An Analysis of the Multifaceted Roles of Heme in the Pathogenesis of Cancer and Related Diseases
Abstract
:Simple Summary
Abstract
1. Introduction
2. Heme and Cancer
2.1. Elevated Heme Levels Promote Lung Tumorigenesis
2.2. Heme Synthesis, Export, and Catabolism, along with Dietary Heme Intake, Play a Role in Pancreatic and Colorectal Cancer
2.3. Paradoxical Roles of HO-1 in Cancer
2.4. Heme Acts as a Regulator of Circadian Rhythm Implicated in Cancer
2.5. Heme Controls the Activities of a Variety of Key Regulators Underlying Diverse Cancers
2.5.1. BACH1
2.5.2. PGRMC1
2.5.3. P53
2.5.4. CBS
2.5.5. NO Signaling Related Hemoproteins
2.6. Heme Promotes Angiogenesis Implicated in Tumorigenesis
3. Diseases and Conditions Associated with Elevated Heme
3.1. Elevated Heme Levels Underly Lung Injury
3.2. Elevated Heme Levels Affect Cardiac Physiology
3.3. Role of Heme as a Pro-Inflammatory Influencer and Hb-Derived DAMP
3.4. Elevated Heme Metabolism Promotes Neurodegenerative Functions in the Nervous System but Is Perturbed in Alzheimer’s Dementia
3.5. Elevated Heme Is Associated with Impaired Glucose Tolerance and Insulin Resistance in Type II Diabetes Mellitus while Intracellular Heme Deficiency Attenuates Mitochondrial Activity and Impairs Glucose Metabolism
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, L. Heme Biology: Heme Acts as a Versatile Signaling Molecule Regulating Diverse Biological Processes; World Scientific: Singapore, 2020. [Google Scholar]
- Ponka, P. Cell biology of heme. Am. J. Med. Sci. 1999, 318, 241–256. [Google Scholar] [CrossRef]
- Bilska-Wilkosz, A.; Iciek, M.; Górny, M.; Kowalczyk-Pachel, D. The role of hemoproteins: Hemoglobin, myoglobin and neuroglobin in endogenous thiosulfate production processes. Int. J. Mol. Sci. 2017, 18, 1315. [Google Scholar] [CrossRef] [Green Version]
- Moore, G.R.; Pettigrew, G.W. Cytochromes C: Evolutionary, Structural and Physicochemical Aspects; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Manikandan, P.; Nagini, S. Cytochrome P450 structure, function and clinical significance: A review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef]
- Everse, J. Heme Proteins. In Encyclopedia of Biological Chemistry; Academic Press: Cambridge, MA, USA, 2013; pp. 532–538. [Google Scholar] [CrossRef]
- Kim, H.J.; Khalimonchuk, O.; Smith, P.M.; Winge, D.R. Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta BBA Mol. Cell Res. 2012, 1823, 1604–1616. [Google Scholar] [CrossRef] [Green Version]
- McEwen, E.; Kedersha, N.; Song, B.; Scheuner, D.; Gilks, N.; Han, A.; Chen, J.J.; Anderson, P.; Kaufman, R.J. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 2005, 280, 16925–16933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mense, S.M.; Zhang, L. Heme: A versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006, 16, 681–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshavsky, A. The ubiquitin system, autophagy, and regulated protein degradation. Annu. Rev. Biochem. 2017, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Zenke-Kawasaki, Y.; Dohi, Y.; Katoh, Y.; Ikura, T.; Ikura, M.; Asahara, T.; Tokunaga, F.; Iwai, K.; Igarashi, K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 2007, 27, 6962–6971. [Google Scholar] [CrossRef] [Green Version]
- Chiabrando, D.; Vinchi, F.; Fiorito, V.; Mercurio, S.; Tolosano, E. Heme in pathophysiology: A matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 2014, 5, 61. [Google Scholar] [CrossRef] [Green Version]
- Larsen, R.; Gouveia, Z.; Soares, M.P.; Gozzelino, R. Heme cytotoxicity and the pathogenesis of immune-mediated inflammatory diseases. Front. Pharmacol. 2012, 3, 77. [Google Scholar] [CrossRef] [Green Version]
- Hopp, M.T.; Schmalohr, B.F.; Kuhl, T.; Detzel, M.S.; Wissbrock, A.; Imhof, D. Heme Determination and Quantification Methods and Their Suitability for Practical Applications and Everyday Use. Anal. Chem. 2020, 92, 9429–9440. [Google Scholar] [CrossRef]
- Khan, A.A.; Quigley, J.G. Control of intracellular heme levels: Heme transporters and heme oxygenases. Biochim. Biophys. Acta BBA Mol. Cell Res. 2011, 1813, 668–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanna, D.A.; Harvey, R.M.; Martinez-Guzman, O.; Yuan, X.; Chandrasekharan, B.; Raju, G.; Outten, F.W.; Hamza, I.; Reddi, A.R. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. USA 2016, 113, 7539–7544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascaro, M.; Alonso, E.N.; Alonso, E.G.; Lacunza, E.; Curino, A.C.; Facchinetti, M.M. Nuclear Localization of Heme Oxygenase-1 in Pathophysiological Conditions: Does It Explain the Dual Role in Cancer? Antioxidants 2021, 10, 87. [Google Scholar] [CrossRef]
- Fiorito, V.; Chiabrando, D.; Petrillo, S.; Bertino, F.; Tolosano, E. The Multifaceted Role of Heme in Cancer. Front. Oncol. 2019, 9, 1540. [Google Scholar] [CrossRef] [Green Version]
- Fiorito, V.; Allocco, A.L.; Petrillo, S.; Gazzano, E.; Torretta, S.; Marchi, S.; Destefanis, F.; Pacelli, C.; Audrito, V.; Provero, P.; et al. The heme synthesis-export system regulates the tricarboxylic acid cycle flux and oxidative phosphorylation. Cell Rep. 2021, 35, 109252. [Google Scholar] [CrossRef]
- Kalainayakan, S.P.; FitzGerald, K.E.; Konduri, P.C.; Vidal, C.; Zhang, L. Essential roles of mitochondrial and heme function in lung cancer bioenergetics and tumorigenesis. Cell Biosci. 2018, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Sohoni, S.; Ghosh, P.; Wang, T.; Kalainayakan, S.P.; Vidal, C.; Dey, S.; Konduri, P.C.; Zhang, L. Elevated Heme Synthesis and Uptake Underpin Intensified Oxidative Metabolism and Tumorigenic Functions in Non-Small Cell Lung Cancer Cells. Cancer Res. 2019, 79, 2511–2525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashton, T.M.; McKenna, W.G.; Kunz-Schughart, L.A.; Higgins, G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 2018, 24, 2482–2490. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, P.; Vidal, C.; Dey, S.; Zhang, L. Mitochondria Targeting as an Effective Strategy for Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 3363. [Google Scholar] [CrossRef]
- Sandoval-Acuna, C.; Torrealba, N.; Tomkova, V.; Jadhav, S.B.; Blazkova, K.; Merta, L.; Lettlova, S.; Adamcova, M.K.; Rosel, D.; Brabek, J.; et al. Targeting Mitochondrial Iron Metabolism Suppresses Tumor Growth and Metastasis by Inducing Mitochondrial Dysfunction and Mitophagy. Cancer Res. 2021, 81, 2289–2303. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Guo, Y.; Ashrafi, A.; Chen, J.; Dey, S.; Zhong, S.; Liu, J.; Campbell, J.; Konduri, P.C.; Gerberich, J.; et al. Oxygen-Enhanced Optoacoustic Tomography Reveals the Effectiveness of Targeting Heme and Oxidative Phosphorylation at Normalizing Tumor Vascular Oxygenation. Cancer Res. 2020, 80, 3542–3555. [Google Scholar] [CrossRef] [PubMed]
- Hooda, J.; Cadinu, D.; Alam, M.M.; Shah, A.; Cao, T.M.; Sullivan, L.A.; Brekken, R.; Zhang, L. Enhanced heme function and mitochondrial respiration promote the progression of lung cancer cells. PLoS ONE 2013, 8, e63402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalainayakan, S.P.; Ghosh, P.; Dey, S.; Fitzgerald, K.E.; Sohoni, S.; Konduri, P.C.; Garrossian, M.; Liu, L.; Zhang, L. Cyclopamine tartrate, a modulator of hedgehog signaling and mitochondrial respiration, effectively arrests lung tumor growth and progression. Sci. Rep. 2019, 9, 1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.G.; Chudnovskiy, A.; Baudrier, L.; Prizer, B.; Liu, Y.; Ostendorf, B.N.; Yamaguchi, N.; Arab, A.; Tavora, B.; Timson, R.; et al. Functional Genomics In Vivo Reveal Metabolic Dependencies of Pancreatic Cancer Cells. Cell Metab. 2021, 33, 211–221. [Google Scholar] [CrossRef]
- Biancur, D.E.; Kapner, K.S.; Yamamoto, K.; Banh, R.S.; Neggers, J.E.; Sohn, A.S.W.; Wu, W.; Manguso, R.T.; Brown, A.; Root, D.E.; et al. Functional Genomics Identifies Metabolic Vulnerabilities in Pancreatic Cancer. Cell Metab. 2021, 33, 199–210. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Red Meat and Processed Meat; International Agency for Research on Cancer: Lyon, France, 2018. [Google Scholar]
- Cross, A.J.; Sinha, R. Meat-related mutagens/carcinogens in the etiology of colorectal cancer. Environ. Mol. Mutagenesis 2004, 44, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Felton, J.S.; Knize, M.G.; Wu, R.W.; Colvin, M.E.; Hatch, F.T.; Malfatti, M.A. Mutagenic potency of food-derived heterocyclic amines. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2007, 616, 90–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunter, M.J.; Divi, R.L.; Kulldorff, M.; Vermeulen, R.; Haverkos, K.J.; Kuo, M.M.; Strickland, P.; Poirier, M.C.; Rothman, N.; Sinha, R. Leukocyte polycyclic aromatic hydrocarbon–DNA adduct formation and colorectal adenoma. Carcinogenesis 2007, 28, 1426–1429. [Google Scholar] [CrossRef] [Green Version]
- Samraj, A.; Läubli, H.; Varki, N.; Varki, A. Involvement of a non-human sialic acid in human cancer. Front. Oncol. 2014, 4, 33. [Google Scholar] [CrossRef] [Green Version]
- Hedlund, M.; Padler-Karavani, V.; Varki, N.M.; Varki, A. Evidence for a human-specific mechanism for diet and antibody-mediated inflammation in carcinoma progression. Proc. Natl. Acad. Sci. USA 2008, 105, 18936–18941. [Google Scholar] [CrossRef] [Green Version]
- Samraj, A.N.; Pearce, O.M.; Läubli, H.; Crittenden, A.N.; Bergfeld, A.K.; Banda, K.; Gregg, C.J.; Bingman, A.E.; Secrest, P.; Diaz, S.L. A red meat-derived glycan promotes inflammation and cancer progression. Proc. Natl. Acad. Sci. USA 2015, 112, 542–547. [Google Scholar] [CrossRef] [Green Version]
- Keller, J.; Chevolleau, S.; Noguer-Meireles, M.H.; Pujos-Guillot, E.; Delosiere, M.; Chantelauze, C.; Joly, C.; Blas, Y.E.F.; Jouanin, I.; Durand, D.; et al. Heme-Iron-Induced Production of 4-Hydroxynonenal in Intestinal Lumen May Have Extra-Intestinal Consequences through Protein-Adduct Formation. Antioxidants 2020, 9, 1293. [Google Scholar] [CrossRef]
