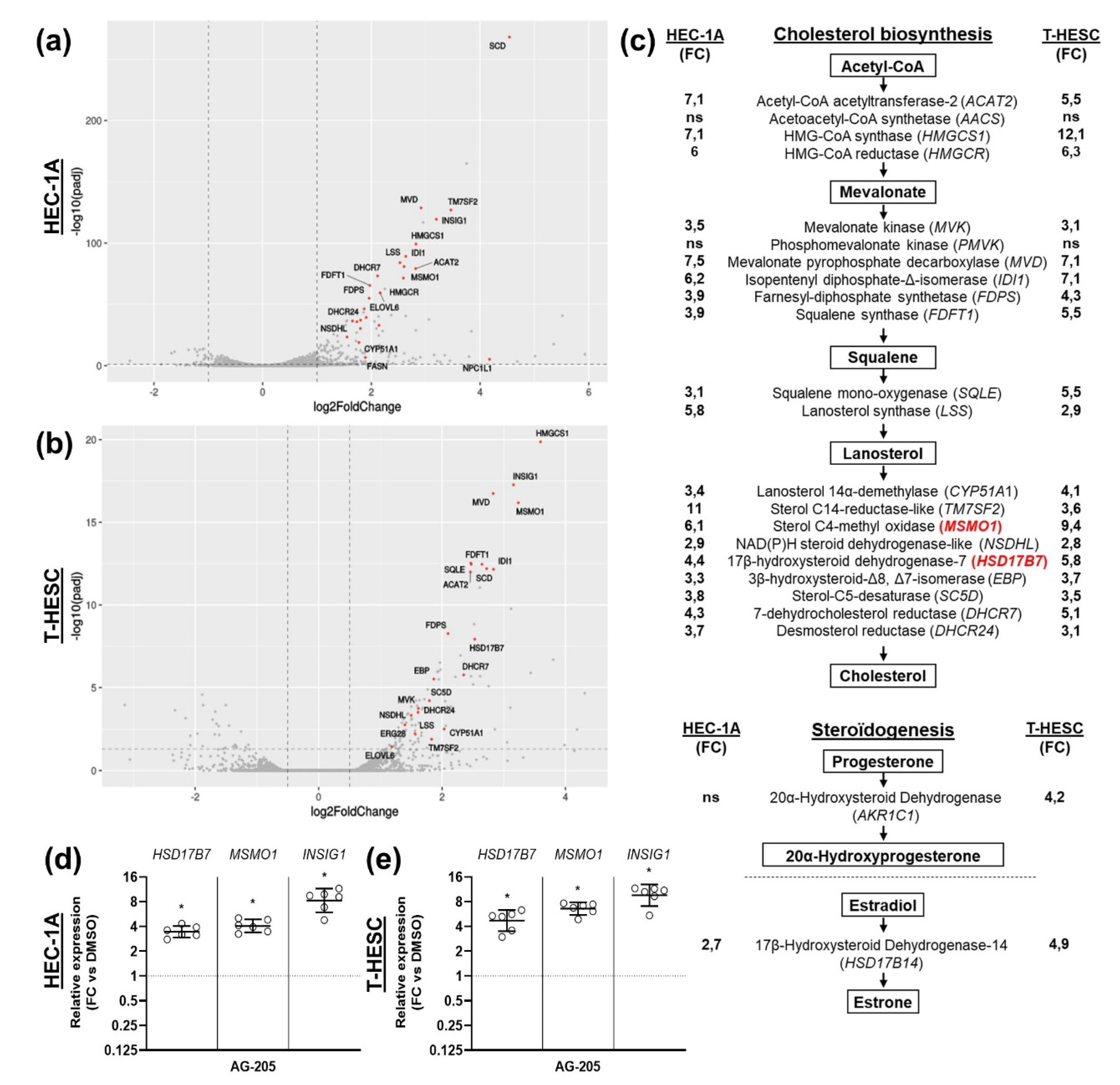
3.2. Effects of AG-205 in Endometrial Cell Culture
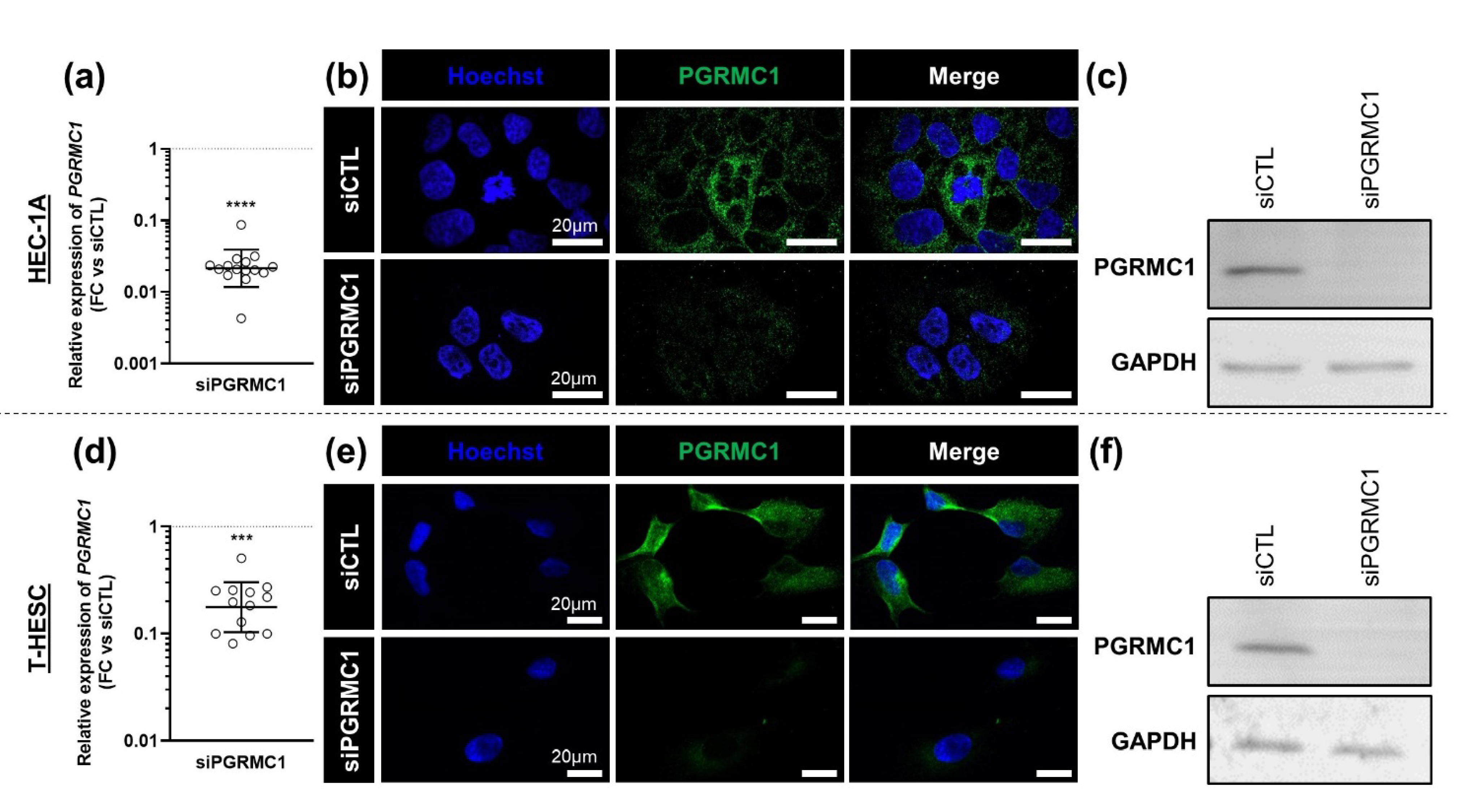
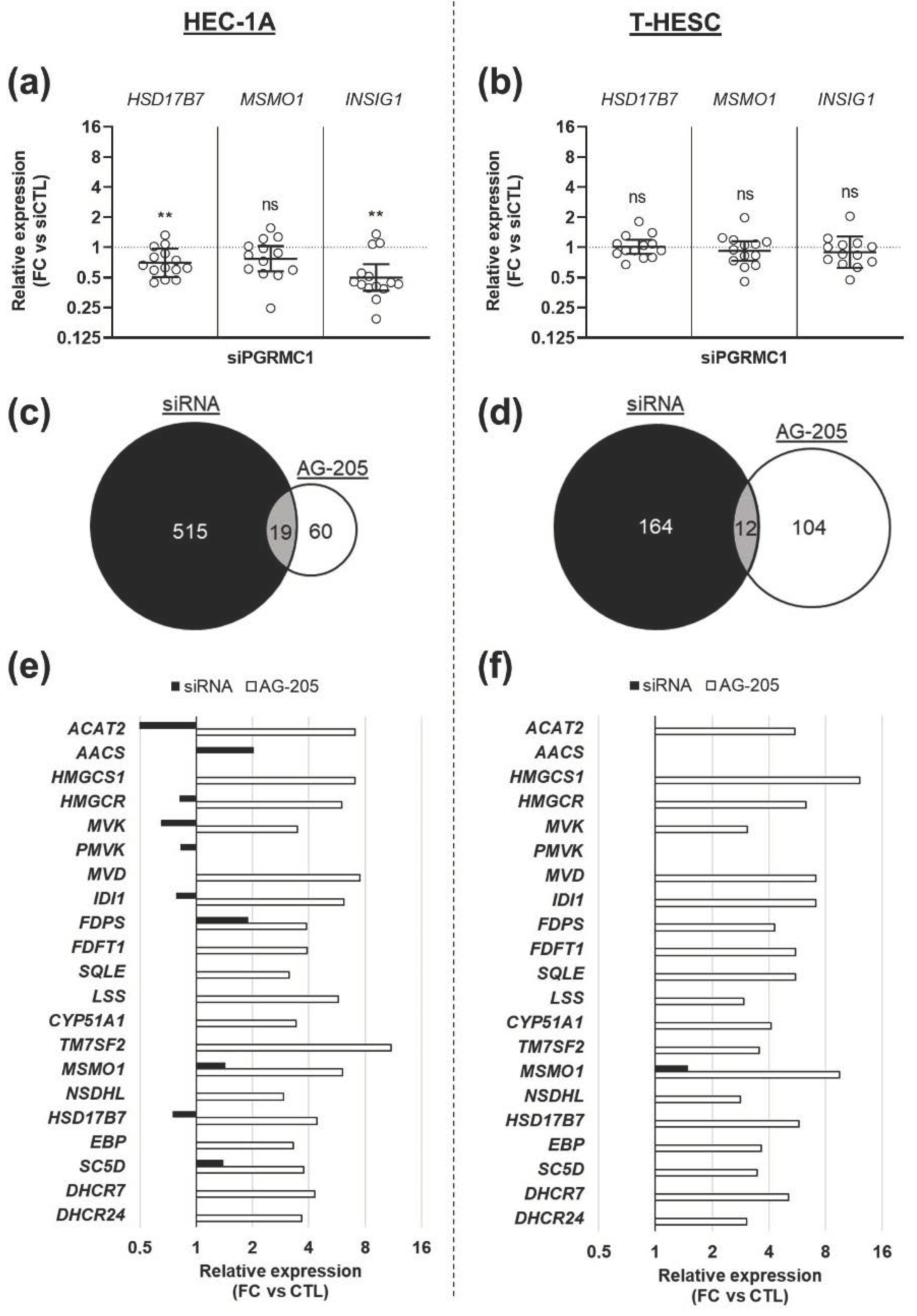
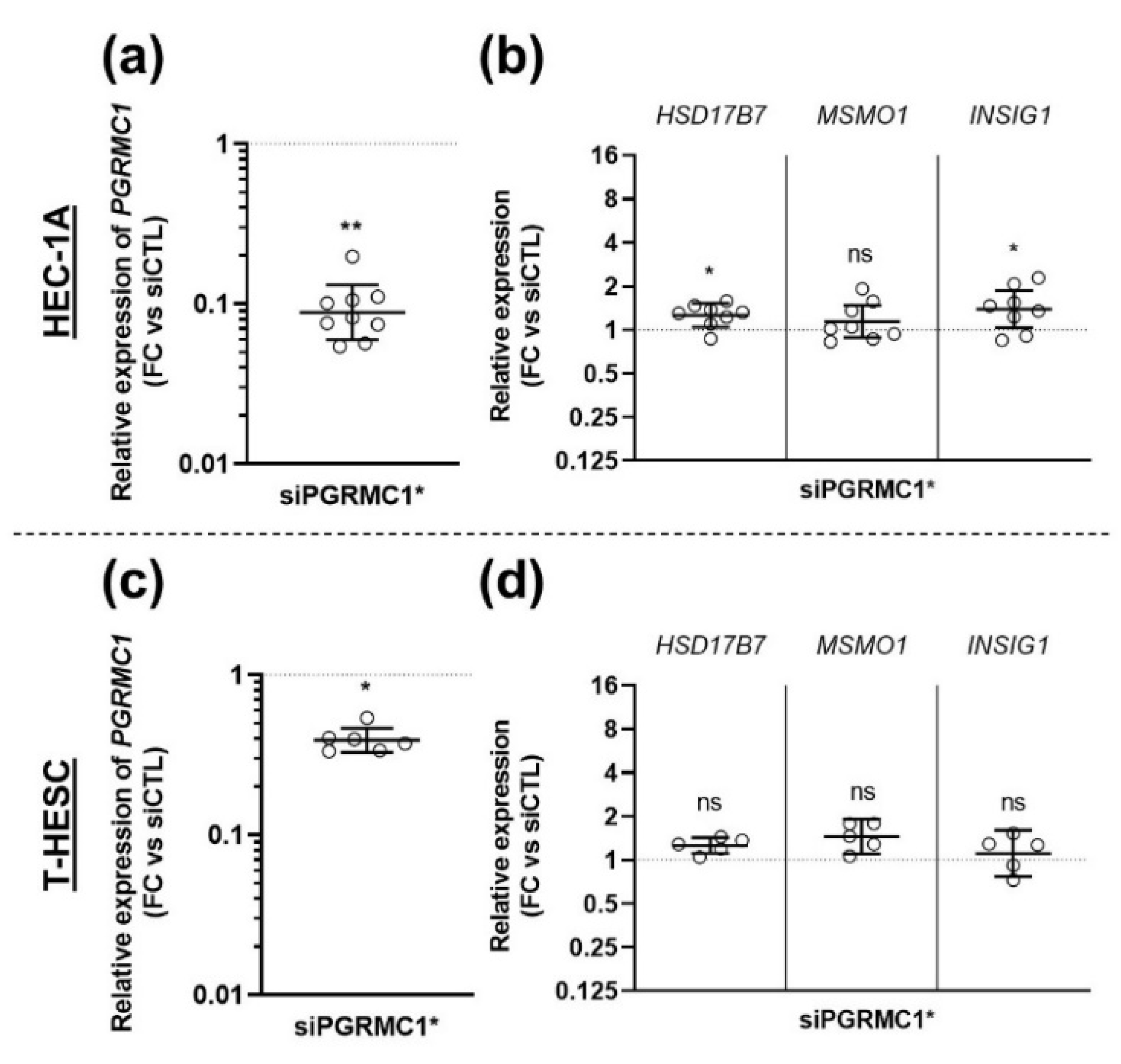
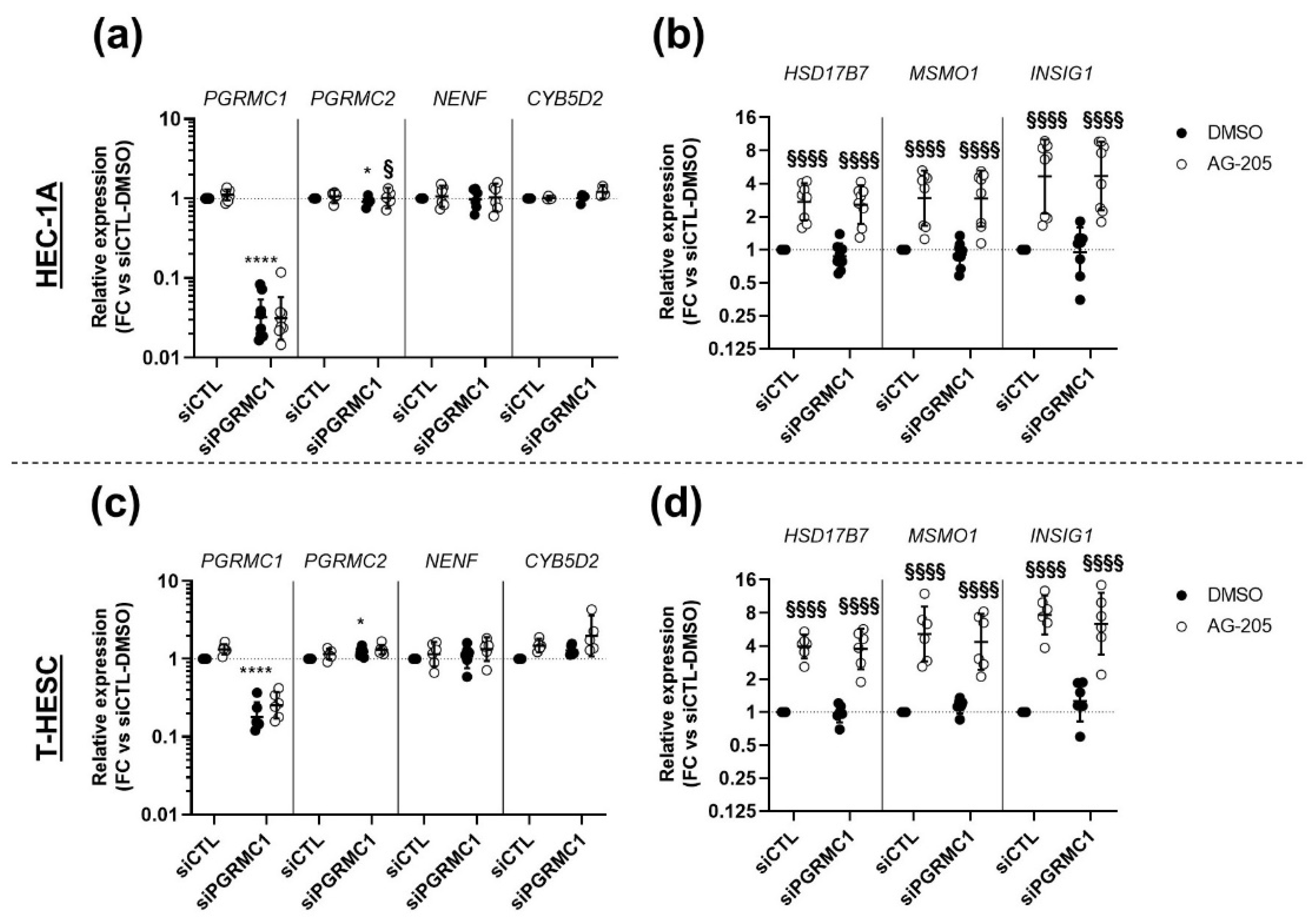
3.3. Effects of AG-205 Are Not Mimicked by Downregulation of PGRMC1 Expression
3.4. Effects of AG-205 Are Independent of PGRMC1
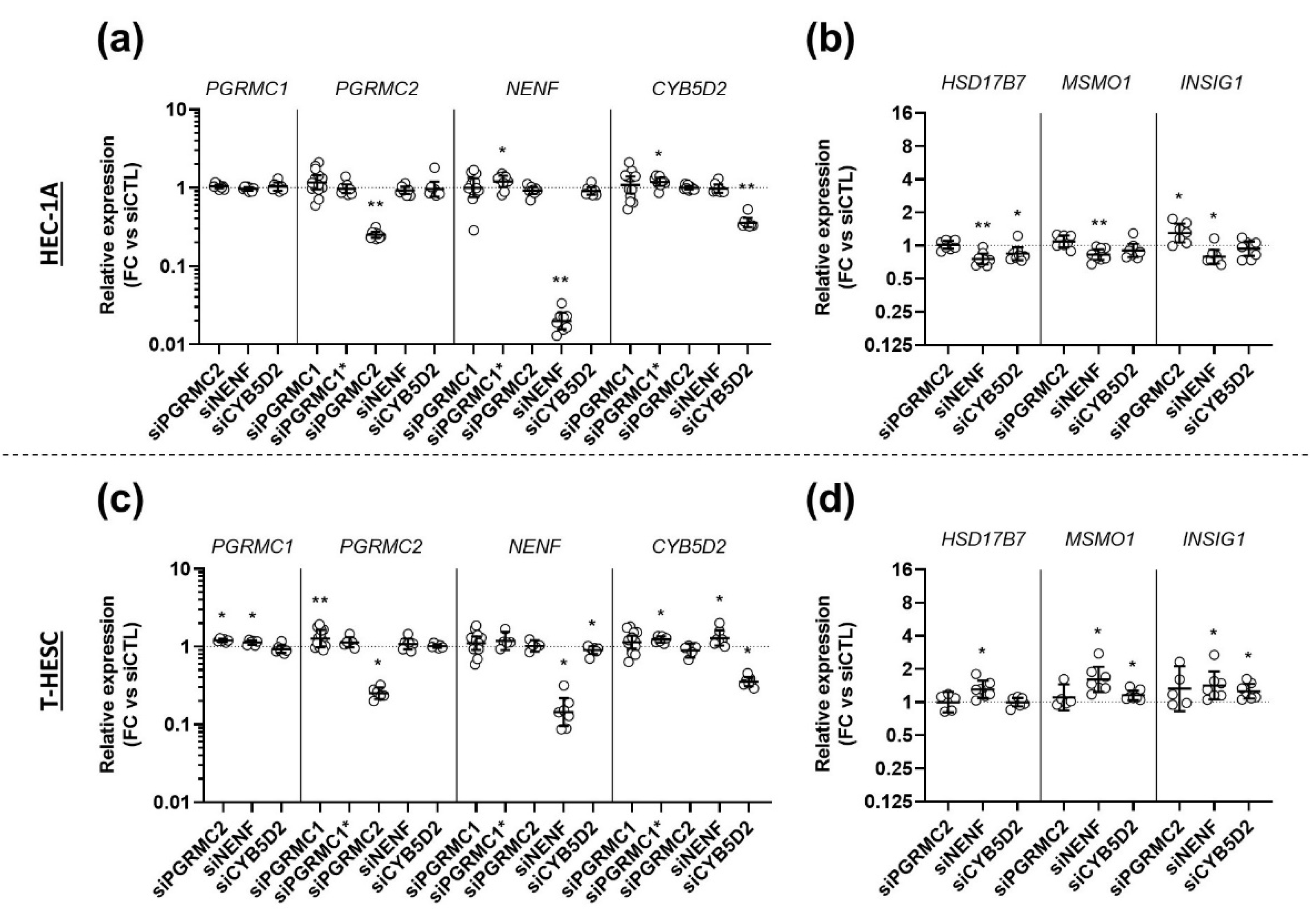
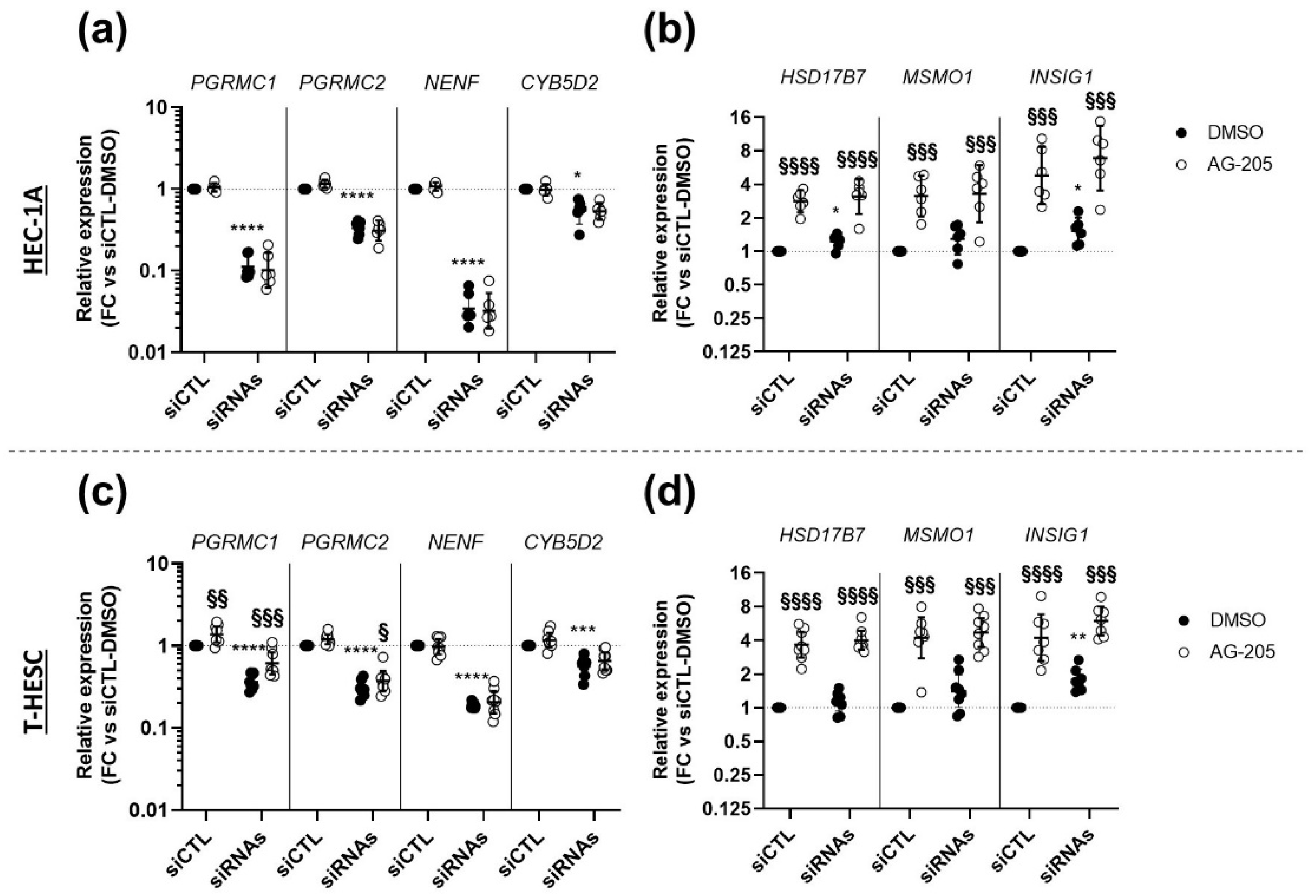
3.5. Effects of AG-205 Are Independent of All Four MAPRs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kimura, I.; Nakayama, Y.; Konishi, M.; Terasawa, K.; Ohta, M.; Itoh, N.; Fujimoto, M. Functions of MAPR (membrane-associated progesterone receptor) family members as heme/steroid-binding proteins. Curr. Protein Pept. Sci. 2012, 13, 687–696. [Google Scholar] [CrossRef]
- Cahill, M.A.; Jazayeri, J.A.; Catalano, S.M.; Toyokuni, S.; Kovacevic, Z.; Richardson, D.R. The emerging role of progesterone receptor membrane component 1 (PGRMC1) in cancer biology. Biochim. Biophys. Acta 2016, 1866, 339–349. [Google Scholar] [CrossRef]
- Wu, X.J.; Zhu, Y. Downregulation of nuclear progestin receptor (Pgr) and subfertility in double knockouts of progestin receptor membrane component 1 (pgrmc1) and pgrmc2 in zebrafish. Gen. Comp. Endocrinol. 2020, 285, 113275. [Google Scholar] [CrossRef]
