Targeting S1PR1 May Result in Enhanced Migration of Cancer Cells in Bladder Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Specimens and Clinical Data
2.2. Gene Expression Database Collection and Analysis
2.3. Evaluation of Tumor Purity and Immune Cell Simulated Infiltration
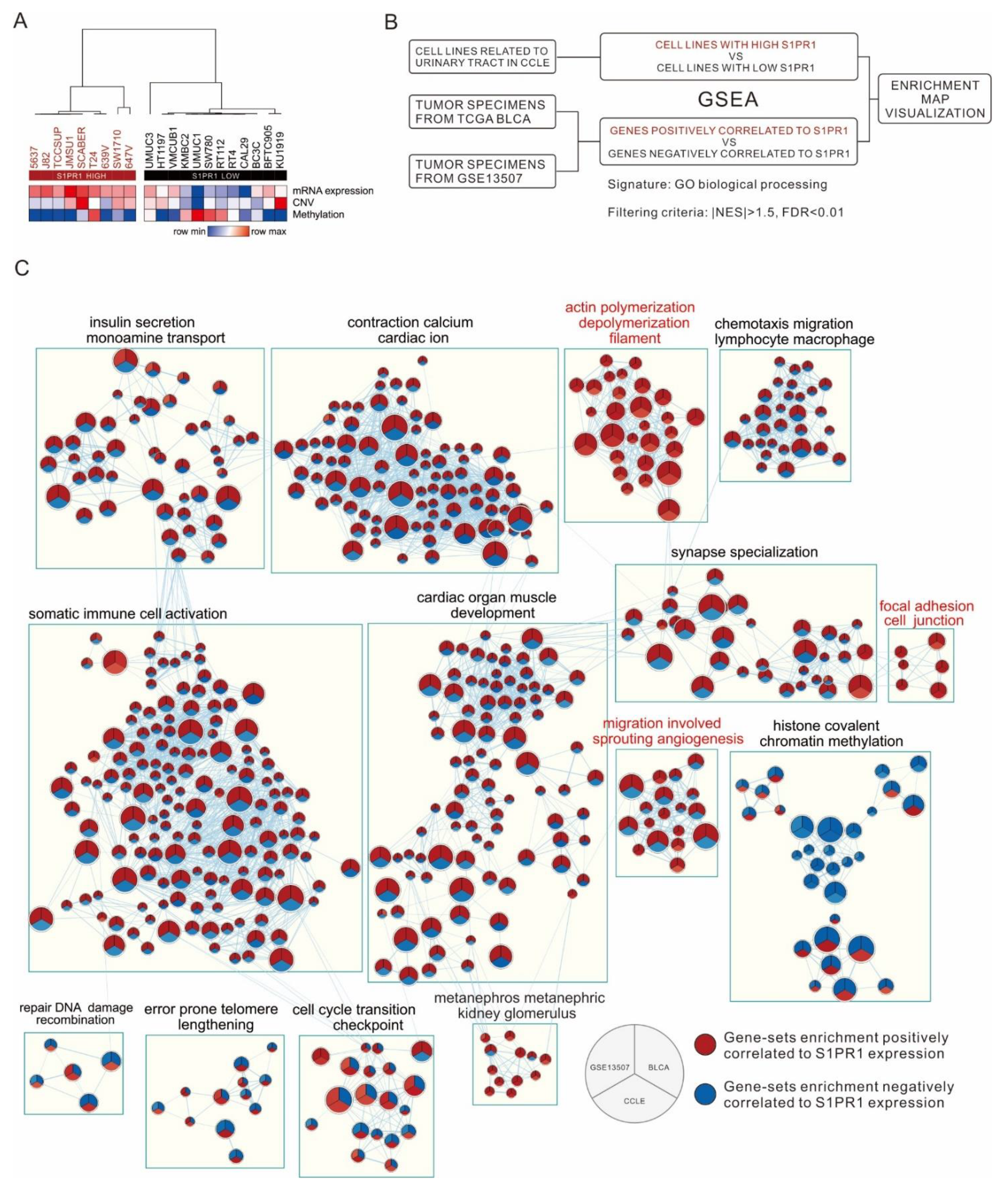
2.4. Enrichment Map Visualization
2.5. Messenger RNA Expression Analysis
2.6. Immunoblotting
2.7. Cell Culture and Establishment of Stably Expressed shRNA Cell Lines
2.8. Wound Healing Assay and LIVE Cell Imaging
2.9. Transwell Migration Assay
2.10. Patient-Derived Tumor Primary Culture (PDC)
2.11. Statistical Analysis
3. Results
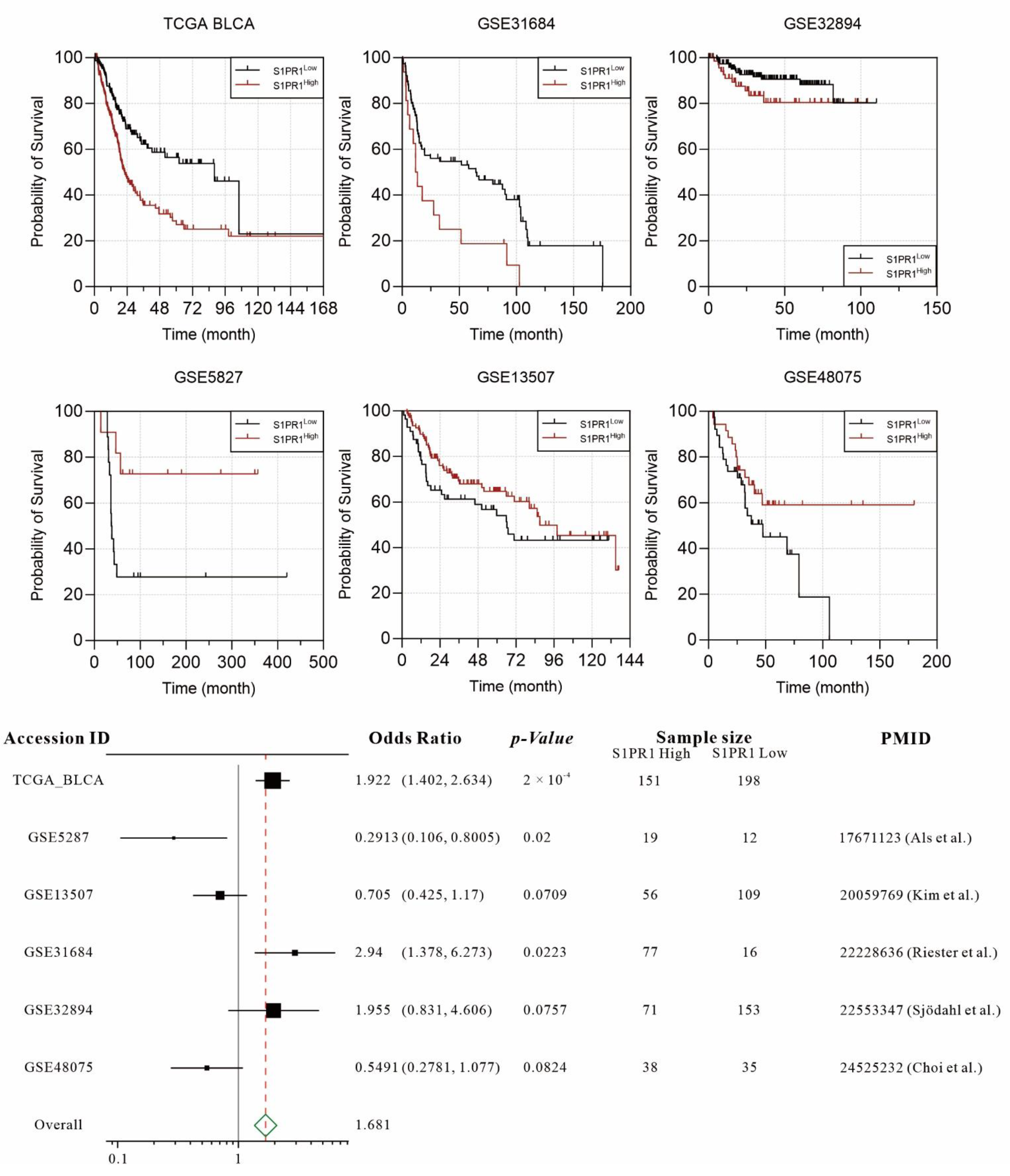
3.1. Retrospective Evaluation of the Association between S1PR1 Expression and Bladder Cancer Prognosis Shows Divergent Results in Different Databases
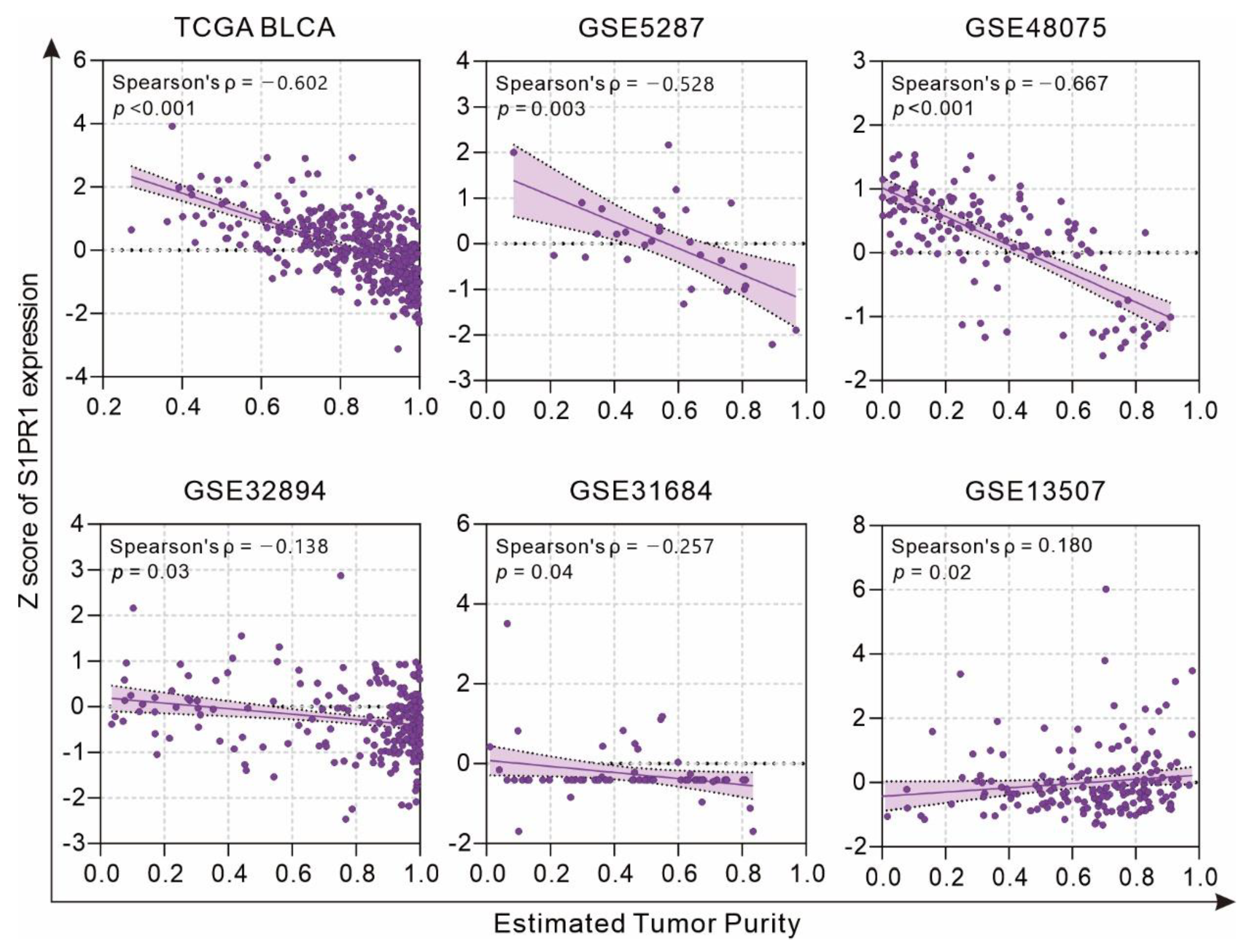
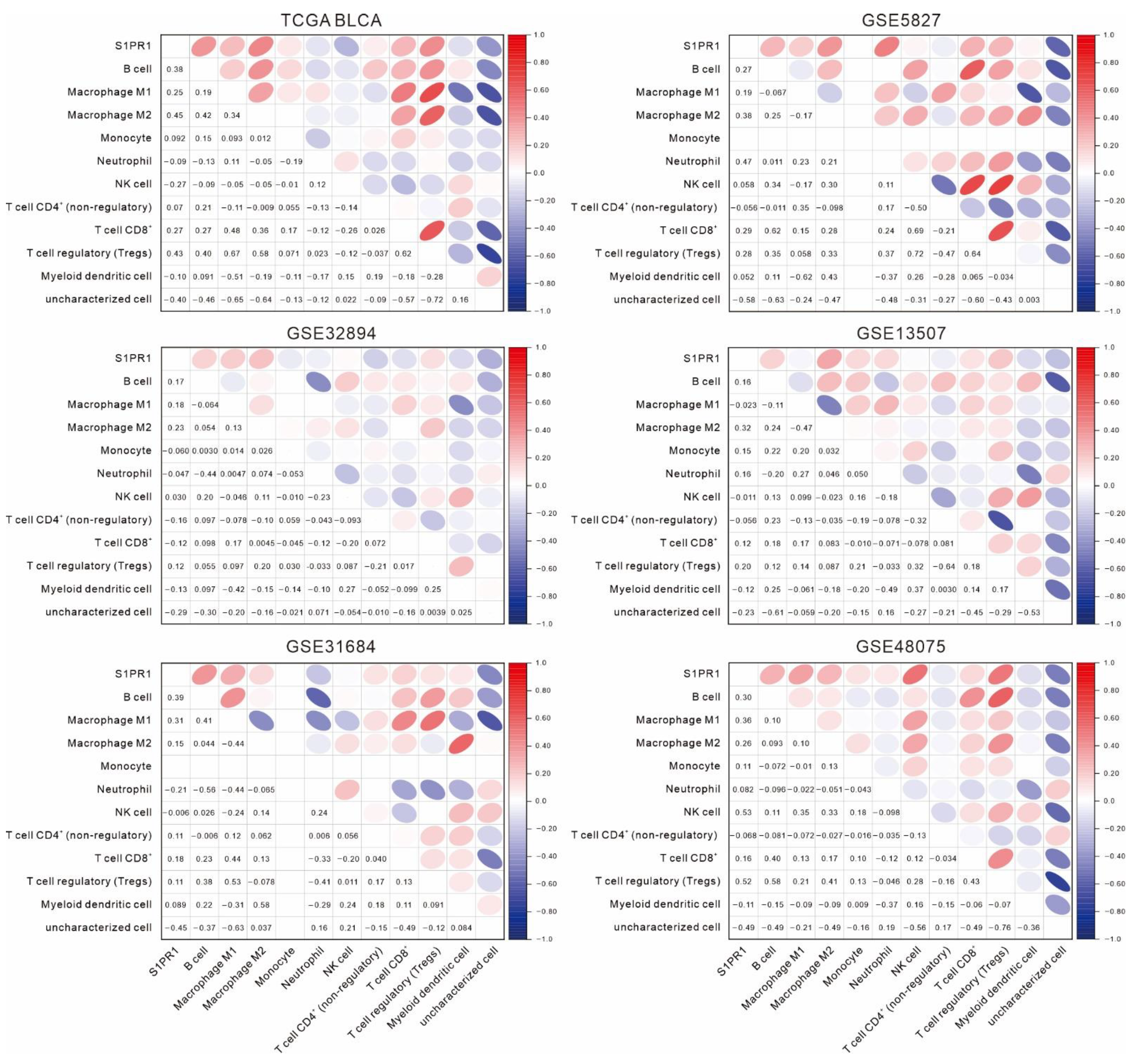
3.2. Differences in S1PR1 Expression and Prognosis of Patients with Bladder Cancer May Be Related to the Degree of Neutrophil Infiltration
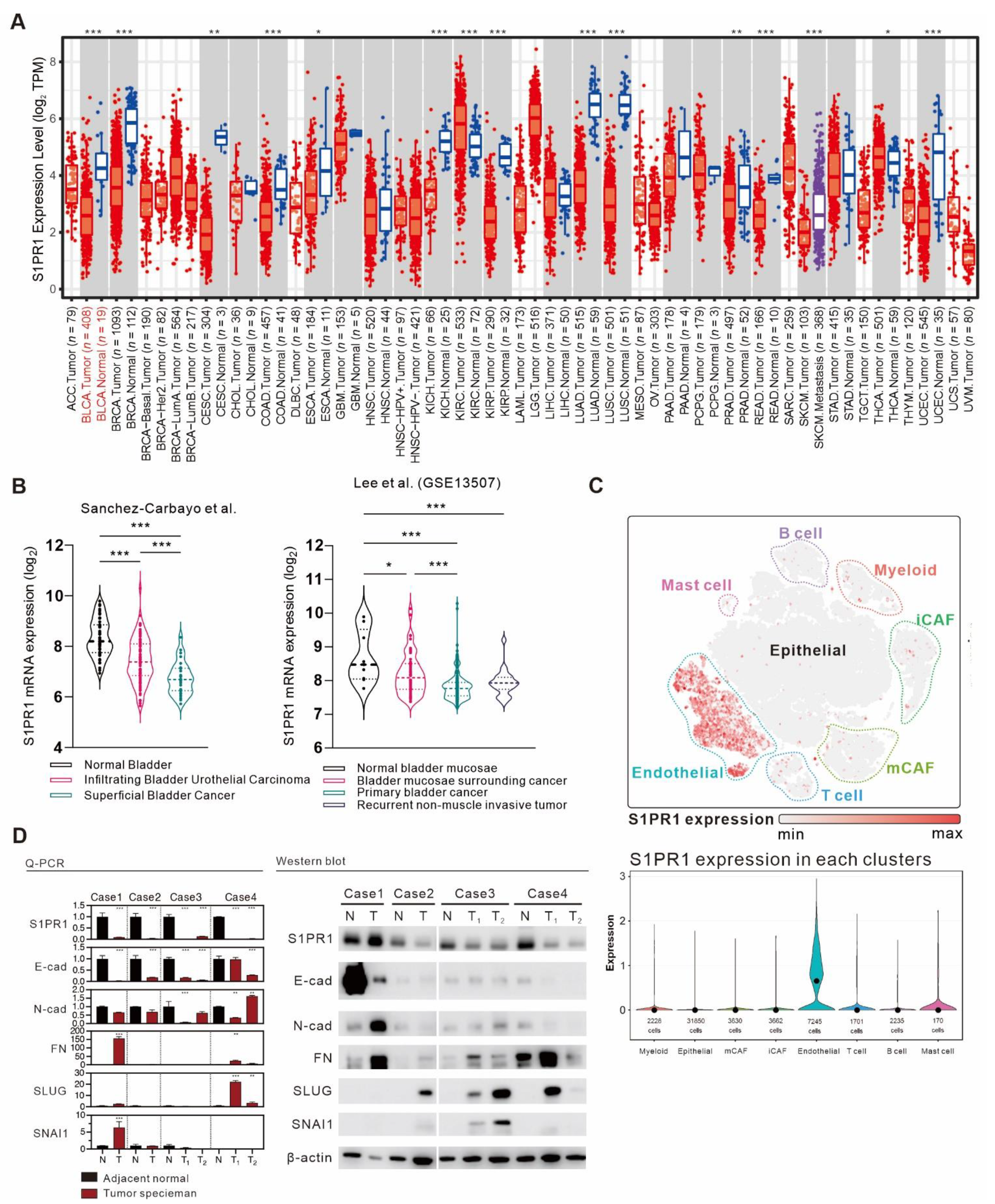
3.3. Comprehensive Assessment of S1PR1 Expression Differences between Bladder Cancer Tumors and Normal Tissue
3.4. Comparison of S1PR1 Expression in Bladder Cancer Cell Lines with Clinical Databases Reveals its Potential Function in Cell Adhesion
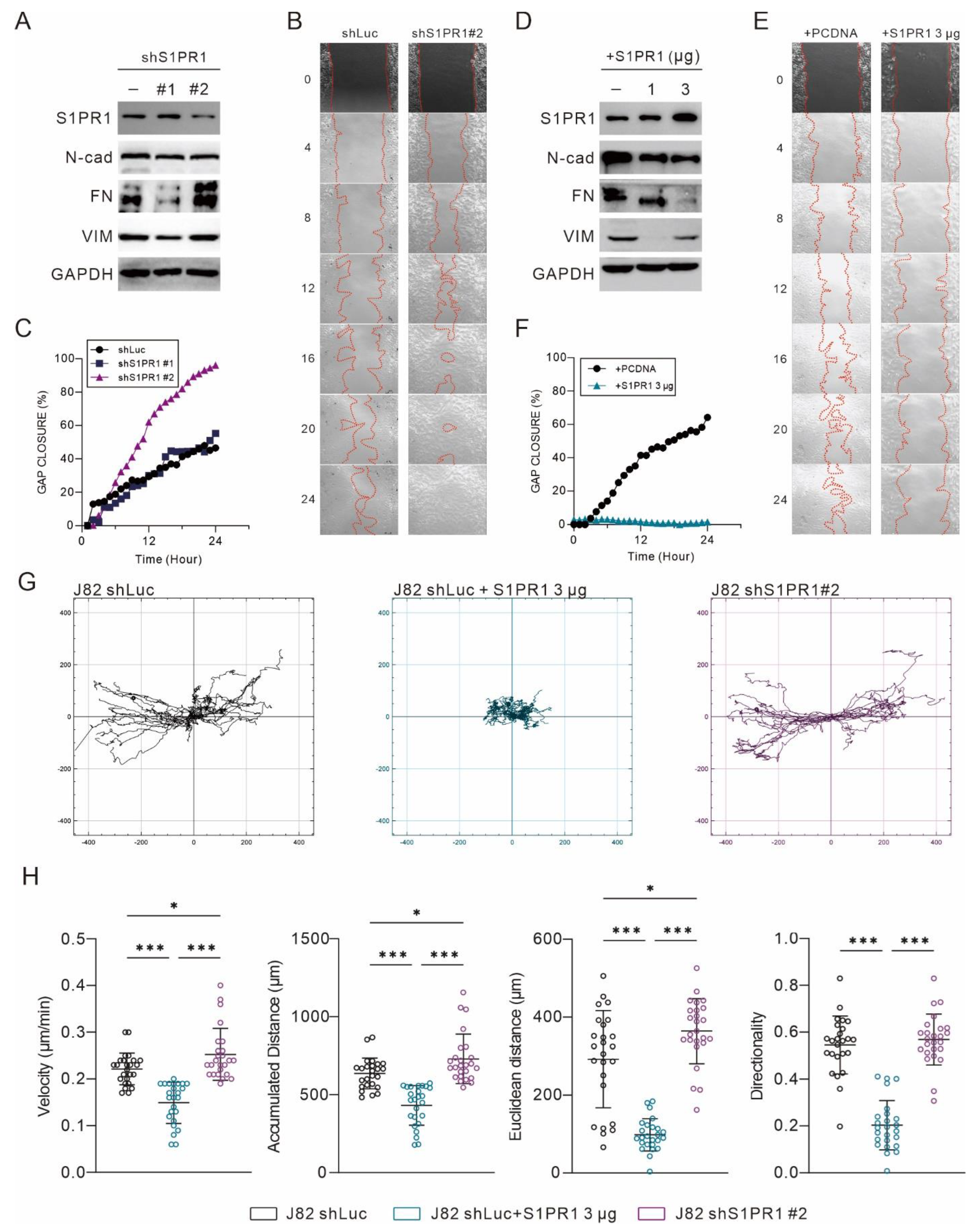
3.5. S1PR1 Expression Shows an Opposite Association with the Promoting of Epithelial–Mesenchymal Transition
3.6. The Administration of FTY-720 Promotes EMT in Bladder Carcinoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lenis, A.T.; Lec, P.M.; Chamie, K.; Mshs, M.D. Bladder Cancer: A Review. JAMA 2020, 324, 1980–1991. [Google Scholar] [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [Green Version]
- Richters, A.; Aben, K.K.H.; Kiemeney, L. The global burden of urinary bladder cancer: An update. World J. Urol. 2020, 38, 1895–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witjes, J.A.; Comperat, E.; Cowan, N.C.; De Santis, M.; Gakis, G.; Lebret, T.; Ribal, M.J.; Van der Heijden, A.G.; Sherif, A. EAU guidelines on muscle-invasive and metastatic bladder cancer: Summary of the 2013 guidelines. Eur. Urol. 2014, 65, 778–792. [Google Scholar] [CrossRef]
