Genetic Variation of Native Perilla Germplasms Collected from South Korea Using Simple Sequence Repeat (SSR) Markers and Morphological Characteristics
Abstract
:1. Introduction
2. Results
2.1. Morphological Variation in the Accessions of Perilla Frutescens Var. Frutescens of Perilla Crop
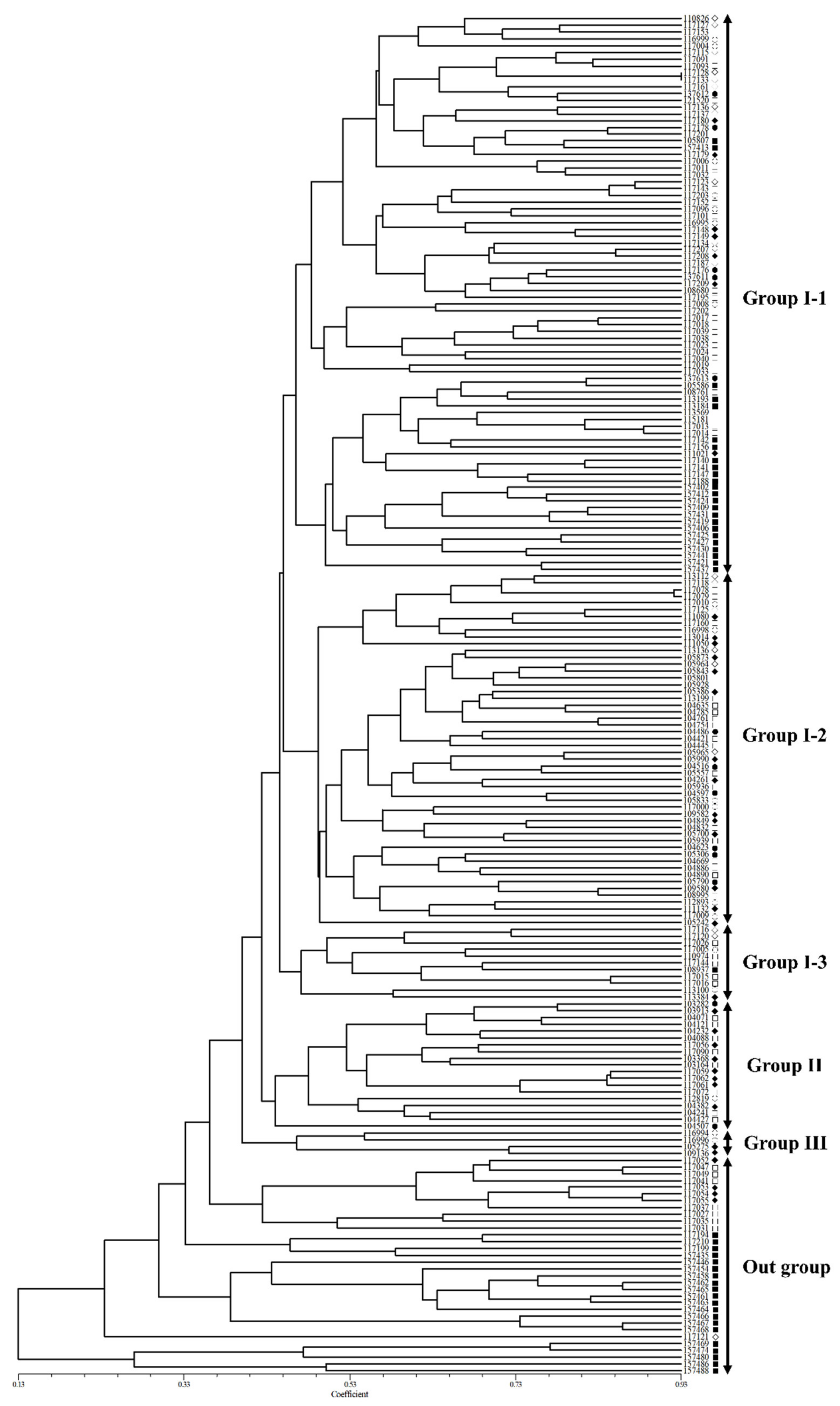
2.2. Cluster and Correlation Analysis among 183 Accessions of Native Perilla Frutescens Var. Frutescens Using Morphological Characteristics
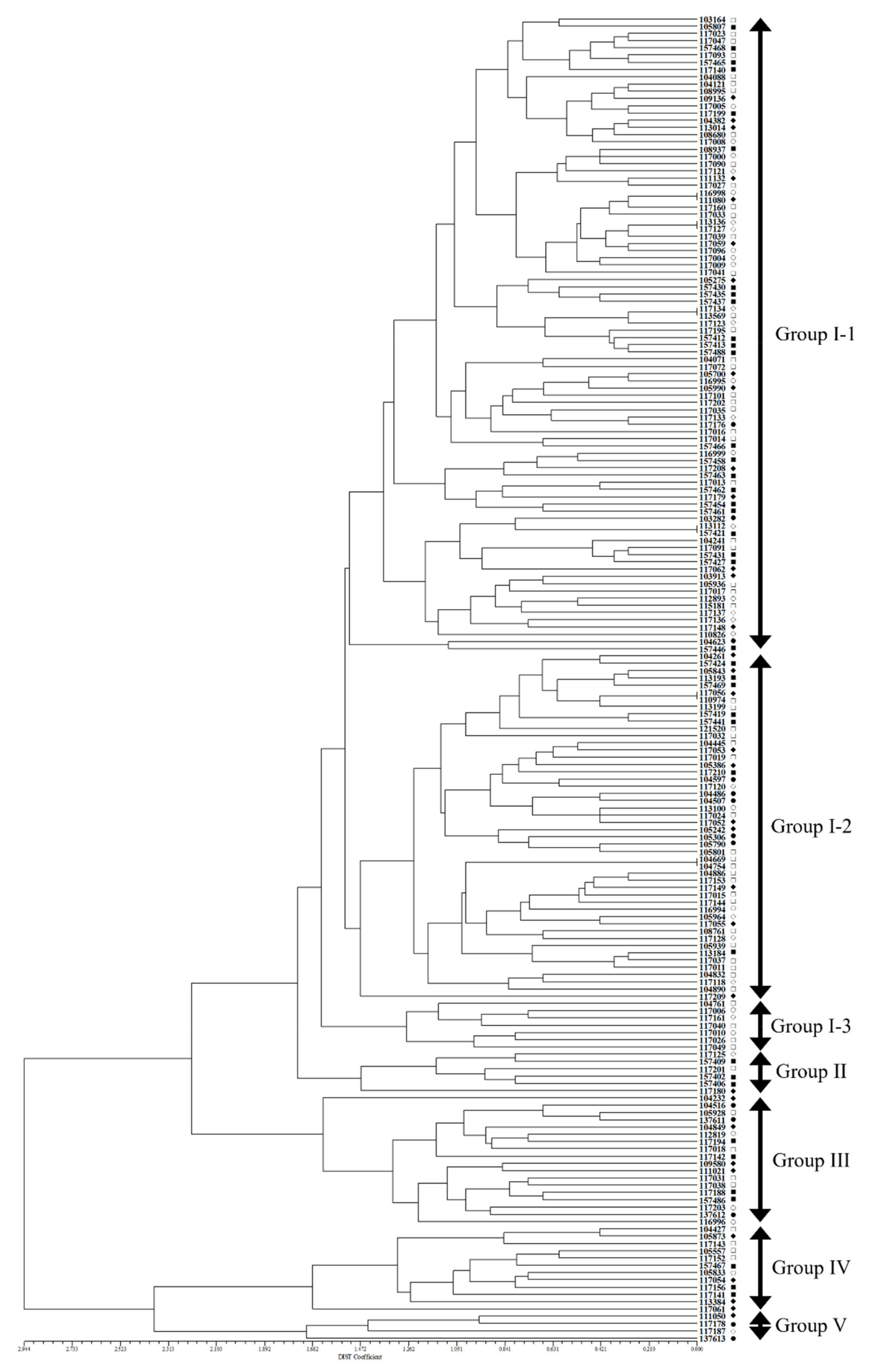
2.3. SSR Variation in Accessions of Native Perilla Frutescens Var. Frutescens of Perilla Crop
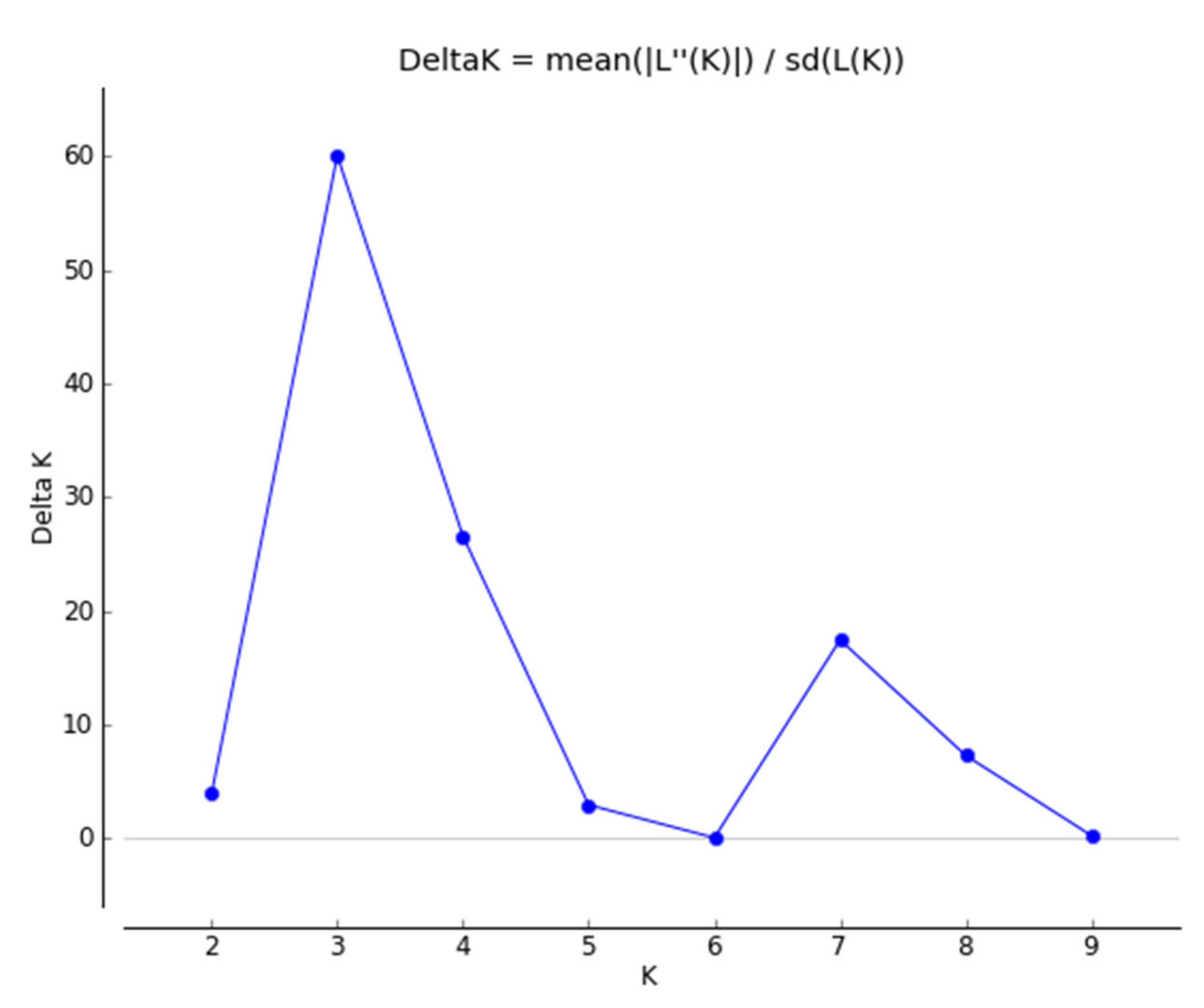
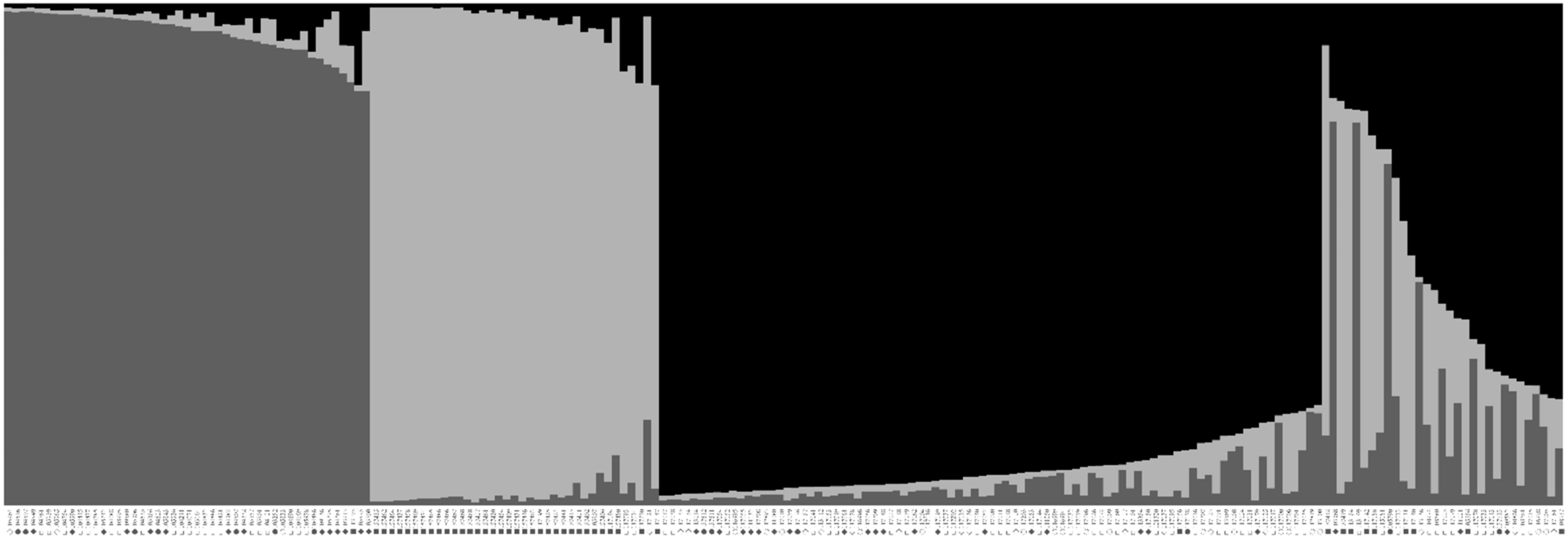
2.4. Population Structure and Genetic Relationships among Accessions of Native Perilla Frutescens Var. Frutescens from Five Regions of South Korea and Other Region
2.5. Correlation between Perilla SSR Markers and Perilla Morphological Traits
3. Discussion
3.1. Morphological Variation of Native Accessions of Perilla Frutescens Var. Frutescens in South Korea
3.2. Genetic Variation and Population Structure of Cultivated P. Frutescens Var. frutescens
4. Materials and Methods
4.1. Plant Materials
4.2. Morphological Characteristics Analyzed
4.3. SSR Analysis and DNA Electrophoresis
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makino, T. Makino’s New Illustrated Flora of Japan; Hokuryukan Co.: Tokyo, Japan, 1961. (In Japanese) [Google Scholar]
- Lee, J.K.; Ohnishi, O. Geographical differentiation of morphological characters among Perilla crops and their weedy types in East Asia. Breed. Sci. 2001, 51, 247–255. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.K.; Ohnishi, O. Genetic relationships among cultivated types of Perilla Frutescens and their weedy types in East Asia revealed by AFLP markers. Genet. Resour. Crop. Evol. 2003, 50, 65–74. [Google Scholar] [CrossRef]
- Nitta, M. Origin Perilla Crops and Their Weedy Type. Ph.D. Thesis, Kyoto University, Kyoto, Japan, 2001; p. 78. [Google Scholar]
- Lee, J.K.; Nitta, M.; Kim, N.S.; Park, C.H.; Yoon, K.M.; Shin, Y.B.; Ohnishi, O. Genetic diversity of Perilla and related weedy types in Korea determined by AFLP analyses. Crop. Sci. 2002, 42, 2161–2166. [Google Scholar] [CrossRef]