- McCallum, M.L.; Pru, C.A.; Niikura, Y.; Yee, S.P.; Lydon, J.P.; Peluso, J.J.; Pru, J.K. Conditional Ablation of Progesterone Receptor Membrane Component 1 Results in Subfertility in the Female and Development of Endometrial Cysts. Endocrinology 2016, 157, 3309–3319. [Google Scholar] [CrossRef]
- Allen, T.K.; Feng, L.; Nazzal, M.; Grotegut, C.A.; Buhimschi, I.A.; Murtha, A.P. The Effect of Progestins on Tumor Necrosis Factor α-Induced Matrix Metalloproteinase-9 Activity and Gene Expression in Human Primary Amnion and Chorion Cells In Vitro. Anesth. Analg. 2015, 120, 1085–1094. [Google Scholar] [CrossRef]
- Yoshitani, N.; Satou, K.; Saito, K.; Suzuki, S.; Hatanaka, H.; Seki, M.; Shinozaki, K.; Hirota, H.; Yokoyama, S. A structure-based strategy for discovery of small ligands binding to functionally unknown proteins: Combination of in silico screening and surface plasmon resonance measurements. Proteomics 2005, 5, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Strushkevich, N.V.; Harnastai, I.N.; Iwamoto, H.; Gilep, A.A.; Takemori, H.; Usanov, S.A.; Nonaka, Y.; Hori, H.; Vinson, G.P.; et al. Molecular identification of adrenal inner zone antigen as a heme-binding protein. FEBS J. 2005, 272, 5832–5843. [Google Scholar] [CrossRef]