- Brausi, M.; Witjes, J.A.; Lamm, D.; Persad, R.; Palou, J.; Colombel, M.; Buckley, R.; Soloway, M.; Akaza, H.; Bohle, A. A review of current guidelines and best practice recommendations for the management of nonmuscle invasive bladder cancer by the International Bladder Cancer Group. J. Urol. 2011, 186, 2158–2167. [Google Scholar] [CrossRef]
- Cartier, A.; Hla, T. Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science 2019, 366. [Google Scholar] [CrossRef]
- Riese, J.; Gromann, A.; Luhrs, F.; Kleinwort, A.; Schulze, T. Sphingosine-1-Phosphate Receptor Type 4 (S1P4) Is Differentially Regulated in Peritoneal B1 B Cells upon TLR4 Stimulation and Facilitates the Egress of Peritoneal B1a B Cells and Subsequent Accumulation of Splenic IRA B Cells under Inflammatory Conditions. Int. J. Mol. Sci. 2021, 22, 3465. [Google Scholar] [CrossRef] [PubMed]
- Abarca-Zabalia, J.; Garcia, M.I.; Lozano Ros, A.; Marin-Jimenez, I.; Martinez-Gines, M.L.; Lopez-Cauce, B.; Martin-Barbero, M.L.; Salvador-Martin, S.; Sanjurjo-Saez, M.; Garcia-Dominguez, J.M.; et al. Differential Expression of SMAD Genes and S1PR1 on Circulating CD4+ T Cells in Multiple Sclerosis and Crohn’s Disease. Int. J. Mol. Sci. 2020, 21, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allende, M.L.; Tuymetova, G.; Lee, B.G.; Bonifacino, E.; Wu, Y.P.; Proia, R.L. S1P1 receptor directs the release of immature B cells from bone marrow into blood. J. Exp. Med. 2010, 207, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benechet, A.P.; Menon, M.; Xu, D.; Samji, T.; Maher, L.; Murooka, T.T.; Mempel, T.R.; Sheridan, B.S.; Lemoine, F.M.; Khanna, K.M. T cell-intrinsic S1PR1 regulates endogenous effector T-cell egress dynamics from lymph nodes during infection. Proc. Natl. Acad. Sci. USA 2016, 113, 2182–2187. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, L.; Qian, A.S.; Tahir, U.; Yu, P.; Trigatti, B.L. Sphingosine-1-Phosphate Receptor 1, Expressed in Myeloid Cells, Slows Diet-Induced Atherosclerosis and Protects against Macrophage Apoptosis in Ldlr KO Mice. Int. J. Mol. Sci. 2017, 18, 2721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryan, A.M.; Del Poeta, M. Sphingosine-1-phosphate receptors and innate immunity. Cell Microbiol. 2018, 20, e12836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, H.; Kim, P.J.; Jeon, Y.K.; Cho, Y.M.; Kim, K.; Park, B.H.; Ku, J.Y. Sphingosine-1-phosphate receptor 1 (S1PR1) expression in non-muscle invasive urothelial carcinoma: Association with poor clinical outcome and potential therapeutic target. Eur. J. Cancer 2015, 51, 1937–1945. [Google Scholar] [CrossRef]