- Nitta, M.; Lee, J.K.; Ohnishi, O. Asian Perilla crops and their weedy forms: Their cultivation, utilization and genetic relationships. Econ. Bot. 2003, 57, 245–253. [Google Scholar] [CrossRef]
- Ma, S.J.; Lee, J.K. Morphological variation of two cultivated types of Perilla crop from different areas of China. Korean J. Hortic. Sci. Biotechnol. 2017, 35, 510–522. [Google Scholar]
- Li, H.L. The vegetables of ancient china. Econ. Bot. 1969, 23, 235–260. [Google Scholar] [CrossRef]
- Sa, K.J.; Choi, S.H.; Ueno, M.; Park, K.C.; Park, Y.J.; Ma, K.H.; Lee, J.K. Identification of genetic variations of cultivated and weedy types of Perilla species in Korea and Japan using morphological and SSR markers. Genes Genom. 2013, 35, 649–659. [Google Scholar] [CrossRef]
- Lee, J.I.; Bang, J.K.; Lee, B.H.; Kim, K.H. Quality improvement in Perilla. I. Varietal differences of oil content and fatty acid composition. Korean J. Crop. Sci. 1991, 36, 48–61. [Google Scholar]
- Asif, M. Health effects of omega-3,6,9 fatty acids: Perilla frutescens is a good example of plant oils. Orient Pharm. Exp. Med. 2011, 11, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lee, M.H.; Cho, E.J.; Lee, S. High-yield methods for purification of a-linolenic acid from Perilla frutescens var. japonica oil. Appl. Biol. Chem. 2016, 59, 89–94. [Google Scholar] [CrossRef]
- Park, H.; Sa, K.J.; Hyun, D.Y.; Lee, S.; Lee, J.K. Identifying SSR markers related to seed fatty acid content in Perilla crop (Perilla frutescens L.). Plants 2021, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.K.; Sa, K.J.; Hyun, D.Y.; Cho, G.T.; Lee, J.K. Assessment of genetic diversity and population structure among a collection of Korean Perilla germplasms based on SSR markers. Genes Genom. 2020, 42, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, H.G. Germplasm preservation: Objectives and need. In Biotic Diversity and Germplasm Preservation, Global Imperatives; Knutson, L., Stoner, A.K., Eds.; Kluwer Academic Press: Dordrecht, The Netherlands, 1989; pp. 13–41. [Google Scholar]
- Williams, J.T. Plant genetic resources: Some new directions. Adv. Agron. 1991, 45, 61–91. [Google Scholar]