- Ahmed, I.S.; Rohe, H.J.; Twist, K.E.; Mattingly, M.N.; Craven, R.J. Progesterone receptor membrane component 1 (Pgrmc1): A heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J. Pharmacol. Exp. Ther. 2010, 333, 564–573. [Google Scholar] [CrossRef]
- Will, E.A.; Liu, X.; Peluso, J.J. AG 205, a progesterone receptor membrane component 1 antagonist, ablates progesterone’s ability to block oxidative stress-induced apoptosis of human granulosa/luteal cells†. Biol. Reprod. 2017, 96, 843–854. [Google Scholar] [CrossRef]
- Pedroza, D.A.; Rajamanickam, V.; Subramani, R.; Bencomo, A.; Galvez, A.; Lakshmanaswamy, R. Progesterone receptor membrane component 1 promotes the growth of breast cancers by altering the phosphoproteome and augmenting EGFR/PI3K/AKT signalling. Br. J. Cancer 2020, 123, 1326–1335. [Google Scholar] [CrossRef]
- Aizen, J.; Thomas, P. Role of Pgrmc1 in estrogen maintenance of meiotic arrest in zebrafish oocytes through Gper/Egfr. J. Endocrinol. 2015, 225, 59–68. [Google Scholar] [CrossRef]
- Terzaghi, L.; Tessaro, I.; Raucci, F.; Merico, V.; Mazzini, G.; Garagna, S.; Zuccotti, M.; Franciosi, F.; Lodde, V. PGRMC1 participates in late events of bovine granulosa cells mitosis and oocyte meiosis. Cell Cycle 2016, 15, 2019–2032. [Google Scholar] [CrossRef][Green Version]
- Craven, R.J. AG-205 for the Treatment of Breast Cancer. U.S. Patent 972,433,7B2, 8 August 2017. [Google Scholar]