- Pierre, F.; Tache, S.; Gueraud, F.; Rerole, A.L.; Jourdan, M.L.; Petit, C. Apc mutation induces resistance of colonic cells to lipoperoxide-triggered apoptosis induced by faecal water from haem-fed rats. Carcinogenesis 2007, 28, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Bastide, N.M.; Chenni, F.; Audebert, M.; Santarelli, R.L.; Tache, S.; Naud, N.; Baradat, M.; Jouanin, I.; Surya, R.; Hobbs, D.A.; et al. A central role for heme iron in colon carcinogenesis associated with red meat intake. Cancer Res. 2015, 75, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Surya, R.; Helies-Toussaint, C.; Martin, O.C.; Gauthier, T.; Gueraud, F.; Tache, S.; Naud, N.; Jouanin, I.; Chantelauze, C.; Durand, D.; et al. Red meat and colorectal cancer: Nrf2-dependent antioxidant response contributes to the resistance of preneoplastic colon cells to fecal water of hemoglobin- and beef-fed rats. Carcinogenesis 2016, 37, 635–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodagoda Gamage, S.M.; Cheng, T.; Lee, K.T.; Dissabandara, L.; Lam, A.K.; Gopalan, V. Hemin, a major heme molecule, induced cellular and genetic alterations in normal colonic and colon cancer cells. Pathol.-Res. Pract. 2021, 224, 153530. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.K.; Chen, S.E.; Chang, L.C. A Dual Role of Heme Oxygenase-1 in Cancer Cells. Int. J. Mol. Sci. 2018, 20, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chau, L.Y. Heme oxygenase-1: Emerging target of cancer therapy. J. Biomed. Sci. 2015, 22, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitti, M.; Ivaldo, C.; Traverso, N.; Furfaro, A.L. Clinical Significance of Heme Oxygenase 1 in Tumor Progression. Antioxidants 2021, 10, 789. [Google Scholar] [CrossRef]
- Luu Hoang, K.N.; Anstee, J.E.; Arnold, J.N. The Diverse Roles of Heme Oxygenase-1 in Tumor Progression. Front. Immunol. 2021, 12, 658315. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, Y.; Xu, Y.; Ma, Q.; Guo, F.; Zhao, Y.; Tao, Y.; Li, M.; Guo, J. Nrf2/HO-1 Axis Regulates the Angiogenesis of Gastric Cancer via Targeting VEGF. Cancer Manag. Res. 2021, 13, 3155–3169. [Google Scholar] [CrossRef]
- Dulak, J.; Józkowicz, A.; Foresti, R.; Kasza, A.; Frick, M.; Huk, I.; Green, C.J.; Pachinger, O.; Weidinger, F.; Motterlini, R. Heme Oxygenase Activity Modulates Vascular Endothelial Growth Factor Synthesis in Vascular Smooth Muscle Cells. Antioxid. Redox Signal. 2002, 4, 229–240. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, K.-S.; Lee, D.-K.; Kim, J.; Kwak, S.-N.; Ha, K.-S.; Choe, J.; Won, M.-H.; Cho, B.-R.; Jeoung, D.; et al. Hypoxia-Responsive MicroRNA-101 Promotes Angiogenesis via Heme Oxygenase-1/Vascular Endothelial Growth Factor Axis by Targeting Cullin 3. Antioxid. Redox Signal. 2014, 21, 2469–2482. [Google Scholar] [CrossRef] [Green Version]
- Birrane, G.; Li, H.; Yang, S.; Tachado, S.D.; Seng, S. Cigarette smoke induces nuclear translocation of heme oxygenase 1 (HO-1) in prostate cancer cells: Nuclear HO-1 promotes vascular endothelial growth factor secretion. Int. J. Oncol. 2013, 42, 1919–1928. [Google Scholar] [CrossRef] [Green Version]
- Miyake, M.; Fujimoto, K.; Anai, S.; Ohnishi, S.; Kuwada, M.; Nakai, Y.; Inoue, T.; Matsumura, Y.; Tomioka, A.; Ikeda, T.; et al. Heme oxygenase-1 promotes angiogenesis in urothelial carcinoma of the urinary bladder. Oncol. Rep. 2011, 25, 653–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Yang, Y.; Huang, Y.; Ma, Q.; Shang, J.; Guo, J.; Cao, X.; Wang, X.; Li, M. Inhibition of Nrf2/HO-1 signaling pathway by Dextran Sulfate suppresses angiogenesis of Gastric Cancer. J. Cancer 2021, 12, 1042–1060. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Kim, C.-K.; Lee, H.; Jeoung, D.; Ha, K.-S.; Kwon, Y.-G.; Kim, K.-W.; Kim, Y.-M. Carbon monoxide promotes VEGF expression by increasing HIF-1alpha protein level via two distinct mechanisms, translational activation and stabilization of HIF-1alpha protein. J. Biol. Chem. 2010, 285, 32116–32125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.-C.; Guan, S.-S.; Yang, H.-J.; Chang, C.-C.; Luo, T.-Y.; Chang, J.; Ho, A.-S. Blocking heme oxygenase-1 by zinc protoporphyrin reduces tumor hypoxia-mediated VEGF release and inhibits tumor angiogenesis as a potential therapeutic agent against colorectal cancer. J. Biomed. Sci. 2016, 23, 18. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.K.; Kim, Y.M. Regulation of Endothelial and Vascular Functions by Carbon Monoxide via Crosstalk With Nitric Oxide. Front. Cardiovasc. Med. 2021, 8, 649630. [Google Scholar] [CrossRef]
- Alaluf, E.; Vokaer, B.; Detavernier, A.; Azouz, A.; Splittgerber, M.; Carrette, A.; Boon, L.; Libert, F.; Soares, M.; Le Moine, A.; et al. Heme oxygenase-1 orchestrates the immunosuppressive program of tumor-associated macrophages. JCI Insight 2020, 5, e133929. [Google Scholar] [CrossRef]
- Chauveau, C.; Rémy, S.; Royer, P.J.; Hill, M.; Tanguy-Royer, S.; Hubert, F.-X.; Tesson, L.; Brion, R.; Beriou, G.; Gregoire, M.; et al. Heme oxygenase-1 expression inhibits dendritic cell maturation and proinflammatory function but conserves IL-10 expression. Blood 2005, 106, 1694–1702. [Google Scholar] [CrossRef] [Green Version]
- Rémy, S.; Blancou, P.; Tesson, L.; Tardif, V.; Brion, R.; Royer, P.J.; Motterlini, R.; Foresti, R.; Painchaut, M.; Pogu, S.; et al. Carbon Monoxide Inhibits TLR-Induced Dendritic Cell Immunogenicity. J. Immunol. 2009, 182, 1877–1884. [Google Scholar] [CrossRef] [Green Version]
- Moreau, A.; Hill, M.; Thébault, P.; Deschamps, J.Y.; Chiffoleau, E.; Chauveau, C.; Moullier, P.; Anegon, I.; Alliot-Licht, B.; Cuturi, M.C. Tolerogenic dendritic cells actively inhibit T cells through heme oxygenase-1 in rodents and in nonhuman primates. FASEB J. 2009, 23, 3070–3077. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.K.; Fitzgerald, H.K.; Dunne, A. Regulation of inflammation by the antioxidant haem oxygenase 1. Nat. Rev. Immunol. 2021, 21, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Jiang, J.; Ma, Q.; Wu, Z.; Wang, Z. The inhibition of heme oxygenase-1 enhances the chemosensitivity and suppresses the proliferation of pancreatic cancer cells through the SHH signaling pathway. Int. J. Oncol. 2018, 52, 2101–2109. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.J.; Kim, K.M.; Jang, K.Y. Individual and co-expression patterns of nerve growth factor and heme oxygenase-1 predict shorter survival of gastric carcinoma patients. Diagn. Pathol. 2017, 12, 48. [Google Scholar] [CrossRef]
- Smith, A.G.; Raven, E.L.; Chernova, T. The regulatory role of heme in neurons. Metallomics 2011, 3, 955–962. [Google Scholar] [CrossRef]
- Gozzelino, R. The Pathophysiology of Heme in the Brain. Curr. Alzheimer. Res. 2016, 13, 174–184. [Google Scholar] [CrossRef]
- Deininger, M.H.; Meyermann, R.; Trautmann, K.; Duffner, F.; Grote, E.H.; Wickboldt, J.; Schluesener, H.J. Heme oxygenase (HO)-1 expressing macrophages/microglial cells accumulate during oligodendroglioma progression. Brain Res. 2000, 882, 1–8. [Google Scholar] [CrossRef]
- Hsu, F.F.; Yeh, C.T.; Sun, Y.J.; Chiang, M.T.; Lan, W.M.; Li, F.A.; Lee, W.H.; Chau, L.Y. Signal peptide peptidase-mediated nuclear localization of heme oxygenase-1 promotes cancer cell proliferation and invasion independent of its enzymatic activity. Oncogene 2015, 34, 2360–2370. [Google Scholar] [CrossRef] [PubMed]
- Consonni, F.M.; Bleve, A.; Totaro, M.G.; Storto, M.; Kunderfranco, P.; Termanini, A.; Pasqualini, F.; Ali, C.; Pandolfo, C.; Sgambelluri, F.; et al. Heme catabolism by tumor-associated macrophages controls metastasis formation. Nat. Immunol. 2021, 22, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Gueron, G.; De Siervi, A.; Ferrando, M.; Salierno, M.; De Luca, P.; Elguero, B.; Meiss, R.; Navone, N.; Vazquez, E.S. Critical Role of Endogenous Heme Oxygenase 1 as a Tuner of the Invasive Potential of Prostate Cancer Cells. Mol. Cancer Res. 2009, 7, 1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrzypek, K.; Tertil, M.; Golda, S.; Ciesla, M.; Weglarczyk, K.; Collet, G.; Guichard, A.; Kozakowska, M.; Boczkowski, J.; Was, H.; et al. Interplay between heme oxygenase-1 and miR-378 affects non-small cell lung carcinoma growth, vascularization, and metastasis. Antioxid. Redox Signal. 2013, 19, 644–660. [Google Scholar] [CrossRef] [Green Version]
- Tertil, M.; Golda, S.; Skrzypek, K.; Florczyk, U.; Weglarczyk, K.; Kotlinowski, J.; Maleszewska, M.; Czauderna, S.; Pichon, C.; Kieda, C.; et al. Nrf2-heme oxygenase-1 axis in mucoepidermoid carcinoma of the lung: Antitumoral effects associated with down-regulation of matrix metalloproteinases. Free Radic. Biol. Med. 2015, 89, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.; Zou, C.; Cheng, W.; Li, Q.; Han, Z.; Wang, X.; Jin, J.; Zou, J.; Liu, Z.; Zhou, Z.; et al. Heme oxygenase-1 retards hepatocellular carcinoma progression through the microRNA pathway. Oncol. Rep. 2016, 36, 2715–2722. [Google Scholar] [CrossRef] [Green Version]
- Hill, M.; Pereira, V.; Chauveau, C.; Zagani, R.; Remy, S.; Tesson, L.; Mazal, D.; Ubillos, L.; Brion, R.; Ashgar, K.; et al. Heme oxygenase-1 inhibits rat and human breast cancer cell proliferation: Mutual cross inhibition with indoleamine 2,3-dioxygenase. FASEB J. 2005, 19, 1957–1968. [Google Scholar] [CrossRef]
- Ferrando, M.; Gueron, G.; Elguero, B.; Giudice, J.; Salles, A.; Leskow, F.C.; Jares-Erijman, E.A.; Colombo, L.; Meiss, R.; Navone, N.; et al. Heme oxygenase 1 (HO-1) challenges the angiogenic switch in prostate cancer. Angiogenesis 2011, 14, 467–479. [Google Scholar] [CrossRef]
- Podkalicka, P.; Mucha, O.; Józkowicz, A.; Dulak, J.; Łoboda, A. Heme oxygenase inhibition in cancers: Possible tools and targets. Contemp. Oncol. 2018, 22, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Gueron, G.; Giudice, J.; Valacco, P.; Paez, A.; Elguero, B.; Toscani, M.; Jaworski, F.; Leskow, F.C.; Cotignola, J.; Marti, M.; et al. Heme-oxygenase-1 implications in cell morphology and the adhesive behavior of prostate cancer cells. Oncotarget 2014, 5, 4087–4102. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.N.; Park, G.H.; Park, S.B.; Kim, J.D.; Eo, H.J.; Son, H.J.; Song, J.H.; Jeong, J.B. Extracts from Sageretia thea reduce cell viability through inducing cyclin D1 proteasomal degradation and HO-1 expression in human colorectal cancer cells. BMC Complement. Altern. Med. 2019, 19, 43. [Google Scholar] [CrossRef]