- Patmanathan, S.N.; Wang, W.; Yap, L.F.; Herr, D.R.; Paterson, I.C. Mechanisms of sphingosine 1-phosphate receptor signalling in cancer. Cell Signal. 2017, 34, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.N.; Zhang, H.; Zhang, L.; Cai, T.T.; Huang, D.J.; He, J.; Ni, H.H.; Zhou, F.J.; Zhang, X.S.; Li, J. Sphingosine 1 phosphate receptor-1 (S1P1) promotes tumor-associated regulatory T cell expansion: Leading to poor survival in bladder cancer. Cell Death Dis. 2019, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, J.; Hartung, H.P. Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin. Neuropharmacol. 2010, 33, 91–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiba, K. FTY720, a new class of immunomodulator, inhibits lymphocyte egress from secondary lymphoid tissues and thymus by agonistic activity at sphingosine 1-phosphate receptors. Pharmacol. Ther. 2005, 108, 308–319. [Google Scholar] [CrossRef]
- Azuma, H.; Takahara, S.; Horie, S.; Muto, S.; Otsuki, Y.; Katsuoka, Y. Induction of apoptosis in human bladder cancer cells in vitro and in vivo caused by FTY720 treatment. J. Urol. 2003, 169, 2372–2377. [Google Scholar] [CrossRef]
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable continuous data distribution system for determining survival in Kaplan-Meier estimator. Comput. Methods Programs Biomed. 2019, 177, 133–139. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, L.; Liu, L.; Hou, Y.; Xiong, M.; Yang, Y.; Hu, J.; Chen, K. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020, 11, 5077. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Broadinstitute. Morpheus. Available online: https://software.broadinstitute.org/morpheus (accessed on 20 August 2021).
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- Sturm, G.; Finotello, F.; List, M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol. Biol. 2020, 2120, 223–232. [Google Scholar] [CrossRef]
- Wu, Y.Y.; Wang, S.H.; Wu, C.H.; Yen, L.C.; Lai, H.F.; Ho, C.L.; Chiu, Y.L. In silico immune infiltration profiling combined with functional enrichment analysis reveals a potential role for naive B cells as a trigger for severe immune responses in the lungs of COVID-19 patients. PLoS ONE 2020, 15, e0242900. [Google Scholar] [CrossRef]