- Salsano, S.; González-Martín, R.; Quiñonero, A.; Pérez-Debén, S.; Domínguez, F. Deciphering the Role of PGRMC1 during Human Decidualization Using an In Vitro Approach. J. Clin. Endocrinol. Metab. 2021, 106, 2313–2327. [Google Scholar] [CrossRef]
- Cahill, M.A.; Medlock, A.E. Thoughts on interactions between PGRMC1 and diverse attested and potential hydrophobic ligands. J. Steroid Biochem. Mol. Biol. 2017, 171, 11–33. [Google Scholar] [CrossRef]
- Wang-Eckhardt, L.; Eckhardt, M. A progesterone receptor membrane component 1 antagonist induces large vesicles independent of progesterone receptor membrane component 1 expression. Biol. Chem. 2020, 401, 1093–1099. [Google Scholar] [CrossRef]
- Kabe, Y.; Koike, I.; Yamamoto, T.; Hirai, M.; Kanai, A.; Furuhata, R.; Tsugawa, H.; Harada, E.; Sugase, K.; Hanadate, K.; et al. Glycyrrhizin Derivatives Suppress Cancer Chemoresistance by Inhibiting Progesterone Receptor Membrane Component 1. Cancers 2021, 13, 3265. [Google Scholar] [CrossRef]
- Krikun, G.; Mor, G.; Alvero, A.; Guller, S.; Schatz, F.; Sapi, E.; Rahman, M.; Caze, R.; Qumsiyeh, M.; Lockwood, C.J. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology 2004, 145, 2291–2296. [Google Scholar] [CrossRef]
- Kuramoto, H.; Tamura, S.; Notake, Y. Establishment of a cell line of human endometrial adenocarcinoma in vitro. Am. J. Obstet. Gynecol. 1972, 114, 1012–1019. [Google Scholar] [CrossRef]