- Nitta, M.; Ohnishi, O. Genetic relationships among two Perilla crops, shiso and egoma, and the weedy type revealed by RAPD markers. Jpn. J. Genet. 1999, 74, 43–48. [Google Scholar] [CrossRef]
- Park, Y.J.; Dixit, A.; Ma, K.H.; Lee, J.K.; Lee, M.H.; Chung, C.S.; Nitta, M.; Okuno, K.; Kim, T.S.; Cho, E.G.; et al. Evaluation of genetic diversity and relationships within an on-farm collection of Perilla frutescens (L.) Britt. using microsatellite markers. Genet. Resour. Crop. Evol. 2008, 55, 523–535. [Google Scholar] [CrossRef]
- Sa, K.J.; Choi, I.K.; Park, K.C.; Lee, J.K. Genetic diversity and population structure among accessions of Perilla frutescens (L.) Britton in East Asia using new developed microsatellite markers. Genes Genom. 2018, 40, 1319–1329. [Google Scholar] [CrossRef]
- Ha, Y.J.; Sa, K.J.; Lee, J.K. Identifying SSR markers associated with seed characteristics in Perilla (Perilla frutescens L.). Physiol. Mol. Biol. Plants 2021, 27, 93–105. [Google Scholar] [CrossRef]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogelet, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, J.K.; Kim, N.S. Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules 2009, 14, 4546–4569. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Sa, K.Y.; Ha, Y.J.; Lee, J.K. Genetic variation and association mapping in F2 population of Perilla crop (Perilla frutescens L.) using new developed Perilla SSR markers. Euphytica 2021, 217, 135. [Google Scholar] [CrossRef]
- Schwanitz, F. The Origin of Cultivated Plants; Harvard University Press: Cambridge, MA, USA, 1966. [Google Scholar]
- Gould, S.J.; Johnston, R.F. Geographic variation. Ann. Rev. Ecol. Syst. 1972, 3, 457–498. [Google Scholar] [CrossRef]
- Harlan, J.R. Origins and processes of domestication. In Grass Evolution and Domestication; Chapman, G.P., Ed.; Cambridge University Press: Cambridge, UK, 1992; pp. 159–175. [Google Scholar]
- Harlan, J.R.; de Wet, J.M.J.; Price, G. Comparative evolution of cereals. Evolution 1973, 27, 322–325. [Google Scholar] [CrossRef]
- Wyatt, R.; Antonovics, J. Butterfly weed re-revisited: Spatial and temporal patterns of leaf shape variation in Asclepias tuberosa. Evolution 1981, 35, 529–542. [Google Scholar]