- Bi, W.; He, C.N.; Li, X.X.; Zhou, L.Y.; Liu, R.J.; Zhang, S.; Li, G.Q.; Chen, Z.C.; Zhang, P.F. Ginnalin A from Kujin tea (Acer tataricum subsp. ginnala) exhibits a colorectal cancer chemoprevention effect via activation of the Nrf2/HO-1 signaling pathway. Food Funct. 2018, 9, 2809–2819. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-F.; Chen, J.-H.; Chang, C.-N.; Lu, D.-Y.; Chang, P.-C.; Wang, S.-L.; Yeh, W.-L. Fisetin inhibits cell migration via inducing HO-1 and reducing MMPs expression in breast cancer cell lines. Food Chem. Toxicol. 2018, 120, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Sferrazzo, G.; Di Rosa, M.; Barone, E.; Li Volti, G.; Musso, N.; Tibullo, D.; Barbagallo, I. Heme Oxygenase-1 in Central Nervous System Malignancies. J. Clin. Med. 2020, 9, 1562. [Google Scholar] [CrossRef] [PubMed]
- Waza, A.A.; Hamid, Z.; Ali, S.; Bhat, S.A.; Bhat, M.A. A review on heme oxygenase-1 induction: Is it a necessary evil. Inflamm. Res. 2018, 67, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Suttner, D.M.; Dennery, P.A. Reversal of HO-1 related cytoprotection with increased expression is due to reactive iron. FASEB J. 1999, 13, 1800–1809. [Google Scholar] [CrossRef]
- Kwon, M.-Y.; Park, E.; Lee, S.-J.; Chung, S.W. Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget 2015, 6, 24393–24403. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Zhao, Y.; Wang, J.; Yang, X.; Li, S.; Wang, Y.; Yang, X.; Fei, J.; Hao, X.; Zhao, Y.; et al. Tagitinin C induces ferroptosis through PERK-Nrf2-HO-1 signaling pathway in colorectal cancer cells. Int. J. Biol. Sci. 2021, 17, 2703–2717. [Google Scholar] [CrossRef]
- Malfa, G.A.; Tomasello, B.; Acquaviva, R.; Genovese, C.; La Mantia, A.; Cammarata, F.P.; Ragusa, M.; Renis, M.; Di Giacomo, C. Betula etnensis Raf. (Betulaceae) Extract Induced HO-1 Expression and Ferroptosis Cell Death in Human Colon Cancer Cells. Int. J. Mol. Sci. 2019, 20, 2723. [Google Scholar] [CrossRef] [Green Version]
- Gandini, N.A.; Alonso, E.N.; Fermento, M.E.; Mascaró, M.; Abba, M.C.; Coló, G.P.; Arévalo, J.; Ferronato, M.J.; Guevara, J.A.; Núñez, M.; et al. Heme Oxygenase-1 Has an Antitumor Role in Breast Cancer. Antioxid. Redox Signal. 2019, 30, 2030–2049. [Google Scholar] [CrossRef]
- Alex, A.; Luo, Q.; Mathew, D.; Di, R.; Bhatwadekar, A.D. Metformin Corrects Abnormal Circadian Rhythm and Kir4.1 Channels in Diabetes. Investig. Ophthalmol. Vis. Sci. 2020, 61, 46. [Google Scholar] [CrossRef]
- Gu, F.; Zhang, H.; Hyland, P.L.; Berndt, S.; Gapstur, S.M.; Wheeler, W.; Consortium, T.E.; Amos, C.I.; Bezieau, S.; Bickeböller, H.; et al. Inherited variation in circadian rhythm genes and risks of prostate cancer and three other cancer sites in combined cancer consortia. Int. J. Cancer 2017, 141, 1794–1802. [Google Scholar] [CrossRef]
- Hou, T.; Su, W.; Guo, Z.; Gong, M.C. A Novel Diabetic Mouse Model for Real-Time Monitoring of Clock Gene Oscillation and Blood Pressure Circadian Rhythm. J. Biol. Rhythm. 2019, 34, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Hudec, M.; Dankova, P.; Solc, R.; Bettazova, N.; Cerna, M. Epigenetic Regulation of Circadian Rhythm and Its Possible Role in Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 3005. [Google Scholar] [CrossRef] [PubMed]
- Mason, I.C.; Qian, J.; Adler, G.K.; Scheer, F.A.J.L. Impact of circadian disruption on glucose metabolism: Implications for type 2 diabetes. Diabetologia 2020, 63, 462–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simcox, J.A.; Mitchell, T.C.; Gao, Y.; Just, S.F.; Cooksey, R.; Cox, J.; Ajioka, R.; Jones, D.; Lee, S.-h.; King, D.; et al. Dietary Iron Controls Circadian Hepatic Glucose Metabolism Through Heme Synthesis. Diabetes 2015, 64, 1108–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordina-Duverger, E.; Menegaux, F.; Popa, A.; Rabstein, S.; Harth, V.; Pesch, B.; Brüning, T.; Fritschi, L.; Glass, D.C.; Heyworth, J.S.; et al. Night shift work and breast cancer: A pooled analysis of population-based case-control studies with complete work history. Eur. J. Epidemiol. 2018, 33, 369–379. [Google Scholar] [CrossRef]
- de Assis, L.V.M.; Kinker, G.S.; Moraes, M.N.; Markus, R.P.; Fernandes, P.A.; Castrucci, A.M.d.L. Expression of the Circadian Clock Gene BMAL1 Positively Correlates With Antitumor Immunity and Patient Survival in Metastatic Melanoma. Front. Oncol. 2018, 8, 185. [Google Scholar] [CrossRef] [Green Version]
- Huisman, S.A.; Ahmadi, A.R.; Ijzermans, J.N.M.; Verhoef, C.; van der Horst, G.T.J.; de Bruin, R.W.F. Disruption of clock gene expression in human colorectal liver metastases. Tumor Biol. 2016, 37, 13973–13981. [Google Scholar] [CrossRef] [Green Version]
- Leung, L.; Grundy, A.; Siemiatycki, J.; Arseneau, J.; Gilbert, L.; Gotlieb, W.H.; Provencher, D.M.; Aronson, K.J.; Koushik, A. Shift Work Patterns, Chronotype, and Epithelial Ovarian Cancer Risk. Cancer Epidemiol. Prev. Biomark. 2019, 28, 987–995. [Google Scholar] [CrossRef] [Green Version]
- Papantoniou, K.; Devore, E.E.; Massa, J.; Strohmaier, S.; Vetter, C.; Yang, L.; Shi, Y.; Giovannucci, E.; Speizer, F.; Schernhammer, E.S. Rotating night shift work and colorectal cancer risk in the nurses’ health studies. Int. J. Cancer 2018, 143, 2709–2717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reszka, E.; Przybek, M.; Muurlink, O.; Pepłonska, B. Circadian gene variants and breast cancer. Cancer Lett. 2017, 390, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Salamanca-Fernández, E.; Rodríguez-Barranco, M.; Guevara, M.; Ardanaz, E.; Olry de Labry Lima, A.; Sánchez, M.J. Night-shift work and breast and prostate cancer risk: Updating the evidence from epidemiological studies. An. Del Sist. Sanit. De Navar. 2018, 41, 211–226. [Google Scholar] [CrossRef] [Green Version]
- Soták, M.; Polidarová, L.; Ergang, P.; Sumová, A.; Pácha, J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int. J. Cancer 2013, 132, 1032–1041. [Google Scholar] [CrossRef]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef] [PubMed]
- Wendeu-Foyet, M.G.; Bayon, V.; Cénée, S.; Trétarre, B.; Rébillard, X.; Cancel-Tassin, G.; Cussenot, O.; Lamy, P.-J.; Faraut, B.; Ben Khedher, S.; et al. Night work and prostate cancer risk: Results from the EPICAP Study. Occup. Environ. Med. 2018, 75, 573–581. [Google Scholar] [CrossRef]
- Group, I.M.V. Carcinogenicity of night shift work. Lancet Oncol. 2019, 20, 1058–1059. [Google Scholar] [CrossRef]
- Freeman, S.L.; Kwon, H.; Portolano, N.; Parkin, G.; Girija, U.V.; Basran, J.; Fielding, A.J.; Fairall, L.; Svistunenko, D.A.; Moody, P.C. Heme binding to human CLOCK affects interactions with the E-box. Proc. Natl. Acad. Sci. USA 2019, 116, 19911–19916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astone, M.; Santoro, M.M. Time to fight: Targeting the circadian clock molecular machinery in cancer therapy. Drug Discov. Today 2021, 26, 1164–1184. [Google Scholar] [CrossRef]
- Airola, M.V.; Du, J.; Dawson, J.H.; Crane, B.R. Heme Binding to the Mammalian Circadian Clock Protein Period 2 is Non-Specific. Biochemistry 2010, 49, 4327–4338. [Google Scholar] [CrossRef] [Green Version]
- Kaasik, K.; Lee, C.C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 2004, 430, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Lukat-Rodgers, G.S.; Correia, C.; Botuyan, M.V.; Mer, G.; Rodgers, K.R. Heme-based sensing by the mammalian circadian protein CLOCK. Inorg. Chem. 2010, 49, 6349–6365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Wu, N.; Lazar, M.A. Nuclear Receptor Rev-Erbα: A Heme Receptor that Coordinates Circadian Rhythm and Metabolism. Nucl. Recept. Signal. 2010, 8, nrs.08001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghuram, S.; Stayrook, K.R.; Huang, P.; Rogers, P.M.; Nosie, A.K.; McClure, D.B.; Burris, L.L.; Khorasanizadeh, S.; Burris, T.P.; Rastinejad, F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol. 2007, 14, 1207–1213. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Wu, N.; Curtin, J.C.; Qatanani, M.; Szwergold, N.R.; Reid, R.A.; Waitt, G.M.; Parks, D.J.; Pearce, K.H.; Wisely, G.B.; et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 2007, 318, 1786–1789. [Google Scholar] [CrossRef]
- Matta-Camacho, E.; Banerjee, S.; Hughes, T.S.; Solt, L.A.; Wang, Y.; Burris, T.P.; Kojetin, D.J. Structure of REV-ERBβ ligand-binding domain bound to a porphyrin antagonist. J. Biol. Chem. 2014, 289, 20054–20066. [Google Scholar] [CrossRef] [Green Version]
- Fleischhacker, A.S.; Carter, E.L.; Ragsdale, S.W. Redox Regulation of Heme Oxygenase-2 and the Transcription Factor, Rev-Erb, Through Heme Regulatory Motifs. Antioxid. Redox Signal. 2018, 29, 1841–1857. [Google Scholar] [CrossRef]
- Klemz, R.; Reischl, S.; Wallach, T.; Witte, N.; Jürchott, K.; Klemz, S.; Lang, V.; Lorenzen, S.; Knauer, M.; Heidenreich, S.; et al. Reciprocal regulation of carbon monoxide metabolism and the circadian clock. Nat. Struct. Mol. Biol. 2017, 24, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liu, A.; Lin, Z.; Wang, B.; Chai, X.; Chen, S.; Lu, W.; Zheng, M.; Cao, T.; Zhong, M.; et al. Downregulation of the circadian rhythm regulator HLF promotes multiple-organ distant metastases in non-small cell lung cancer through PPAR/NF-κb signaling. Cancer Lett. 2020, 482, 56–71. [Google Scholar] [CrossRef]
- Burgermeister, E.; Battaglin, F.; Eladly, F.; Wu, W.; Herweck, F.; Schulte, N.; Betge, J.; Hartel, N.; Kather, J.N.; Weis, C.A.; et al. Aryl hydrocarbon receptor nuclear translocator-like (ARNTL/BMAL1) is associated with bevacizumab resistance in colorectal cancer via regulation of vascular endothelial growth factor A. EBioMedicine 2019, 45, 139–154. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Bonkovsky, H.L.; Guo, J.-t. Structural analysis of heme proteins: Implications for design and prediction. BMC Struct. Biol. 2011, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Syllwasschy, B.F.; Beck, M.S.; Druzeta, I.; Hopp, M.T.; Ramoji, A.; Neugebauer, U.; Nozinovic, S.; Menche, D.; Willbold, D.; Ohlenschlager, O.; et al. High-affinity binding and catalytic activity of His/Tyr-based sequences: Extending heme-regulatory motifs beyond CP. Biochim. Biophys. Acta BBA Gen. Subj. 2020, 1864, 129603. [Google Scholar] [CrossRef]