- Bunch, K.; Tinnemore, D.; Huff, S.; Hoffer, Z.S.; Burney, R.O.; Stallings, J.D. Expression patterns of progesterone receptor membrane components 1 and 2 in endometria from women with and without endometriosis. Reprod. Sci. 2014, 21, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 April 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Powell, D.W.; Bard, M.; Eckstein, J.; Barbuch, R.; Link, A.J.; Espenshade, P.J. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 2007, 5, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Hand, R.A.; Jia, N.; Bard, M.; Craven, R.J. Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot. Cell 2003, 2, 306–317. [Google Scholar] [CrossRef]
- Mallory, J.C.; Crudden, G.; Johnson, B.L.; Mo, C.; Pierson, C.A.; Bard, M.; Craven, R.J. Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005, 25, 1669–1679. [Google Scholar] [CrossRef]
- Asperger, H.; Stamm, N.; Gierke, B.; Pawlak, M.; Hofmann, U.; Zanger, U.M.; Marton, A.; Katona, R.L.; Buhala, A.; Vizler, C.; et al. Progesterone receptor membrane component 1 regulates lipid homeostasis and drives oncogenic signaling resulting in breast cancer progression. Breast Cancer Res. BCR 2020, 22, 75. [Google Scholar] [CrossRef]
- Suchanek, M.; Radzikowska, A.; Thiele, C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2005, 2, 261–267. [Google Scholar] [CrossRef]