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Purugganan, M.D. Evolutionary insights into the nature of plant domestication. Curr. Biol. 2019, 29, R705–R714. [Google Scholar] [CrossRef] [Green Version]
- Vigouroux, Y.; Barnaud, A.; Scarcelli, N.; Thuillet, A.C. Biodiversity, evolution and adaptation of cultivated crops. C. R. Biol. 2011, 334, 450–457. [Google Scholar] [CrossRef]
- Lee, J.K.; Kim, N.S.; Ohnishi, O. Perilla crop and related weedy types collected in Korea. Korean J. Breed. Sci. 2007, 39, 316–323. [Google Scholar]
- Ma, S.J.; Sa, K.J.; Hong, T.K.; Lee, J.K. Genetic diversity and population structure analysis in Perilla crop and their weedy types from northern and southern areas of China based on simple sequence repeat (SSRs). Genes Genom. 2019, 41, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.E.; Sa, K.J.; Lee, J.K. Bulk segregant analysis identifies SSR markers associated with leaf-and seed related traits in Perilla crop (Perilla frutescens L.). Genes Genom. 2021, 43, 323–332. [Google Scholar] [CrossRef]
- Hamza, S.; Hamida, W.B.; Rebaï, A.; Harrabi, M. SSR-based genetic diversity assessment among tunisian winter barley and relationship with morphological traits. Euphytica 2004, 135, 107–118. [Google Scholar] [CrossRef]
- Peng, J.; Bai, Y.; Haley, S.; Lapitan, N. Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 2009, 135, 95–122. [Google Scholar] [CrossRef] [PubMed]
- Tantasawat, P.; Trongchun, J.; Prajongjai, T.; Jenweerawat, S.; Chaowiset, W. SSR analysis of soybean (Glycine max (L.) Merr.) genetic relationship and variety identification in Thailand. Aust. J. Crop. Sci. 2011, 5, 283–290. [Google Scholar]
- Kimaro, D.; Melis, R.; Sibiya, J.; Shimelis, H.; Shayanowako, A. Analysis of genetic diversity and population structure of pigeonpea [Cajanus cajan (L.) Millsp] accessions using SSR markers. Plants 2020, 9, 1643. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Lee, J.K.; Kim, N.S.; Yu, J.W.; Dixit, A.; Cho, E.G.; Park, Y.J. Isolation and characterization of SSR. markers in Perilla frutescens Britt. Mol. Eco. Notes 2005, 5, 454–456. [Google Scholar] [CrossRef]