- Zhang, L.; Guarente, L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995, 14, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Girvan, H.M.; Munro, A.W. Heme sensor proteins. J. Biol. Chem. 2013, 288, 13194–13203. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, K.; Sun, J.; Taketani, S.; Nakajima, O.; Nishitani, C.; Sassa, S.; Hayashi, N.; Yamamoto, M.; Shibahara, S.; Fujita, H. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001, 20, 2835–2843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe-Matsui, M.; Matsumoto, T.; Matsui, T.; Ikeda-Saito, M.; Muto, A.; Murayama, K.; Igarashi, K. Heme binds to an intrinsically disordered region of Bach2 and alters its conformation. Arch. Biochem. Biophys. 2015, 565, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, K. Human tryptophanyl-tRNA synthetase binds with heme to enhance its aminoacylation activity. Biochemistry 2007, 46, 11291–11298. [Google Scholar] [CrossRef]
- Yang, F.; Xia, X.; Lei, H.-Y.; Wang, E.-D. Hemin binds to human cytoplasmic arginyl-tRNA synthetase and inhibits its catalytic activity. J. Biol. Chem. 2010, 285, 39437–39446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partin, A.C.; Ngo, T.D.; Herrell, E.; Jeong, B.-C.; Hon, G.; Nam, Y. Heme enables proper positioning of Drosha and DGCR8 on primary microRNAs. Nat. Commun. 2017, 8, 1737. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Mitra, S.; Wu, G.; Berka, V.; Song, J.; Yu, Y.; Poget, S.; Wang, D.-N.; Tsai, A.-L.; Zhou, M. Six-transmembrane epithelial antigen of prostate 1 (STEAP1) has a single b heme and is capable of reducing metal ion complexes and oxygen. Biochemistry 2016, 55, 6673–6684. [Google Scholar] [CrossRef] [PubMed]
- Kleven, M.D.; Dlakić, M.; Lawrence, C.M. Characterization of a single b-type heme, FAD, and metal binding sites in the transmembrane domain of six-transmembrane epithelial antigen of the prostate (STEAP) family proteins. J. Biol. Chem. 2015, 290, 22558–22569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganasen, M.; Togashi, H.; Takeda, H.; Asakura, H.; Tosha, T.; Yamashita, K.; Hirata, K.; Nariai, Y.; Urano, T.; Yuan, X. Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate. Commun. Biol. 2018, 1, 120. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, Y.; Okutani, H.; Takeda, Y.; Uchida, T.; Iwai, K.; Ishimori, K. Specific heme binding to heme regulatory motifs in iron regulatory proteins and its functional significance. J. Inorg. Biochem. 2019, 198, 110726. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Nomura, K.; Katoh, Y.; Yamashita, R.; Kaneko, K.; Furuyama, K. Novel mechanisms for heme-dependent degradation of ALAS1 protein as a component of negative feedback regulation of heme biosynthesis. J. Biol. Chem. 2016, 291, 20516–20529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Dumbrepatil, A.B.; Fleischhacker, A.S.; Marsh, E.N.G.; Ragsdale, S.W. Heme oxygenase-2 is post-translationally regulated by heme occupancy in the catalytic site. J. Biol. Chem. 2020, 295, 17227–17240. [Google Scholar] [CrossRef]
- Uchida, T.; Sagami, I.; Shimizu, T.; Ishimori, K.; Kitagawa, T. Effects of the bHLH domain on axial coordination of heme in the PAS-A domain of neuronal PAS domain protein 2 (NPAS2): Conversion from His119/Cys170 coordination to His119/His171 coordination. J. Inorg. Biochem. 2012, 108, 188–195. [Google Scholar] [CrossRef]
- Yang, J.; Kim, K.D.; Lucas, A.; Drahos, K.E.; Santos, C.S.; Mury, S.P.; Capelluto, D.G.; Finkielstein, C.V. A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol. Cell. Biol. 2008, 28, 4697–4711. [Google Scholar] [CrossRef] [Green Version]
- Okano, S.; Akashi, M.; Hayasaka, K.; Nakajima, O. Unusual circadian locomotor activity and pathophysiology in mutant CRY1 transgenic mice. Neurosci. Lett. 2009, 451, 246–251. [Google Scholar] [CrossRef]
- Zhao, L.N.; Mu, Y.; Chew, L.Y. Heme prevents amyloid beta peptide aggregation through hydrophobic interaction based on molecular dynamics simulation. Phys. Chem. Chem. Phys. 2013, 15, 14098–14106. [Google Scholar] [CrossRef] [Green Version]
- Wißbrock, A.; Goradia, N.B.; Kumar, A.; George, A.A.P.; Kühl, T.; Bellstedt, P.; Ramachandran, R.; Hoffmann, P.; Galler, K.; Popp, J. Structural insights into heme binding to IL-36α proinflammatory cytokine. Sci. Rep. 2019, 9, 16893. [Google Scholar] [CrossRef]
- Wiel, C.; Le Gal, K.; Ibrahim, M.X.; Jahangir, C.A.; Kashif, M.; Yao, H.; Ziegler, D.V.; Xu, X.; Ghosh, T.; Mondal, T.; et al. BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 2019, 178, 330–345.e22. [Google Scholar] [CrossRef]
- Lee, J.; Yesilkanal, A.E.; Wynne, J.P.; Frankenberger, C.; Liu, J.; Yan, J.; Elbaz, M.; Rabe, D.C.; Rustandy, F.D.; Tiwari, P.; et al. Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature 2019, 568, 254–258. [Google Scholar] [CrossRef]
- Kabe, Y.; Nakane, T.; Koike, I.; Yamamoto, T.; Sugiura, Y.; Harada, E.; Sugase, K.; Shimamura, T.; Ohmura, M.; Muraoka, K. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 2016, 7, 11030. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Sheng, X.; Chang, Z.; Wu, Q.; Wang, S.; Xuan, Z.; Li, D.; Wu, Y.; Shang, Y.; Kong, X. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014, 7, 180–193. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Wißbrock, A.; Goradia, N.; Bellstedt, P.; Ramachandran, R.; Imhof, D.; Ohlenschläger, O. Heme interaction of the intrinsically disordered N-terminal peptide segment of human cystathionine-β-synthase. Sci. Rep. 2018, 8, 2474. [Google Scholar] [CrossRef] [Green Version]
- Sotolongo, A.; Monica, F.Z.; Kots, A.; Xiao, H.; Liu, J.; Seto, E.; Bian, K.; Murad, F. Epigenetic regulation of soluble guanylate cyclase (sGC) beta1 in breast cancer cells. FASEB J. 2016, 30, 3171–3180. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhou, S.; Pang, L.; Yang, J.; Li, H.J.; Huo, X.; Qian, S.Y. Celastrol suppresses nitric oxide synthases and the angiogenesis pathway in colorectal cancer. Free Radic. Res. 2019, 53, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Segawa, K.; Watanabe-Matsui, M.; Matsui, T.; Igarashi, K.; Murayama, K. Functional Heme Binding to the Intrinsically Disordered C-Terminal Region of Bach1, a Transcriptional Repressor. Tohoku J. Exp. Med. 2019, 247, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jian, L.; Mei, Y.; Xing, C.; Rongdi, Y. Haem relieves hyperoxia-mediated inhibition of HMEC-1 cell proliferation, migration and angiogenesis by inhibiting BACH1 expression. BMC Ophthalmol. 2021, 21, 104. [Google Scholar] [CrossRef] [PubMed]
- Yusoff, F.M.; Maruhashi, T.; Kawano, K.I.; Nakashima, A.; Chayama, K.; Tashiro, S.; Igarashi, K.; Higashi, Y. Bach1 plays an important role in angiogenesis through regulation of oxidative stress. Microvasc. Res. 2021, 134, 104126. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Strushkevich, N.V.; Harnastai, I.N.; Iwamoto, H.; Gilep, A.A.; Takemori, H.; Usanov, S.A.; Nonaka, Y.; Hori, H.; Vinson, G.P.; et al. Molecular identification of adrenal inner zone antigen as a heme-binding protein. FEBS J. 2005, 272, 5832–5843. [Google Scholar] [CrossRef] [PubMed]
- Kaluka, D.; Batabyal, D.; Chiang, B.Y.; Poulos, T.L.; Yeh, S.R. Spectroscopic and mutagenesis studies of human PGRMC1. Biochemistry 2015, 54, 1638–1647. [Google Scholar] [CrossRef]
- Meier, M.; Janosik, M.; Kery, V.; Kraus, J.P.; Burkhard, P. Structure of human cystathionine β-synthase: A unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 2001, 20, 3910–3916. [Google Scholar] [CrossRef] [Green Version]
- Taoka, S.; Lepore, B.W.; Kabil, Ö.; Ojha, S.; Ringe, D.; Banerjee, R. Human cystathionine β-synthase is a heme sensor protein. Evidence that the redox sensor is heme and not the vicinal cysteines in the CXXC motif seen in the crystal structure of the truncated enzyme. Biochemistry 2002, 41, 10454–10461. [Google Scholar] [CrossRef] [PubMed]
- Horst, B.G.; Marletta, M.A. Physiological activation and deactivation of soluble guanylate cyclase. Nitric Oxide 2018, 77, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Allerston, C.K.; von Delft, F.; Gileadi, O. Crystal structures of the catalytic domain of human soluble guanylate cyclase. PLoS ONE 2013, 8, e57644. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; Liu, R.; Wu, J.X.; Chen, L. Structural insights into the mechanism of human soluble guanylate cyclase. Nature 2019, 574, 206–210. [Google Scholar] [CrossRef]
- Klatt, P.; Pfeiffer, S.; List, B.M.; Lehner, D.; Glatter, O.; Bachinger, H.P.; Werner, E.R.; Schmidt, K.; Mayer, B. Characterization of heme-deficient neuronal nitric-oxide synthase reveals a role for heme in subunit dimerization and binding of the amino acid substrate and tetrahydrobiopterin. J. Biol. Chem. 1996, 271, 7336–7342. [Google Scholar] [CrossRef] [Green Version]
- List, B.M.; Klosch, B.; Volker, C.; Gorren, A.C.; Sessa, W.C.; Werner, E.R.; Kukovetz, W.R.; Schmidt, K.; Mayer, B. Characterization of bovine endothelial nitric oxide synthase as a homodimer with down-regulated uncoupled NADPH oxidase activity: Tetrahydrobiopterin binding kinetics and role of haem in dimerization. Biochem. J. 1997, 323 Pt 1, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Bender, A.T.; Nakatsuka, M.; Osawa, Y. Heme insertion, assembly, and activation of apo-neuronal nitric-oxide synthase in vitro. J. Biol. Chem. 2000, 275, 26018–26023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Guo, J.; Wei, X.; Niu, C.; Jia, M.; Li, Q.; Meng, D. Bach1: Function, Regulation, and Involvement in Disease. Oxidative Med. Cell. Longev. 2018, 2018, 1347969. [Google Scholar] [CrossRef] [PubMed]
- Kitamuro, T.; Takahashi, K.; Ogawa, K.; Udono-Fujimori, R.; Takeda, K.; Furuyama, K.; Nakayama, M.; Sun, J.; Fujita, H.; Hida, W.; et al. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J. Biol. Chem. 2003, 278, 9125–9133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hira, S.; Tomita, T.; Matsui, T.; Igarashi, K.; Ikeda-Saito, M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life 2007, 59, 542–551. [Google Scholar] [CrossRef]