- Hampton, K.K.; Stewart, R.; Napier, D.; Claudio, P.P.; Craven, R.J. PGRMC1 Elevation in Multiple Cancers and Essential Role in Stem Cell Survival. Adv. Lung Cancer 2015, 4, 37–51. [Google Scholar] [CrossRef]
- Teakel, S.L.; Ludescher, M.; Thejer, B.M.; Poschmann, G.; Forwood, J.K.; Neubauer, H.; Cahill, M.A. Protein complexes including PGRMC1 and actin-associated proteins are disrupted by AG-205. Biochem. Biophys. Res. Commun. 2020, 524, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Kabe, Y.; Nakane, T.; Koike, I.; Yamamoto, T.; Sugiura, Y.; Harada, E.; Sugase, K.; Shimamura, T.; Ohmura, M.; Muraoka, K.; et al. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 2016, 7, 11030. [Google Scholar] [CrossRef] [PubMed]
- Pedroza, D.A.; Subramani, R.; Tiula, K.; Do, A.; Rashiraj, N.; Galvez, A.; Chatterjee, A.; Bencomo, A.; Rivera, S.; Lakshmanaswamy, R. Crosstalk between progesterone receptor membrane component 1 and estrogen receptor α promotes breast cancer cell proliferation. Lab. Investig. J. Tech. Methods Pathol. 2021, 101, 733–744. [Google Scholar] [CrossRef]
- Bashour, N.M.; Wray, S. Progesterone directly and rapidly inhibits GnRH neuronal activity via progesterone receptor membrane component 1. Endocrinology 2012, 153, 4457–4469. [Google Scholar] [CrossRef][Green Version]