- Su, H.C.; Wu, S.C.; Yen, L.C.; Chiao, L.K.; Wang, J.K.; Chiu, Y.L.; Ho, C.L.; Huang, S.M. Gene expression profiling identifies the role of Zac1 in cervical cancer metastasis. Sci. Rep. 2020, 10, 11837. [Google Scholar] [CrossRef]
- van de Merbel, A.F.; van der Horst, G.; van der Mark, M.H.; van Uhm, J.I.M.; van Gennep, E.J.; Kloen, P.; Beimers, L.; Pelger, R.C.M.; van der Pluijm, G. An ex vivo Tissue Culture Model for the Assessment of Individualized Drug Responses in Prostate and Bladder Cancer. Front. Oncol. 2018, 8, 400. [Google Scholar] [CrossRef]
- Als, A.B.; Dyrskjot, L.; von der Maase, H.; Koed, K.; Mansilla, F.; Toldbod, H.E.; Jensen, J.L.; Ulhoi, B.P.; Sengelov, L.; Jensen, K.M.; et al. Emmprin and survivin predict response and survival following cisplatin-containing chemotherapy in patients with advanced bladder cancer. Clin. Cancer Res. 2007, 13, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.J.; Kim, E.J.; Kim, S.K.; Kim, Y.J.; Ha, Y.S.; Jeong, P.; Kim, M.J.; Yun, S.J.; Lee, K.M.; Moon, S.K.; et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol. Cancer 2010, 9, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riester, M.; Taylor, J.M.; Feifer, A.; Koppie, T.; Rosenberg, J.E.; Downey, R.J.; Bochner, B.H.; Michor, F. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clin. Cancer Res. 2012, 18, 1323–1333. [Google Scholar] [CrossRef] [Green Version]
- Sjodahl, G.; Lauss, M.; Lovgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Ferno, M.; Ringner, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Carbayo, M.; Socci, N.D.; Lozano, J.; Saint, F.; Cordon-Cardo, C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006, 24, 778–789. [Google Scholar] [CrossRef]
- Lee, J.S.; Leem, S.H.; Lee, S.Y.; Kim, S.C.; Park, E.S.; Kim, S.B.; Kim, S.K.; Kim, Y.J.; Kim, W.J.; Chu, I.S. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J. Clin. Oncol. 2010, 28, 2660–2667. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Nojima, H.; Kuboki, S.; Yoshitomi, H.; Furukawa, K.; Takayashiki, T.; Takano, S.; Ohtsuka, M. Sphingosine-1-phosphate Receptor-1 Promotes Vascular Invasion and EMT in Hepatocellular Carcinoma. J. Surg. Res. 2021, 259, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, A.; Luo, G.; Ma, F.; Wei, X.; Bi, Y. S1P1 receptor inhibits kidney