- Suzuki, H.; Tashiro, S.; Hira, S.; Sun, J.; Yamazaki, C.; Zenke, Y.; Ikeda-Saito, M.; Yoshida, M.; Igarashi, K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004, 23, 2544–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lignitto, L.; LeBoeuf, S.E.; Homer, H.; Jiang, S.; Askenazi, M.; Karakousi, T.R.; Pass, H.I.; Bhutkar, A.J.; Tsirigos, A.; Ueberheide, B.; et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell 2019, 178, 316–329.e18. [Google Scholar] [CrossRef]
- Sun, J.; Hoshino, H.; Takaku, K.; Nakajima, O.; Muto, A.; Suzuki, H.; Tashiro, S.; Takahashi, S.; Shibahara, S.; Alam, J.; et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002, 21, 5216–5224. [Google Scholar] [CrossRef]
- Cahill, M.A. The evolutionary appearance of signaling motifs in PGRMC1. Biosci. Trends 2017, 11, 179–192. [Google Scholar] [CrossRef] [Green Version]
- Cahill, M.A.; Neubauer, H. PGRMC Proteins Are Coming of Age: A Special Issue on the Role of PGRMC1 and PGRMC2 in Metabolism and Cancer Biology. Cancers 2021, 13, 512. [Google Scholar] [CrossRef]
- Zhang, D.; Xia, X.; Wang, X.; Zhang, P.; Lu, W.; Yu, Y.; Deng, S.; Yang, H.; Zhu, H.; Xu, N.; et al. PGRMC1 Is a Novel Potential Tumor Biomarker of Human Renal Cell Carcinoma Based on Quantitative Proteomic and Integrative Biological Assessments. PLoS ONE 2017, 12, e0170453. [Google Scholar] [CrossRef]
- He, Y.; Zhang, P.; Zhang, D.; Xia, Z.; Wang, X.; Deng, S.; Li, H.; Zhu, H.; Xu, N.; Liang, S. Combined assessment of low PGRMC1/positive ATP1A1 levels has enhanced prognostic value for renal cell carcinoma. Oncol. Rep. 2018, 40, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Mir, S.U.; Ahmed, I.S.; Arnold, S.; Craven, R.J. Elevated progesterone receptor membrane component 1/sigma-2 receptor levels in lung tumors and plasma from lung cancer patients. Int. J. Cancer 2012, 131, E1–E9. [Google Scholar] [CrossRef] [PubMed]
- Hampton, K.K.; Stewart, R.; Napier, D.; Claudio, P.P.; Craven, R.J. PGRMC1 elevation in multiple cancers and essential role in stem cell survival. Adv. Lung Cancer 2015, 4, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, C.-C.; Chou, H.-C.; Chen, Y.-J.; Kuo, W.-H.; Chan, C.-H.; Lin, Y.-C.; Liao, E.-C.; Chang, S.-J.; Chan, H.-L. Role of PGRMC1 in cell physiology of cervical cancer. Life Sci. 2019, 231, 116541. [Google Scholar] [CrossRef]
- Cai, G.; Ruan, X.; Gu, M.; Zhao, Y.; Wang, Y.; Mueck, A.O. PGRMC1 in animal breast cancer tissue and blood is associated with increased tumor growth with norethisterone in contrast to progesterone and dydrogesterone: Four-arm randomized placebo-controlled xenograft study. Gynecol. Endocrinol. 2020, 36, 1024–1027. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, H.; Clare, S.E.; Wozny, W.; Schwall, G.P.; Poznanović, S.; Stegmann, W.; Vogel, U.; Sotlar, K.; Wallwiener, D.; Kurek, R. Breast cancer proteomics reveals correlation between estrogen receptor status and differential phosphorylation of PGRMC1. Breast Cancer Res. 2008, 10, R85. [Google Scholar] [CrossRef] [Green Version]
- Ruan, X.; Cai, G.; Wei, Y.; Gu, M.; Zhang, Y.; Zhao, Y.; Mueck, A.O. Association of circulating Progesterone Receptor Membrane Component-1 (PGRMC1) with breast tumor characteristics and comparison with known tumor markers. Menopause 2020, 27, 183–193. [Google Scholar] [CrossRef]
- Zhao, Y.; Ruan, X. Identification of PGRMC1 as a Candidate Oncogene for Head and Neck Cancers and Its Involvement in Metabolic Activities. Front. Bioeng. Biotechnol. 2020, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahill, M.A.; Medlock, A.E. Thoughts on interactions between PGRMC1 and diverse attested and potential hydrophobic ligands. J. Steroid Biochem. Mol. Biol. 2017, 171, 11–33. [Google Scholar] [CrossRef]
- Cahill, M.A.; Jazayeri, J.A.; Kovacevic, Z.; Richardson, D.R. PGRMC1 regulation by phosphorylation: Potential new insights in controlling biological activity! Oncotarget 2016, 7, 50822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piel, R.B., III; Shiferaw, M.T.; Vashisht, A.A.; Marcero, J.R.; Praissman, J.L.; Phillips, J.D.; Wohlschlegel, J.A.; Medlock, A.E. A novel role for progesterone receptor membrane component 1 (PGRMC1): A partner and regulator of ferrochelatase. Biochemistry 2016, 55, 5204–5217. [Google Scholar] [CrossRef] [Green Version]
- Strachan, T.; Read, A. Human Molecular Genetics; Garland Science: New York, NY, USA, 2003; p. 418. [Google Scholar]
- Bunz, F.; Dutriaux, A.; Lengauer, C.; Waldman, T.; Zhou, S.; Brown, J.; Sedivy, J.; Kinzler, K.W.; Vogelstein, B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998, 282, 1497–1501. [Google Scholar] [CrossRef]
- Toshiyuki, M.; Reed, J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995, 80, 293–299. [Google Scholar] [CrossRef] [Green Version]
- Teodoro, J.G.; Evans, S.K.; Green, M.R. Inhibition of tumor angiogenesis by p53: A new role for the guardian of the genome. J. Mol. Med. 2007, 85, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Hu, W.; Feng, Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015, 356, 197–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toro, A.; Anselmino, N.; Solari, C.; Francia, M.; Oses, C.; Sanchis, P.; Bizzotto, J.; Vazquez Echegaray, C.; Petrone, M.V.; Levi, V.; et al. Novel Interplay between p53 and HO-1 in Embryonic Stem Cells. Cells 2020, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, S.Y.; Choi, H.S.; Kim, M.K.; Lee, H.M.; Jang, Y.J.; Ryu, C.J. Progesterone Receptor Membrane Component 1 suppresses the p53 and Wnt/beta-catenin pathways to promote human pluripotent stem cell self-renewal. Sci. Rep. 2018, 8, 3048. [Google Scholar] [CrossRef] [PubMed]
- Dohi, Y.; Ikura, T.; Hoshikawa, Y.; Katoh, Y.; Ota, K.; Nakanome, A.; Muto, A.; Omura, S.; Ohta, T.; Ito, A.; et al. Bach1 inhibits oxidative stress-induced cellular senescence by impeding p53 function on chromatin. Nat. Struct. Mol. Biol. 2008, 15, 1246–1254. [Google Scholar] [CrossRef]
- Nishizawa, H.; Ota, K.; Dohi, Y.; Ikura, T.; Igarashi, K. Bach1-mediated suppression of p53 is inhibited by p19(ARF) independently of MDM2. Cancer Sci. 2012, 103, 897–903. [Google Scholar] [CrossRef]
- Kery, V.; Bukovska, G.; Kraus, J.P. Transsulfuration depends on heme in addition to pyridoxal 5’-phosphate. Cystathionine beta-synthase is a heme protein. J. Biol. Chem. 1994, 269, 25283–25288. [Google Scholar] [CrossRef]
- Watanabe, M.; Osada, J.; Aratani, Y.; Kluckman, K.; Reddick, R.; Malinow, M.R.; Maeda, N. Mice deficient in cystathionine beta-synthase: Animal models for mild and severe homocyst (e) inemia. Proc. Natl. Acad. Sci. USA 1995, 92, 1585–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Padovani, D.; Leslie, R.A.; Chiku, T.; Banerjee, R. Relative contributions of cystathionine β-synthase and γ-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 2009, 284, 22457–22466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, W.; Wang, J.; Qian, J.; Wang, M.; Wu, P.; Chen, F.; Yan, S. Allosteric control of human cystathionine β-synthase activity by a redox active disulfide bond. J. Biol. Chem. 2018, 293, 2523–2533. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.D.; Snyder, S.H.; Kashfi, K. Effects of hydrogen sulfide on mitochondrial function and cellular bioenergetics. Redox Biol. 2021, 38, 101772. [Google Scholar] [CrossRef] [PubMed]
- Pogribna, M.; Melnyk, S.; Pogribny, I.; Chango, A.; Yi, P.; James, S.J. Homocysteine metabolism in children with Down syndrome: In vitro modulation. Am. J. Hum. Genet. 2001, 69, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamoun, P.; Belardinelli, M.C.; Chabli, A.; Lallouchi, K.; Chadefaux-Vekemans, B. Endogenous hydrogen sulfide overproduction in Down syndrome. Am. J. Med. Genet. A 2003, 116A, 310–311. [Google Scholar] [CrossRef]
- Panagaki, T.; Randi, E.B.; Augsburger, F.; Szabo, C. Overproduction of H2S, generated by CBS, inhibits mitochondrial Complex IV and suppresses oxidative phosphorylation in Down syndrome. Proc. Natl. Acad. Sci. USA 2019, 116, 18769–18771. [Google Scholar] [CrossRef] [Green Version]
- Szabo, C.; Coletta, C.; Chao, C.; Módis, K.; Szczesny, B.; Papapetropoulos, A.; Hellmich, M.R. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 12474–12479. [Google Scholar] [CrossRef] [Green Version]
- Druzhyna, N.; Szczesny, B.; Olah, G.; Módis, K.; Asimakopoulou, A.; Pavlidou, A.; Szoleczky, P.; Gerö, D.; Yanagi, K.; Törö, G. Screening of a composite library of clinically used drugs and well-characterized pharmacological compounds for cystathionine β-synthase inhibition identifies benserazide as a drug potentially suitable for repurposing for the experimental therapy of colon cancer. Pharmacol. Res. 2016, 113, 18–37. [Google Scholar]
- Untereiner, A.A.; Pavlidou, A.; Druzhyna, N.; Papapetropoulos, A.; Hellmich, M.R.; Szabo, C. Drug resistance induces the upregulation of H2S-producing enzymes in HCT116 colon cancer cells. Biochem. Pharmacol. 2018, 149, 174–185. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Saha, S.; Giri, K.; Lanza, I.R.; Nair, K.S.; Jennings, N.B.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Basal, E.; Weaver, A.L. Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS ONE 2013, 8, e79167. [Google Scholar] [CrossRef] [PubMed]
- Kumar Chakraborty, P.; Murphy, B.; Banerjee Mustafi, S.; Dey, A.; Xiong, X.; Rao, G.; Naz, S.; Zhang, M.; Yang, D.; Dhanasekaran, D.N. Cystathionine β-synthase regulates mitochondrial morphogenesis in ovarian cancer. FASEB J. 2018, 32, 4145–4157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, S.; Kawahara, B.; Gupta, D.; Tsai, R.; Khachatryan, M.; Roy-Chowdhuri, S.; Bose, S.; Yoon, A.; Faull, K.; Farias-Eisner, R. Role of cystathionine β-synthase in human breast Cancer. Free Radic. Biol. Med. 2015, 86, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Wu, B.; Cao, Q.; Wu, L.; Yang, G. Hydrogen sulfide mediates the anti-survival effect of sulforaphane on human prostate cancer cells. Toxicol. Appl. Pharmacol. 2011, 257, 420–428. [Google Scholar] [CrossRef]