- Sa, K.J.; Lim, S.E.; Choi, I.K.; Park, K.C.; Lee, J.K. Development and characterization of new microsatellite markers for Perilla frutescens (L.) Britton. Am. J. Plant. Sci. 2019, 10, 1623–1630. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Rohlf, F. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System; Version Exter Software: Setauket, NY, USA, 2000. [Google Scholar]
- Pritchard, J.K.; Wen, W.; Falush, D. Documentation for Structure Software: Version 2.3. 2003. Available online: http://pritc.h.bsd.uchicago.edu/structure.html (accessed on 10 December 2020).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantel, N. The detection of disease clustering and a generalized repression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
| QL2 | QL3 | QL4 | QL5 | QL6 | QL7 | QL8 | QN1 | QN2 | QN3 | |
|---|---|---|---|---|---|---|---|---|---|---|
| QL1 | −0.133 | 1.000 ** | 0.011 | 0.109 | −0.088 | 0.051 | −0.012 | 0.139 | 0.104 | −0.049 |
| QL2 | −0.133 | 0.095 | −0.030 | −0.120 | −0.045 | −0.084 | 0.013 | −0.071 | 0.011 | |
| QL3 | 0.011 | 0.109 | −0.088 | 0.051 | −0.012 | 0.139 | 0.104 | −0.049 | ||
| QL4 | 0.191 ** | −0.018 | 0.050 | 0.026 | 0.239 ** | 0.146 * | −0.032 | |||
| QL5 | 0.073 | −0.047 | −0.057 | 0.031 | 0.032 | −0.153 * | ||||
| QL6 | 0.155 * | 0.151 * | 0.050 | 0.036 | 0.166 * | |||||
| QL7 | 0.070 | −0.019 | −0.065 | −0.087 | ||||||
| QL8 | −0.052 | −0.041 | 0.043 | |||||||
| QN1 | 0.534 ** | 0.172 * | ||||||||
| QN2 | 0.162 * |
| SSR Loci | Allele Size | No. of Allele | MAF | GD | PIC |
|---|---|---|---|---|---|
| KNUPF1 | 176–182 | 5 | 0.835 | 0.290 | 0.272 |
| KNUPF2 | 156–166 | 9 | 0.385 | 0.726 | 0.680 |
| KNUPF4 | 158–168 | 7 | 0.370 | 0.708 | 0.654 |
| KNUPF5 | 177–183 | 6 | 0.470 | 0.693 | 0.651 |
| KNUPF10 | 195–220 | 13 | 0.565 | 0.648 | 0.626 |
| KNUPF16 | 141–190 | 13 | 0.225 | 0.828 | 0.805 |
| KNUPF23 | 204–214 | 4 | 0.565 | 0.592 | 0.532 |
| KNUPF25 | 179–190 | 5 | 0.485 | 0.629 | 0.561 |
| KNUPF26 | 153–264 | 3 | 0.570 | 0.571 | 0.501 |
| KNUPF28 | 174–192 | 6 | 0.660 | 0.522 | 0.484 |
| KNUPF33 | 186–213 | 11 | 0.335 | 0.788 | 0.760 |
| KNUPF36 | 170–180 | 6 | 0.390 | 0.749 | 0.712 |
| KNUPF37 | 256–259 | 4 | 0.490 | 0.641 | 0.577 |
| KNUPF39 | 196–223 | 9 | 0.410 | 0.711 | 0.662 |
| KNUPF43 | 127–155 | 10 | 0.255 | 0.791 | 0.759 |