epithelial mesenchymal transition triggered by ischemia/reperfusion injury via the PI3K/Akt pathway. Acta Biochim. Biophys. Sin. 2018, 50, 651–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masana, M.I.; Brown, R.C.; Pu, H.; Gurney, M.E.; Dubocovich, M.L. Cloning and characterization of a new member of the G-protein coupled receptor EDG family. Recept. Channels 1995, 3, 255–262. [Google Scholar]
- Zondag, G.C.; Postma, F.R.; Etten, I.V.; Verlaan, I.; Moolenaar, W.H. Sphingosine 1-phosphate signalling through the G-protein-coupled receptor Edg-1. Biochem. J. 1998, 330 Pt 2, 605–609. [Google Scholar] [CrossRef]
- Lee, M.J.; Thangada, S.; Claffey, K.P.; Ancellin, N.; Liu, C.H.; Kluk, M.; Volpi, M.; Sha’afi, R.I.; Hla, T. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell 1999, 99, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wada, R.; Yamashita, T.; Mi, Y.; Deng, C.X.; Hobson, J.P.; Rosenfeldt, H.M.; Nava, V.E.; Chae, S.S.; Lee, M.J.; et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Investig. 2000, 106, 951–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, M.; Aoki, H.; Ramanathan, R.; Hait, N.C.; Takabe, K. Sphingosine-1-Phosphate Signaling in Immune Cells and Inflammation: Roles and Therapeutic Potential. Mediat. Inflamm. 2016, 2016, 8606878. [Google Scholar] [CrossRef] [Green Version]
- Chi, H.; Flavell, R.A. Cutting edge: Regulation of T cell trafficking and primary immune responses by sphingosine 1-phosphate receptor 1. J. Immunol. 2005, 174, 2485–2488. [Google Scholar] [CrossRef] [Green Version]
- Roviezzo, F.; Del Galdo, F.; Abbate, G.; Bucci, M.; D’Agostino, B.; Antunes, E.; De Dominicis, G.; Parente, L.; Rossi, F.; Cirino, G.; et al. Human eosinophil chemotaxis and selective in vivo recruitment by sphingosine 1-phosphate. Proc. Natl. Acad. Sci. USA 2004, 101, 11170–11175. [Google Scholar] [CrossRef] [Green Version]
- Chu, P.; Pardo, J.; Zhao, H.; Li, C.C.; Pali, E.; Shen, M.M.; Qu, K.; Yu, S.X.; Huang, B.C.; Yu, P.; et al. Systematic identification of regulatory proteins critical for T-cell activation. J. Biol. 2003, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Dorsam, G.; Graeler, M.H.; Seroogy, C.; Kong, Y.; Voice, J.K.; Goetzl, E.J. Transduction of multiple effects of sphingosine 1-phosphate (S1P) on T cell functions by the S1P1 G protein-coupled receptor. J. Immunol. 