- Baykov, A.A.; Tuominen, H.K.; Lahti, R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem. Biol. 2011, 6, 1156–1163. [Google Scholar] [CrossRef]
- Weeks, C.L.; Singh, S.; Madzelan, P.; Banerjee, R.; Spiro, T.G. Heme regulation of human cystathionine beta-synthase activity: Insights from fluorescence and Raman spectroscopy. J. Am. Chem. Soc. 2009, 131, 12809–12816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicente, J.B.; Colaço, H.G.; Mendes, M.I.; Sarti, P.; Leandro, P.; Giuffrè, A. NO* binds human cystathionine β-synthase quickly and tightly. J. Biol. Chem. 2014, 289, 8579–8587. [Google Scholar] [CrossRef] [Green Version]
- Taoka, S.; Ohja, S.; Shan, X.; Kruger, W.D.; Banerjee, R. Evidence for heme-mediated redox regulation of human cystathionine β-synthase activity. J. Biol. Chem. 1998, 273, 25179–25184. [Google Scholar] [CrossRef] [Green Version]
- Benchoam, D.; Cuevasanta, E.; Julio Plana, L.; Capece, L.; Banerjee, R.; Alvarez, B. Heme-Thiolate Perturbation in Cystathionine beta-Synthase by Mercury Compounds. ACS Omega 2021, 6, 2192–2205. [Google Scholar] [CrossRef]
- Mohammadoo-Khorasani, M.; Karami Tehrani, F.; Atri, M. Soluble guanylate cyclase isoenzymes: The expression of alpha1, alpha2, beta1, and beta2 subunits in the benign and malignant breast tumors. J. Cell. Physiol. 2020, 235, 1358–1365. [Google Scholar] [CrossRef]
- Stasch, J.P.; Pacher, P.; Evgenov, O.V. Soluble guanylate cyclase as an emerging therapeutic target in cardiopulmonary disease. Circulation 2011, 123, 2263–2273. [Google Scholar] [CrossRef] [Green Version]
- Sandner, P. From molecules to patients: Exploring the therapeutic role of soluble guanylate cyclase stimulators. Biol. Chem. 2018, 399, 679–690. [Google Scholar] [CrossRef]
- Wen, H.C.; Chuu, C.P.; Chen, C.Y.; Shiah, S.G.; Kung, H.J.; King, K.L.; Su, L.C.; Chang, S.C.; Chang, C.H. Elevation of soluble guanylate cyclase suppresses proliferation and survival of human breast cancer cells. PLoS ONE 2015, 10, e0125518. [Google Scholar] [CrossRef] [Green Version]
- Mohammadoo Khorasani, M.; Karami Tehrani, F.; Parizadeh, S.M.R.; Atri, M. Differential expression of alternative transcripts of soluble guanylyl cyclase, GYCY1a3 and GUCY1b3 genes, in the malignant and benign breast tumors. Nitric Oxide 2019, 83, 65–71. [Google Scholar] [CrossRef]
- Tuttle, T.R.; Takiar, V.; Kumar, B.; Kumar, P.; Ben-Jonathan, N. Soluble guanylate cyclase stimulators increase sensitivity to cisplatin in head and neck squamous cell carcinoma cells. Cancer Lett. 2017, 389, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Albakri, Q.A.; Stuehr, D.J. Intracellular assembly of inducible NO synthase is limited by nitric oxide-mediated changes in heme insertion and availability. J. Biol. Chem. 1996, 271, 5414–5421. [Google Scholar] [CrossRef] [Green Version]
- Stuehr, D.J.; Haque, M.M. Nitric oxide synthase enzymology in the 20 years after the Nobel Prize. Br. J. Pharmacol. 2019, 176, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Cianchi, F.; Cortesini, C.; Fantappie, O.; Messerini, L.; Schiavone, N.; Vannacci, A.; Nistri, S.; Sardi, I.; Baroni, G.; Marzocca, C.; et al. Inducible nitric oxide synthase expression in human colorectal cancer: Correlation with tumor angiogenesis. Am. J. Pathol. 2003, 162, 793–801. [Google Scholar] [CrossRef]
- Venema, R.C.; Ju, H.; Zou, R.; Ryan, J.W.; Venema, V.J. Subunit interactions of endothelial nitric-oxide synthase. Comparisons to the neuronal and inducible nitric-oxide synthase isoforms. J. Biol. Chem. 1997, 272, 1276–1282. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Panda, K.; Stuehr, D.J. Control of nitric oxide synthase dimer assembly by a heme-NO-dependent mechanism. Biochemistry 2002, 41, 4618–4625. [Google Scholar] [CrossRef] [PubMed]
- Waheed, S.M.; Ghosh, A.; Chakravarti, R.; Biswas, A.; Haque, M.M.; Panda, K.; Stuehr, D.J. Nitric oxide blocks cellular heme insertion into a broad range of heme proteins. Free Radic. Biol. Med. 2010, 48, 1548–1558. [Google Scholar] [CrossRef] [Green Version]
- Chakravarti, R.; Aulak, K.S.; Fox, P.L.; Stuehr, D.J. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl. Acad. Sci. USA 2010, 107, 18004–18009. [Google Scholar] [CrossRef] [Green Version]
- Fukuhara, H.; Inoue, K.; Kurabayashi, A.; Furihata, M.; Fujita, H.; Utsumi, K.; Sasaki, J.; Shuin, T. The inhibition of ferrochelatase enhances 5-aminolevulinic acid-based photodynamic action for prostate cancer. Photodiagnosis Photodyn. Ther. 2013, 10, 399–409. [Google Scholar] [CrossRef]
- Furukawa, T.; Kohno, H.; Tokunaga, R.; Taketani, S. Nitric oxide-mediated inactivation of mammalian ferrochelatase in vivo and in vitro: Possible involvement of the iron-sulphur cluster of the enzyme. Biochem. J. 1995, 310 Pt 2, 533–538. [Google Scholar] [CrossRef] [Green Version]
- Sellers, V.M.; Johnson, M.K.; Dailey, H.A. Function of the [2FE-2S] cluster in mammalian ferrochelatase: A possible role as a nitric oxide sensor. Biochemistry 1996, 35, 2699–2704. [Google Scholar] [CrossRef] [PubMed]
- Rajnakova, A.; Moochhala, S.; Goh, P.M.; Ngoi, S. Expression of nitric oxide synthase, cyclooxygenase, and p53 in different stages of human gastric cancer. Cancer Lett. 2001, 172, 177–185. [Google Scholar] [CrossRef]
- Basudhar, D.; Glynn, S.A.; Greer, M.; Somasundaram, V.; No, J.H.; Scheiblin, D.A.; Garrido, P.; Heinz, W.F.; Ryan, A.E.; Weiss, J.M.; et al. Coexpression of NOS2 and COX2 accelerates tumor growth and reduces survival in estrogen receptor-negative breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 13030–13035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habibollahi, P.; Jamshidiha, M.; Daryani, N.E.; Jahanzad, I.; Ghahremani, M.H.; Ostad, S.N. Correlation between inducible nitric oxide synthase and cyclooxygenase-2 expression in human colorectal adenocarcinoma: A cross-sectional study. Pathol. Oncol. Res. 2010, 16, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Takahashi, M.; Ochiai, A. Increased protein expression of both inducible nitric oxide synthase and cyclooxygenase-2 in human colon cancers. Cancer Lett. 2006, 239, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.; Doria, D.; Bertazzoni, E.; Benini, A.; Bassi, C. Increased expression of inducible nitric oxide synthase and cyclooxygenase-2 in pancreatic cancer. Prostaglandins Other Lipid Mediat. 2004, 73, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Uotila, P.; Valve, E.; Martikainen, P.; Nevalainen, M.; Nurmi, M.; Harkonen, P. Increased expression of cyclooxygenase-2 and nitric oxide synthase-2 in human prostate cancer. Urol. Res. 2001, 29, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Patel, V.; Banerjee, D. Nitric Oxide and S-Nitrosylation in Cancers: Emphasis on Breast Cancer. Breast Cancer Basic Clin. Res. 2020, 14, 1178223419882688. [Google Scholar] [CrossRef]
- Pacher, P.; Beckman, J.S.; Liaudet, L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007, 87, 315–424. [Google Scholar] [CrossRef] [Green Version]
- Schneiderhan, N.; Budde, A.; Zhang, Y.; Brune, B. Nitric oxide induces phosphorylation of p53 and impairs nuclear export. Oncogene 2003, 22, 2857–2868. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Michael, D.; de Murcia, G.; Oren, M. p53 Activation by nitric oxide involves down-regulation of Mdm2. J. Biol. Chem. 2002, 277, 15697–15702. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, L.M.; Demple, B. Nitric oxide-induced apoptosis in lymphoblastoid and fibroblast cells dependent on the phosphorylation and activation of p53. Cancer Res. 2005, 65, 6097–6104. [Google Scholar] [CrossRef] [Green Version]
- Hess, D.T.; Matsumoto, A.; Kim, S.O.; Marshall, H.E.; Stamler, J.S. Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005, 6, 150–166. [Google Scholar] [CrossRef]
- Vannini, F.; Kashfi, K.; Nath, N. The dual role of iNOS in cancer. Redox Biol. 2015, 6, 334–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.; Zang, G.; Wang, Y.; Sun, Z.; Li, Y.; Lu, C.; Wang, Z. Differences of Angiogenesis Factors in Tumor and Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2021, 14, 3375–3388. [Google Scholar] [CrossRef]
- Dulak, J.; Deshane, J.; Jozkowicz, A.; Agarwal, A. Heme oxygenase-1 and carbon monoxide in vascular pathobiology: Focus on angiogenesis. Circulation 2008, 117, 231–241. [Google Scholar] [CrossRef]
- Zuazo-Gaztelu, I.; Casanovas, O. Unraveling the Role of Angiogenesis in Cancer Ecosystems. Front. Oncol. 2018, 8. [Google Scholar] [CrossRef]
- Vandekeere, S.; Dewerchin, M.; Carmeliet, P. Angiogenesis Revisited: An Overlooked Role of Endothelial Cell Metabolism in Vessel Sprouting. Microcirculation 2015, 22, 509–517. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Jiang, L.; Wei, X.; Niu, C.; Wang, R.; Zhang, J.; Meng, D.; Yao, K. Bach1 Induces Endothelial Cell Apoptosis and Cell-Cycle Arrest through ROS Generation. Oxidative Med. Cell. Longev. 2016, 2016, 6234043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Yin, M.; Wei, X.; Liu, J.; Wang, X.; Niu, C.; Kang, X.; Xu, J.; Zhou, Z.; Sun, S.; et al. Bach1 Represses Wnt/β-Catenin Signaling and Angiogenesis. Circ. Res. 2015, 117, 364–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Yin, M.; Xu, J.; Jia, M.; Sun, S.; Wang, X.; Zhang, J.; Meng, D. The Transcription Factor Bach1 Suppresses the Developmental Angiogenesis of Zebrafish. Oxidative Med. Cell. Longev. 2017, 2017, 2143875. [Google Scholar] [CrossRef]
- Shetty, T.; Corson, T.W. Mitochondrial Heme Synthesis Enzymes as Therapeutic Targets in Vascular Diseases. Front. Pharmacol. 2020, 11, 1015. [Google Scholar] [CrossRef]
- Shetty, T.; Sishtla, K.; Park, B.; Repass, M.J.; Corson, T.W. Heme Synthesis Inhibition Blocks Angiogenesis via Mitochondrial Dysfunction. Iscience 2020, 23, 101391. [Google Scholar] [CrossRef]
- Petrillo, S.; Chiabrando, D.; Genova, T.; Fiorito, V.; Ingoglia, G.; Vinchi, F.; Mussano, F.; Carossa, S.; Silengo, L.; Altruda, F.; et al. Heme accumulation in endothelial cells impairs angiogenesis by triggering paraptosis. Cell Death Differ. 2018, 25, 573–588. [Google Scholar] [CrossRef]