| KNUPF55 | 233–253 | 11 | 0.520 | 0.681 | 0.653 |
| KNUPF59 | 152–170 | 4 | 0.580 | 0.525 | 0.431 |
| KNUPF71 | 182–185 | 4 | 0.435 | 0.645 | 0.572 |
| KNUPF74 | 190–198 | 4 | 0.445 | 0.597 | 0.512 |
| KNUPF77 | 166–174 | 3 | 0.405 | 0.651 | 0.576 |
| Total | 137 | ||||
| Mean | 6.85 | 0.470 | 0.649 | 0.599 |
| Marker | No. of Alleles | Major Allele Frequency | Genetic Diversity | PIC | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | G6 | G1 | G2 | G3 | G4 | G5 | G6 | G1 | G2 | G3 | G4 | G5 | G6 | G1 | G2 | G3 | G4 | G5 | G6 | |
| KNUPF1 | 2 | 2 | 2 | 4 | 3 | 4 | 0.947 | 0.923 | 0.900 | 0.889 | 0.851 | 0.667 | 0.100 | 0.142 | 0.180 | 0.205 | 0.265 | 0.491 | 0.095 | 0.132 | 0.164 | 0.198 | 0.249 | 0.433 |
| KNUPF2 | 6 | 3 | 6 | 6 | 5 | 4 | 0.632 | 0.462 | 0.450 | 0.500 | 0.463 | 0.800 | 0.565 | 0.639 | 0.705 | 0.668 | 0.644 | 0.343 | 0.536 | 0.566 | 0.663 | 0.624 | 0.579 | 0.320 |
| KNUPF4 | 5 | 4 | 5 | 4 | 6 | 6 | 0.474 | 0.538 | 0.450 | 0.472 | 0.418 | 0.511 | 0.676 | 0.627 | 0.630 | 0.597 | 0.664 | 0.674 | 0.627 | 0.576 | 0.560 | 0.515 | 0.600 | 0.637 |
| KNUPF5 | 5 | 4 | 3 | 4 | 6 | 5 | 0.737 | 0.692 | 0.500 | 0.694 | 0.507 | 0.444 | 0.438 | 0.485 | 0.605 | 0.485 | 0.661 | 0.688 | 0.416 | 0.451 | 0.527 | 0.452 | 0.616 | 0.638 |
| KNUPF10 | 5 | 4 | 6 | 6 | 10 | 6 | 0.368 | 0.615 | 0.250 | 0.528 | 0.672 | 0.667 | 0.726 | 0.568 | 0.800 | 0.651 | 0.534 | 0.528 | 0.680 | 0.526 | 0.770 | 0.610 | 0.520 | 0.504 |
| KNUPF16 | 5 | 5 | 5 | 8 | 11 | 7 | 0.316 | 0.385 | 0.400 | 0.250 | 0.284 | 0.689 | 0.765 | 0.722 | 0.715 | 0.823 | 0.811 | 0.505 | 0.726 | 0.676 | 0.668 | 0.799 | 0.785 | 0.485 |
| KNUPF23 | 4 | 2 | 3 | 4 | 3 | 4 | 0.579 | 0.538 | 0.650 | 0.389 | 0.552 | 0.711 | 0.560 | 0.497 | 0.515 | 0.677 | 0.594 | 0.454 | 0.488 | 0.374 | 0.460 | 0.611 | 0.528 | 0.413 |
| KNUPF25 | 4 | 3 | 3 | 4 | 4 | 4 | 0.526 | 0.385 | 0.700 | 0.444 | 0.448 | 0.667 | 0.632 | 0.651 | 0.445 | 0.650 | 0.619 | 0.516 | 0.578 | 0.576 | 0.381 | 0.583 | 0.540 | 0.479 |
| KNUPF26 | 3 | 3 | 3 | 3 | 3 | 3 | 0.474 | 0.462 | 0.750 | 0.444 | 0.537 | 0.711 | 0.637 | 0.615 | 0.395 | 0.634 | 0.591 | 0.440 | 0.565 | 0.535 | 0.347 | 0.558 | 0.517 | 0.386 |