2003, 171, 3500–3507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Piao, W.; Brinkman, C.C.; Li, L.; Kulinski, J.M.; Olivera, A.; Cartier, A.; Hla, T.; Hippen, K.L.; Blazar, B.R.; et al. CD4 T cell sphingosine 1-phosphate receptor (S1PR)1 and S1PR4 and endothelial S1PR2 regulate afferent lymphatic migration. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Resop, R.S.; Douaisi, M.; Craft, J.; Jachimowski, L.C.; Blom, B.; Uittenbogaart, C.H. Sphingosine-1-phosphate/sphingosine-1-phosphate receptor 1 signaling is required for migration of naive human T cells from the thymus to the periphery. J. Allergy Clin. Immunol. 2016, 138, 551–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sassoli, C.; Pierucci, F.; Tani, A.; Frati, A.; Chellini, F.; Matteini, F.; Vestri, A.; Anderloni, G.; Nosi, D.; Zecchi-Orlandini, S.; et al. Sphingosine 1-Phosphate Receptor 1 Is Required for MMP-2 Function in Bone Marrow Mesenchymal Stromal Cells: Implications for Cytoskeleton Assembly and Proliferation. Stem Cells Int. 2018, 2018, 5034679. [Google Scholar] [CrossRef] [Green Version]
- Palangi, A.; Shakhssalim, N.; Parvin, M.; Bayat, S.; Allameh, A. Differential expression of S1P receptor subtypes in human bladder transitional cell carcinoma. Clin. Transl. Oncol. 2019, 21, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wei, Y.; Zhong, Y.; Zhu, D.; Ren, L.; Xu, P.; Zheng, P.; Feng, Q.; Ji, M.; Lv, M.; et al. Aberrant expression of sphingosine-1-phosphate receptor 1 correlates with metachronous liver metastasis and poor prognosis in colorectal cancer. Tumour Biol. 2014, 35, 9743–9750. [Google Scholar] [CrossRef]
- Yuan, L.W.; Liu, D.C.; Yang, Z.L. Correlation of S1P1 and ERp29 expression to progression, metastasis, and poor prognosis of gallbladder adenocarcinoma. Hepatobiliary Pancreat. Dis. Int. 2013, 12, 189–195. [Google Scholar] [CrossRef]
- Liu, S.; Ni, C.; Zhang, D.; Sun, H.; Dong, X.; Che, N.; Liang, X.; Chen, C.; Liu, F.; Bai, J.; et al. S1PR1 regulates the switch of two angiogenic modes by VE-cadherin phosphorylation in breast cancer. Cell Death Dis. 2019, 10, 200. [Google Scholar] [CrossRef]