- Vandekeere, S.; Dubois, C.; Kalucka, J.; Sullivan, M.R.; García-Caballero, M.; Goveia, J.; Chen, R.; Diehl, F.F.; Bar-Lev, L.; Souffreau, J.; et al. Serine Synthesis via PHGDH Is Essential for Heme Production in Endothelial Cells. Cell Metab. 2018, 28, 573–587.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licker, M.; de Perrot, M.; Spiliopoulos, A.; Robert, J.; Diaper, J.; Chevalley, C.; Tschopp, J.-M. Risk factors for acute lung injury after thoracic surgery for lung cancer. Anesth. Analg. 2003, 97, 1558–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, Y.; Kurdowska, A.; Allen, T.C. Acute Lung Injury: A Clinical and Molecular Review. Arch. Pathol. Lab. Med. 2016, 140, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Rubenfeld, G.D.; Caldwell, E.; Peabody, E.; Weaver, J.; Martin, D.P.; Neff, M.; Stern, E.J.; Hudson, L.D. Incidence and outcomes of acute lung injury. N. Engl. J. Med. 2005, 353, 1685–1693. [Google Scholar] [CrossRef] [Green Version]
- Dreyfuss, D.; Ricard, J.-D. Acute lung injury and bacterial infection. Clin. Chest Med. 2005, 26, 105–112. [Google Scholar] [CrossRef]
- Wagener, B.M.; Hu, P.J.; Oh, J.Y.; Evans, C.A.; Richter, J.R.; Honavar, J.; Brandon, A.P.; Creighton, J.; Stephens, S.W.; Morgan, C.; et al. Role of heme in lung bacterial infection after trauma hemorrhage and stored red blood cell transfusion: A preclinical experimental study. PLoS Med. 2018, 15, e1002522. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, S.; Jilling, T.; Doran, S.; Ahmad, I.; Eagen, J.E.; Gu, S.; Gillespie, M.; Albert, C.J.; Ford, D.; Oh, J.Y.; et al. Phosgene inhalation causes hemolysis and acute lung injury. Toxicol. Lett. 2019, 312, 204–213. [Google Scholar] [CrossRef]
- Aggarwal, S.; Lam, A.; Bolisetty, S.; Carlisle, M.A.; Traylor, A.; Agarwal, A.; Matalon, S. Heme Attenuation Ameliorates Irritant Gas Inhalation-Induced Acute Lung Injury. Antioxid. Redox Signal. 2016, 24, 99–112. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, S.; Lazrak, A.; Ahmad, I.; Yu, Z.; Bryant, A.; Mobley, J.; Ford, D.; Matalon, S. Heme Impairs Alveolar Epithelial Sodium Channels Post Toxic Gas Inhalation. BioRxiv 2020. [Google Scholar] [CrossRef]
- Aggarwal, S.; Ahmad, I.; Lam, A.; Carlisle, M.A.; Li, C.; Wells, J.M.; Raju, S.V.; Athar, M.; Rowe, S.M.; Dransfield, M.T.; et al. Heme scavenging reduces pulmonary endoplasmic reticulum stress, fibrosis, and emphysema. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Schmidt, H.M.; Kelley, E.E.; Straub, A.C. The impact of xanthine oxidase (XO) on hemolytic diseases. Redox Biol. 2019, 21, 101072. [Google Scholar] [CrossRef]
- Vinchi, F.; De Franceschi, L.; Ghigo, A.; Townes, T.; Cimino, J.; Silengo, L.; Hirsch, E.; Altruda, F.; Tolosano, E. Hemopexin therapy improves cardiovascular function by preventing heme-induced endothelial toxicity in mouse models of hemolytic diseases. Circulation 2013, 127, 1317–1329. [Google Scholar] [CrossRef] [Green Version]
- Belcher, J.D.; Chen, C.; Nguyen, J.; Abdulla, F.; Zhang, P.; Nguyen, H.; Nguyen, P.; Killeen, T.; Miescher, S.M.; Brinkman, N.; et al. Haptoglobin and hemopexin inhibit vaso-occlusion and inflammation in murine sickle cell disease: Role of heme oxygenase-1 induction. PLoS ONE 2018, 13, e0196455. [Google Scholar] [CrossRef] [Green Version]
- Beckman, J.D.; Abdullah, F.; Chen, C.; Kirchner, R.; Rivera-Rodriguez, D.; Kiser, Z.M.; Nguyen, A.; Zhang, P.; Nguyen, J.; Hebbel, R.P.; et al. Endothelial TLR4 Expression Mediates Vaso-Occlusive Crisis in Sickle Cell Disease. Front. Immunol. 2020, 11, 613278. [Google Scholar] [CrossRef]
- Zhang, P.; Nguyen, J.; Abdulla, F.; Nelson, A.T.; Beckman, J.D.; Vercellotti, G.M.; Belcher, J.D. Soluble MD-2 and Heme in Sickle Cell Disease Plasma Promote Pro-Inflammatory Signaling in Endothelial Cells. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Sawicki, K.T.; Chang, H.C.; Ardehali, H. Role of heme in cardiovascular physiology and disease. J. Am. Heart Assoc. 2015, 4, e001138. [Google Scholar] [CrossRef] [Green Version]
- Khechaduri, A.; Bayeva, M.; Chang, H.C.; Ardehali, H. Heme levels are increased in human failing hearts. J. Am. Coll. Cardiol. 2013, 61, 1884–1893. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Horke, S.; Förstermann, U. Oxidative stress in vascular disease and its pharmacological prevention. Trends Pharmacol. Sci. 2013, 34, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ayer, A.; Zarjou, A.; Agarwal, A.; Stocker, R. Heme Oxygenases in Cardiovascular Health and Disease. Physiol. Rev. 2016, 96, 1449–1508. [Google Scholar] [CrossRef] [PubMed]
- Haines, D.D.; Tosaki, A. Role of Heme Oxygenases in Cardiovascular Syndromes and Co-morbidities. Curr. Pharm. Des. 2018, 24, 2322–2325. [Google Scholar] [CrossRef]
- Suliman, H.B.; Zobi, F.; Piantadosi, C.A. Heme Oxygenase-1/Carbon Monoxide System and Embryonic Stem Cell Differentiation and Maturation into Cardiomyocytes. Antioxid. Redox Signal. 2016, 24, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Jez, M.; Martyniak, A.; Andrysiak, K.; Mucha, O.; Szade, K.; Kania, A.; Chrobok, L.; Palus-Chramiec, K.; Sanetra, A.M.; Lewandowski, M.H.; et al. Role of Heme-Oxygenase-1 in Biology of Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells. Cells 2021, 10, 522. [Google Scholar] [CrossRef]
- Bozza, M.T.; Jeney, V. Pro-inflammatory Actions of Heme and Other Hemoglobin-Derived DAMPs. Front. Immunol. 2020, 11, 1323. [Google Scholar] [CrossRef] [PubMed]
- Godefroy, E.; Liu, Y.; Shi, P.; Mitchell, W.B.; Cohen, D.; Chou, S.T.; Manwani, D.; Yazdanbakhsh, K. Altered heme-mediated modulation of dendritic cell function in sickle cell alloimmunization. Haematologica 2016, 101, 1028–1038. [Google Scholar] [CrossRef] [Green Version]
- Martins, R.; Knapp, S. Heme and hemolysis in innate immunity: Adding insult to injury. Curr. Opin. Immunol. 2018, 50, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Deuel, J.W.; Schaer, C.A.; Boretti, F.S.; Opitz, L.; García-Rubio, I.; Baek, J.; Spahn, D.R.; Buehler, P.W.; Schaer, D.J. Hemoglobinuria-related acute kidney injury is driven by intrarenal oxidative reactions triggering a heme toxicity response. Cell Death Dis. 2016, 7, e2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prestes, E.B.; Alves, L.S.; Rodrigues, D.A.S.; Dutra, F.F.; Fernandez, P.L.; Paiva, C.N.; Kagan, J.C.; Bozza, M.T. Mitochondrial Reactive Oxygen Species Participate in Signaling Triggered by Heme in Macrophages and upon Hemolysis. J. Immunol. 2020, 205, 2795–2805. [Google Scholar] [CrossRef]
- Erdei, J.; Tóth, A.; Balogh, E.; Nyakundi, B.B.; Bányai, E.; Ryffel, B.; Paragh, G.; Cordero, M.D.; Jeney, V. Induction of NLRP3 inflammasome activation by heme in human endothelial cells. Oxidative Med. Cell. Longev. 2018, 2018, 4310816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merle, N.S.; Grunenwald, A.; Rajaratnam, H.; Gnemmi, V.; Frimat, M.; Figueres, M.L.; Knockaert, S.; Bouzekri, S.; Charue, D.; Noe, R.; et al. Intravascular hemolysis activates complement via cell-free heme and heme-loaded microvesicles. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Martins, R.; Maier, J.; Gorki, A.D.; Huber, K.V.; Sharif, O.; Starkl, P.; Saluzzo, S.; Quattrone, F.; Gawish, R.; Lakovits, K.; et al. Heme drives hemolysis-induced susceptibility to infection via disruption of phagocyte functions. Nat. Immunol. 2016, 17, 1361–1372. [Google Scholar] [CrossRef]
- Wu, B.; Wu, Y.; Tang, W. Heme catabolic pathway in inflammation and immune disorders. Front. Pharmacol. 2019, 10, 825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alam, M.Z.; Devalaraja, S.; Haldar, M. The Heme Connection: Linking Erythrocytes and Macrophage Biology. Front. Immunol. 2017, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, P.; Vijayan, V.; Gueler, F.; Immenschuh, S. Interplay of heme with macrophages in homeostasis and inflammation. Int. J. Mol. Sci. 2020, 21, 740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintela-Carvalho, G.; Luz, N.F.; Celes, F.S.; Zanette, D.L.; Andrade, D.; Menezes, D.; Tavares, N.M.; Brodskyn, C.I.; Prates, D.B.; Gonçalves, M.S.; et al. Heme Drives Oxidative Stress-Associated Cell Death in Human Neutrophils Infected with. Front. Immunol. 2017, 8, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laranjeira-Silva, M.F.; Hamza, I.; Pérez-Victoria, J.M. Iron and heme metabolism at the leishmania–host interface. Trends Parasitol. 2020, 36, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Righy, C.; Bozza, M.T.; Oliveira, M.F.; Bozza, F.A. Molecular, Cellular and Clinical Aspects of Intracerebral Hemorrhage: Are the Enemies Within? Curr. Neuropharmacol. 2016, 14, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Chiabrando, D.; Fiorito, V.; Petrillo, S.; Tolosano, E. Unraveling the Role of Heme in Neurodegeneration. Front. Neurosci. 2018, 12, 712. [Google Scholar] [CrossRef]
- Ma, B.; Day, J.P.; Phillips, H.; Slootsky, B.; Tolosano, E.; Doré, S. Deletion of the hemopexin or heme oxygenase-2 gene aggravates brain injury following stroma-free hemoglobin-induced intracerebral hemorrhage. J. Neuroinflammation 2016, 13, 26. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.P.; Gehlhaus, M.; Knoth, R.; Volk, B. Expression and function of cytochrome p450 in brain drug metabolism. Curr. Drug Metab. 2007, 8, 297–306. [Google Scholar] [CrossRef]
- Hayden, E.Y.; Kaur, P.; Williams, T.L.; Matsui, H.; Yeh, S.R.; Rousseau, D.L. Heme Stabilization of α-Synuclein Oligomers during Amyloid Fibril Formation. Biochemistry 2015, 54, 4599–4610. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, S.L.; Kumar, P.T.; McBride, D.; Zeineddine, H.A.; Leclerc, J.; Choi, H.A.; Dash, P.K.; Grotta, J.; Aronowski, J.; Cardenas, J.C.; et al. Unique Contribution of Haptoglobin and Haptoglobin Genotype in Aneurysmal Subarachnoid Hemorrhage. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Song, I.U.; Kim, Y.D.; Chung, S.W.; Cho, H.J. Association between serum haptoglobin and the pathogenesis of Alzheimer’s disease. Intern. Med. 2015, 54, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Immenschuh, S.; Vijayan, V.; Janciauskiene, S.; Gueler, F. Heme as a Target for Therapeutic Interventions. Front. Pharmacol. 2017, 8, 146. [Google Scholar] [CrossRef] [Green Version]