- Gibson, D.A.; Simitsidellis, I.; Collins, F.; Saunders, P.T.K. Endometrial Intracrinology: Oestrogens, Androgens and Endometrial Disorders. Int. J. Mol. Sci. 2018, 19, 3276. [Google Scholar] [CrossRef] [PubMed]
- Reis, F.M.; Coutinho, L.M.; Vannuccini, S.; Batteux, F.; Chapron, C.; Petraglia, F. Progesterone receptor ligands for the treatment of endometriosis: The mechanisms behind therapeutic success and failure. Hum. Reprod. Update 2020, 26, 565–585. [Google Scholar] [CrossRef]
- Donnez, J.; Dolmans, M.-M. Endometriosis and Medical Therapy: From Progestogens to Progesterone Resistance to GnRH Antagonists: A Review. J. Clin. Med. 2021, 10, 1085. [Google Scholar] [CrossRef]
- McKinnon, B.; Mueller, M.; Montgomery, G. Progesterone Resistance in Endometriosis: An Acquired Property? Trends Endocrinol. Metab. TEM 2018, 29, 535–548. [Google Scholar] [CrossRef]
- Smuc, T.; Hevir, N.; Ribic-Pucelj, M.; Husen, B.; Thole, H.; Rizner, T.L. Disturbed estrogen and progesterone action in ovarian endometriosis. Mol. Cell. Endocrinol. 2009, 301, 59–64. [Google Scholar] [CrossRef]
- Thieffry, M.V.W.C.; Aynaci, A.; Maja, M.; Dupuis, C.; Loriot, A.; Marbaix, E.; Henriet, P. Data from: AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Inde-Pendently of PGRMC1 and Related MAPR Proteins. Available online: https://figshare.com/s/256ad96e895af6eaab9c (accessed on 26 May 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thieffry, C.; Van Wynendaele, M.; Aynaci, A.; Maja, M.; Dupuis, C.; Loriot, A.; Marbaix, E.; Henriet, P. AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Independently of PGRMC1 and Related MAPR Proteins. Biomolecules 2021, 11, 1472. https://doi.org/10.3390/biom11101472
Thieffry C, Van Wynendaele M, Aynaci A, Maja M, Dupuis C, Loriot A, Marbaix E, Henriet P. AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Independently of PGRMC1 and Related MAPR Proteins. Biomolecules. 2021; 11(10):1472. https://doi.org/10.3390/biom11101472
Chicago/Turabian StyleThieffry, Charlotte, Marie Van Wynendaele, Asena Aynaci, Mauriane Maja, Caroline Dupuis, Axelle Loriot, Etienne Marbaix, and Patrick Henriet. 2021. "AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Independently of PGRMC1 and Related MAPR Proteins" Biomolecules 11, no. 10: 1472. https://doi.org/10.3390/biom11101472
APA StyleThieffry, C., Van Wynendaele, M., Aynaci, A., Maja, M., Dupuis, C., Loriot, A., Marbaix, E., & Henriet, P. (2021). AG-205 Upregulates Enzymes Involved in Cholesterol Biosynthesis and Steroidogenesis in Human Endometrial Cells Independently of PGRMC1 and Related MAPR Proteins. Biomolecules, 11(10), 1472. https://doi.org/10.3390/biom11101472