- Zhong, L.; Xie, L.; Yang, Z.; Li, L.; Song, S.; Cao, D.; Liu, Y. Prognostic value of S1PR1 and its correlation with immune infiltrates in breast and lung cancers. BMC Cancer 2020, 20, 766. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.N.; Raue, R.; Weigert, A.; von Knethen, A.; Brune, B. Macrophage S1PR1 Signaling Alters Angiogenesis and Lymphangiogenesis During Skin Inflammation. Cells 2019, 8, 785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weichand, B.; Popp, R.; Dziumbla, S.; Mora, J.; Strack, E.; Elwakeel, E.; Frank, A.C.; Scholich, K.; Pierre, S.; Syed, S.N.; et al. S1PR1 on tumor-associated macrophages promotes lymphangiogenesis and metastasis via NLRP3/IL-1beta. J. Exp. Med. 2017, 214, 2695–2713. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xu, L.; Chen, L.; Fu, Q.; Liu, Z.; Chang, Y.; Lin, Z.; Xu, J. Tumor-infiltrating neutrophils predict benefit from adjuvant chemotherapy in patients with muscle invasive bladder cancer. Oncoimmunology 2017, 6, e1293211. [Google Scholar] [CrossRef]
- Mandelli, G.E.; Missale, F.; Bresciani, D.; Gatta, L.B.; Scapini, P.; Caveggion, E.; Roca, E.; Bugatti, M.; Monti, M.; Cristinelli, L.; et al. Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer. Cells 2020, 9, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ubai, T.; Azuma, H.; Kotake, Y.; Inamoto, T.; Takahara, K.; Ito, Y.; Kiyama, S.; Sakamoto, T.; Horie, S.; Muto, S.; et al. FTY720 induced Bcl-associated and Fas-independent apoptosis in human renal cancer cells in vitro and significantly reduced in vivo tumor growth in mouse xenograft. Anticancer Res. 2007, 27, 75–88. [Google Scholar]
- Zhang, L.; Wang, H.; Zhu, J.; Ding, K.; Xu, J. FTY720 reduces migration and invasion of human glioblastoma cell lines via inhibiting the PI3K/AKT/mTOR/p70S6K signaling pathway. Tumour Biol. 2014, 35, 10707–10714. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Wang, J.; Zheng, T.; Liang, Y.; Yin, D.; Song, R.; Pei, T.; Pan, S.; Jiang, H.; Liu, L. FTY720 inhibits proliferation and epithelial-mesenchymal transition in cholangiocarcinoma by inactivating STAT3 signaling. BMC Cancer 2014, 14, 783. [Google Scholar] [CrossRef] [Green Version]
- Nair, A.; Ingram, N.; Verghese, E.T.; Wijetunga, I.; Markham, A.F.; Wyatt, J.; Prasad, K.R.; Coletta, P.L. CD105 is a prognostic marker and valid endothelial target for microbubble platforms in cholangiocarcinoma. Cell Oncol. 2020, 43, 835–845. [Google Scholar] [CrossRef]