| KNUPF28 | 4 | 4 | 3 | 4 | 6 | 5 | 0.684 | 0.462 | 0.800 | 0.611 | 0.642 | 0.711 | 0.493 | 0.627 | 0.335 | 0.560 | 0.532 | 0.443 | 0.456 | 0.556 | 0.303 | 0.508 | 0.486 | 0.392 |
| KNUPF33 | 6 | 4 | 7 | 7 | 7 | 7 | 0.421 | 0.538 | 0.250 | 0.472 | 0.343 | 0.356 | 0.704 | 0.627 | 0.820 | 0.715 | 0.778 | 0.784 | 0.657 | 0.576 | 0.795 | 0.684 | 0.747 | 0.757 |
| KNUPF36 | 4 | 4 | 4 | 5 | 6 | 5 | 0.526 | 0.462 | 0.550 | 0.333 | 0.358 | 0.489 | 0.626 | 0.686 | 0.625 | 0.776 | 0.761 | 0.683 | 0.568 | 0.637 | 0.578 | 0.742 | 0.725 | 0.643 |
| KNUPF37 | 3 | 3 | 3 | 4 | 4 | 4 | 0.474 | 0.462 | 0.800 | 0.500 | 0.582 | 0.578 | 0.632 | 0.639 | 0.335 | 0.619 | 0.578 | 0.591 | 0.558 | 0.566 | 0.303 | 0.549 | 0.519 | 0.537 |
| KNUPF39 | 4 | 4 | 3 | 5 | 6 | 6 | 0.474 | 0.615 | 0.550 | 0.389 | 0.388 | 0.378 | 0.654 | 0.556 | 0.535 | 0.715 | 0.719 | 0.721 | 0.594 | 0.506 | 0.436 | 0.664 | 0.672 | 0.672 |
| KNUPF43 | 6 | 5 | 5 | 5 | 8 | 5 | 0.368 | 0.308 | 0.400 | 0.472 | 0.313 | 0.667 | 0.770 | 0.746 | 0.665 | 0.705 | 0.794 | 0.493 | 0.738 | 0.702 | 0.604 | 0.670 | 0.764 | 0.437 |
| KNUPF55 | 7 | 6 | 6 | 6 | 6 | 6 | 0.263 | 0.308 | 0.450 | 0.389 | 0.627 | 0.689 | 0.798 | 0.781 | 0.710 | 0.764 | 0.566 | 0.502 | 0.769 | 0.748 | 0.670 | 0.733 | 0.533 | 0.479 |
| KNUPF59 | 3 | 3 | 3 | 3 | 4 | 2 | 0.632 | 0.538 | 0.650 | 0.694 | 0.537 | 0.511 | 0.521 | 0.556 | 0.485 | 0.440 | 0.558 | 0.500 | 0.455 | 0.465 | 0.406 | 0.365 | 0.468 | 0.375 |
| KNUPF71 | 3 | 2 | 3 | 4 | 2 | 3 | 0.737 | 0.538 | 0.550 | 0.472 | 0.507 | 0.822 | 0.410 | 0.497 | 0.535 | 0.576 | 0.500 | 0.307 | 0.359 | 0.374 | 0.436 | 0.484 | 0.375 | 0.284 |
| KNUPF74 | 2 | 3 | 3 | 3 | 3 | 4 | 0.632 | 0.538 | 0.600 | 0.444 | 0.507 | 0.489 | 0.465 | 0.556 | 0.560 | 0.610 | 0.589 | 0.595 | 0.357 | 0.465 | 0.499 | 0.527 | 0.506 | 0.513 |
| KNUPF77 | 3 | 3 | 3 | 3 | 3 | 3 | 0.474 | 0.385 | 0.450 | 0.556 | 0.463 | 0.467 | 0.615 | 0.663 | 0.615 | 0.586 | 0.638 | 0.631 | 0.536 | 0.589 | 0.534 | 0.517 | 0.564 | 0.556 |
| Averge | 4 | 4 | 4 | 5 | 5 | 5 | 0.537 | 0.508 | 0.555 | 0.497 | 0.500 | 0.601 | 0.589 | 0.594 | 0.561 | 0.623 | 0.620 | 0.544 | 0.538 | 0.530 | 0.505 | 0.570 | 0.565 | 0.497 |
| Morphological Character | When/how Measured | Category or Unit | |
|---|---|---|---|