- Mei, X.; Chen, Y.S.; Chen, F.R.; Xi, S.Y.; Chen, Z.P. Glioblastoma stem cell differentiation into endothelial cells evidenced through live-cell imaging. Neuro. Oncol. 2017, 19, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhi, Y.; Song, H.; Zong, M.; Yi, J.; Mao, G.; Chen, L.; Huang, G. S1PR1 promotes proliferation and inhibits apoptosis of esophageal squamous cell carcinoma through activating STAT3 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys Acta 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Dai, L.; Qi, Y.; Di, W.; Xia, P. Combination of FTY720 with cisplatin exhibits antagonistic effects in ovarian cancer cells: Role of autophagy. Int. J. Oncol. 2013, 42, 2053–2059. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.N.; Koo, K.H.; Sung, J.Y.; Yun, U.J.; Kim, H. Anoikis resistance: An essential prerequisite for tumor metastasis. Int. J. Cell Biol. 2012, 2012, 306879. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| SLUG | AAGCATTTCAACGCCTCCAAA | GGATCTCTGGTTGTGGTATGACA |
| CDH1 | GCCTCCTGAAAAGAGAGTGGAAG | TGGCAGTGTCTCTCCAAATCCG |
| GAPDH | CATCACTGCCACCCAGAAGACTG | ATGCCAGTGAGCTTCCCGTTCAG |
| CDH2 | TGCGGTACAGTGTAACTGGG | GAAACCGGGCTATCTGCTCG |
| SNAI1 | TCGGAAGCCTAACTACAGCGA | AGATGAGCATTGGCAGCGAG |
| FN1 | CGGTGGCTGTCAGTCAAAG | AAACCTCGGCTTCCTCCATAA |
| S1PR1 | ATCATGGGCTGGAACTGCATCA | CGAGTCCTGACCAAGGAGTAGAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-L.; Meng, E.; Wu, S.-T.; Lai, H.-F.; Lu, Y.-S.; Yang, M.-H.; Tsao, C.-W.; Kao, C.-C.; Chiu, Y.-L. Targeting S1PR1 May Result in Enhanced Migration of Cancer Cells in Bladder Carcinoma. Cancers 2021, 13, 4474. https://doi.org/10.3390/cancers13174474
Chen C-L, Meng E, Wu S-T, Lai H-F, Lu Y-S, Yang M-H, Tsao C-W, Kao C-C, Chiu Y-L. Targeting S1PR1 May Result in Enhanced Migration of Cancer Cells in Bladder Carcinoma. Cancers. 2021; 13(17):4474. https://doi.org/10.3390/cancers13174474
Chicago/Turabian StyleChen, Chin-Li, En Meng, Sheng-Tang Wu, Hsing-Fan Lai, Yi-Shan Lu, Ming-Hsin Yang, Chih-Wei Tsao, Chien-Chang Kao, and Yi-Lin Chiu. 2021. "Targeting S1PR1 May Result in Enhanced Migration of Cancer Cells in Bladder Carcinoma" Cancers 13, no. 17: 4474. https://doi.org/10.3390/cancers13174474
APA StyleChen, C.-L., Meng, E., Wu, S.-T., Lai, H.-F., Lu, Y.-S., Yang, M.-H., Tsao, C.-W., Kao, C.-C., & Chiu, Y.-L. (2021). Targeting S1PR1 May Result in Enhanced Migration of Cancer Cells in Bladder Carcinoma. Cancers, 13(17), 4474. https://doi.org/10.3390/cancers13174474