| QL1 | Color of leaf surface | at flowering stage | 1-light green, 2-green, 3-deep green |
| QL2 | Color of reverse side leaf | at flowering stage | 1-light green, 2-green, 3-deep green |
| QL3 | Stem color | at flowering stage | 1-light green, 2-green, 3-deep green |
| QL4 | Seed color | after harvest | 1-white, 2-gray, 3-brown, 4-dark brown |
| QL5 | Leaf shape | at flowering stage | 1-lanceolate, 2-heart shape, 3-oblong |
| QL6 | Degree of pubescence | at flowering stage | 1-slightly pubescent, 2-normal pubescent, 3-heavily pubescent |
| QL7 | Seed hardness | after harvest | 1-soft, 2-hard |
| QL8 | Plant fragrance | at flowering stage | 1-plant fragrance of var. frutescens, 2-other plant fragrance |
| QN1 | Leaf width | at flowering stage | 1–8.0 cm ≤, 2–8.1~9.0 cm, 3–9.1~10.0 cm, 4–10.1~11.0 cm, 5–11.1~12.0 cm, 6–12.1~13.0 cm, 7–13.1 cm > |
| QN2 | Leaf length | at flowering stage | 1–10.0 cm ≤, 2–10.1~11.0 cm, 3–11.1~12.0 cm, 4–12.1~13.0 cm, 5–13.1~14.0 cm, 6–14.1~15.0 cm, 7–15.1 cm > |
| QN3 | Flowering time | at flowering stage (the day of more than 50% flowering per plant) | 1-early maturing type (flowering days before 15 August), 2-intermediate maturing type (flowering days from 15 August to 5 September), 3-late maturing type (flowering days after 6 September) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.S.; Sa, K.J.; Park, H.; Hyun, D.Y.; Lee, S.; Rhee, J.H.; Lee, J.K. Genetic Variation of Native Perilla Germplasms Collected from South Korea Using Simple Sequence Repeat (SSR) Markers and Morphological Characteristics. Plants 2021, 10, 1764. https://doi.org/10.3390/plants10091764
Oh JS, Sa KJ, Park H, Hyun DY, Lee S, Rhee JH, Lee JK. Genetic Variation of Native Perilla Germplasms Collected from South Korea Using Simple Sequence Repeat (SSR) Markers and Morphological Characteristics. Plants. 2021; 10(9):1764. https://doi.org/10.3390/plants10091764
Chicago/Turabian StyleOh, Jun Seok, Kyu Jin Sa, Hyeon Park, Do Yoon Hyun, Sookyeong Lee, Ju Hee Rhee, and Ju Kyong Lee. 2021. "Genetic Variation of Native Perilla Germplasms Collected from South Korea Using Simple Sequence Repeat (SSR) Markers and Morphological Characteristics" Plants 10, no. 9: 1764. https://doi.org/10.3390/plants10091764