- Parfenova, H.; Leffler, C.W.; Basuroy, S.; Liu, J.; Fedinec, A.L. Antioxidant roles of heme oxygenase, carbon monoxide, and bilirubin in cerebral circulation during seizures. J. Cereb. Blood Flow Metab. 2012, 32, 1024–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, X.D.; Ma, Y.H.; Luo, P.; Cao, L.; Lau, W.B.; Zhao, B.C.; Han, F.; Liu, W.; Ning, W.D.; Su, N.; et al. Up-regulation of heme oxygenase-1 attenuates brain damage after cerebral ischemia via simultaneous inhibition of superoxide production and preservation of NO bioavailability. Exp. Neurol. 2013, 239, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.Y.; Liou, H.C.; Fu, W.M. The mechanism of heme oxygenase-1 action involved in the enhancement of neurotrophic factor expression. Neuropharmacology 2010, 58, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hon, T.; Ye, W.; Zhang, L. Heme deficiency interferes with the Ras-mitogen-activated protein kinase signaling pathway and expression of a subset of neuronal genes. Cell Growth Differ. 2002, 13, 431–439. [Google Scholar]
- Chiabrando, D.; Marro, S.; Mercurio, S.; Giorgi, C.; Petrillo, S.; Vinchi, F.; Fiorito, V.; Fagoonee, S.; Camporeale, A.; Turco, E.; et al. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Investig. 2012, 122, 4569–4579. [Google Scholar] [CrossRef] [Green Version]
- Shaibani, A.; Wong, L.J.; Wei Zhang, V.; Lewis, R.A.; Shinawi, M. Autosomal recessive posterior column ataxia with retinitis pigmentosa caused by novel mutations in the FLVCR1 gene. Int. J. Neurosci. 2015, 125, 43–49. [Google Scholar] [CrossRef]
- Flemmig, J.; Zámocký, M.; Alia, A. Amyloid β and free heme: Bloody new insights into the pathogenesis of Alzheimer’s disease. Neural Regen Res. 2018, 13, 1170–1174. [Google Scholar] [CrossRef]
- Chiziane, E.; Telemann, H.; Krueger, M.; Adler, J.; Arnhold, J.; Alia, A.; Flemmig, J. Free Heme and Amyloid-β: A Fatal Liaison in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 61, 963–984. [Google Scholar] [CrossRef]
- Ashraf, A.; Ashton, N.J.; Chatterjee, P.; Goozee, K.; Shen, K.; Fripp, J.; Ames, D.; Rowe, C.; Masters, C.L.; Villemagne, V.; et al. Plasma transferrin and hemopexin are associated with altered Aβ uptake and cognitive decline in Alzheimer’s disease pathology. Alzheimers Res. Ther. 2020, 12, 72. [Google Scholar] [CrossRef]
- Vidal, C.; Daescu, K.; Fitzgerald, K.E.; Starokadomska, A.; Bezprozvanny, I.; Zhang, L. Amyloid beta perturbs elevated heme flux induced with neuronal development. Alzheimer’s Dement. 2019, 5, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; Hampel, H.; Feldman, H.H.; Scheltens, P.; Aisen, P.; Andrieu, S.; Bakardjian, H.; Benali, H.; Bertram, L.; Blennow, K.; et al. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimer’s Dement. 2016, 12, 292–323. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, B.E.; Smith, M.A.; Richardson, S.L.; Perry, G.; Zhu, X. Down-regulation of aminolevulinate synthase, the rate-limiting enzyme for heme biosynthesis in Alzheimer’s disease. Neurosci. Lett. 2009, 460, 180–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, K.M.; Kócsi, Z.; Stone, J. Microvascular pathology in the aging human brain: Evidence that senile plaques are sites of microhaemorrhages. Neurobiol. Aging 2006, 27, 1786–1796. [Google Scholar] [CrossRef]
- Kikkawa, R. Chronic complications in diabetes mellitus. Br. J. Nutr. 2000, 84 (Suppl. S2), S183–S185. [Google Scholar] [CrossRef]
- Hooda, J.; Shah, A.; Zhang, L. Heme, an Essential Nutrient from Dietary Proteins, Critically Impacts Diverse Physiological and Pathological Processes. Nutrients 2014, 6, 1080–1102. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Cao, J.C.; Arija, V.; Aranda, N.; Bullo, M.; Basora, J.; Martínez-González, M.A.; Díez-Espino, J.; Salas-Salvadó, J. Heme iron intake and risk of new-onset diabetes in a Mediterranean population at high risk of cardiovascular disease: An observational cohort analysis. BMC Public Health 2013, 13, 1042. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R. Body Iron Stores in Relation to Risk of Type 2 Diabetes in Apparently Healthy Women. JAMA 2004, 291, 711. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Folsom, A.R.; Jacobs, D.R. Dietary iron intake and Type 2 diabetes incidence in postmenopausal women: The Iowa Women’s Health Study. Diabetologia 2004, 47, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Pan, A.; Sun, Q.; Bernstein, A.M.; Schulze, M.B.; Manson, J.E.; Willett, W.C.; Hu, F.B. Red meat consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. Am. J. Clin. Nutr. 2011, 94, 1088–1096. [Google Scholar] [CrossRef]
- Rajpathak, S.; Ma, J.; Manson, J.; Willett, W.C.; Hu, F.B. Iron Intake and the Risk of Type 2 Diabetes in Women: A prospective cohort study. Diabetes Care 2006, 29, 1370–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Hu, X.; Yuan, B.; Pan, X.; Meyer, H.E.; Holmboe-Ottesen, G. Association Between Serum Ferritin, Hemoglobin, Iron Intake, and Diabetes in Adults in Jiangsu, China. Diabetes Care 2006, 29, 1878–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, D.L.; Collinson, A. Red Meat, Dietary Heme Iron, and Risk of Type 2 Diabetes: The Involvement of Advanced Lipoxidation Endproducts. Adv. Nutr. 2013, 4, 403–411. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Li, S.; Liu, G.; Yan, F.; Ma, X.; Huang, Z.; Tian, H. Body iron stores and heme-iron intake in relation to risk of type 2 diabetes: A systematic review and meta-analysis. PLoS ONE 2012, 7, e41641. [Google Scholar] [CrossRef] [PubMed]
- Ben, Q.; Xu, M.; Ning, X.; Liu, J.; Hong, S.; Huang, W.; Zhang, H.; Li, Z. Diabetes mellitus and risk of pancreatic cancer: A meta-analysis of cohort studies. Eur. J. Cancer 2011, 47, 1928–1937. [Google Scholar] [CrossRef]
- Friberg, E.; Orsini, N.; Mantzoros, C.S.; Wolk, A. Diabetes mellitus and risk of endometrial cancer: A meta-analysis. Diabetologia 2007, 50, 1365–1374. [Google Scholar] [CrossRef]
- Giovannucci, E.; Harlan, D.M.; Archer, M.C.; Bergenstal, R.M.; Gapstur, S.M.; Habel, L.A.; Pollak, M.; Regensteiner, J.G.; Yee, D. Diabetes and Cancer: A consensus report. Diabetes Care 2010, 33, 1674–1685. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Ben, Q.; Shen, H.; Lu, W.; Zhang, Y.; Zhu, J. Diabetes mellitus and incidence and mortality of colorectal cancer: A systematic review and meta-analysis of cohort studies. Eur. J. Epidemiol. 2011, 26, 863–876. [Google Scholar] [CrossRef]
- Larsson, S.C.; Mantzoros, C.S.; Wolk, A. Diabetes mellitus and risk of breast cancer: A meta-analysis. Int. J. Cancer 2007, 121, 856–862. [Google Scholar] [CrossRef]
- Mitri, J.; Castillo, J.; Pittas, A.G. Diabetes and Risk of Non-Hodgkin’s Lymphoma: A meta-analysis of observational studies. Diabetes Care 2008, 31, 2391–2397. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Wang, X.; Gong, G.; Ben, Q.; Qiu, W.; Chen, Y.; Li, G.; Wang, L. Increased risk of hepatocellular carcinoma in patients with diabetes mellitus: A systematic review and meta-analysis of cohort studies. Int. J. Cancer 2012, 130, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Barone, B.B. Long-term All-Cause Mortality in Cancer Patients With Preexisting Diabetes Mellitus: A Systematic Review and Meta-analysis. JAMA 2008, 300, 2754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renehan, A.G.; Yeh, H.C.; Johnson, J.A.; Wild, S.H.; Gale, E.A.M.; Møller, H.; on behalf of the Diabetes and Cancer Research Consortium. Diabetes and cancer (2): Evaluating the impact of diabetes on mortality in patients with cancer. Diabetologia 2012, 55, 1619–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Navarrete, J.M.; Rodríguez, A.; Ortega, F.; Becerril, S.; Sabater-Masdeu, M.; Latorre, J.; Ricart, W.; Frühbeck, G.; Fernández-Real, J.M. Increased adipose tissue heme levels and exportation are associated with altered systemic glucose metabolism. Sci. Rep. 2017, 7, 5305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simcox, J.A.; McClain, D.A. Iron and diabetes risk. Cell Metab. 2013, 17, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Hoffmeister, P.A.; Storer, B.E.; Sanders, J.E. Diabetes mellitus in long-term survivors of pediatric hematopoietic cell transplantation. J. Pediatr. Hematol. Oncol. 2004, 26, 81–90. [Google Scholar] [CrossRef]
- Baker, K.S.; Ness, K.K.; Steinberger, J.; Carter, A.; Francisco, L.; Burns, L.J.; Sklar, C.; Forman, S.; Weisdorf, D.; Gurney, J.G.; et al. Diabetes, hypertension, and cardiovascular events in survivors of hematopoietic cell transplantation: A report from the bone marrow transplantation survivor study. Blood 2007, 109, 1765–1772. [Google Scholar] [CrossRef] [Green Version]
- Vogiatzi, M.G.; Macklin, E.A.; Trachtenberg, F.L.; Fung, E.B.; Cheung, A.M.; Vichinsky, E.; Olivieri, N.; Kirby, M.; Kwiatkowski, J.L.; Cunningham, M.; et al. Differences in the prevalence of growth, endocrine and vitamin D abnormalities among the various thalassaemia syndromes in North America. Br. J. Haematol. 2009, 146, 546–556. [Google Scholar] [CrossRef] [Green Version]
- Messina, M.F.; Lombardo, F.; Meo, A.; Miceli, M.; Wasniewska, M.; Valenzise, M.; Ruggeri, C.; Arrigo, T.; De Luca, F. Three-year prospective evaluation of glucose tolerance, beta-cell function and peripheral insulin sensitivity in non-diabetic patients with thalassemia major. J. Endocrinol. Investig. 2002, 25, 497–501. [Google Scholar] [CrossRef]
- Min, D.; Brooks, B.; Wong, J.; Aamidor, S.; Seehoo, R.; Sutanto, S.; Harrisberg, B.; Yue, D.K.; Twigg, S.M.; McLennan, S.V. Monocyte CD163 is altered in association with diabetic complications: Possible protective role. J. Leukoc. Biol. 2016, 100, 1375–1383. [Google Scholar] [CrossRef] [Green Version]
- Kawarabayashi, R.; Motoyama, K.; Nakamura, M.; Yamazaki, Y.; Morioka, T.; Mori, K.; Fukumoto, S.; Imanishi, Y.; Shioi, A.; Shoji, T.; et al. The Association between Monocyte Surface CD163 and Insulin Resistance in Patients with Type 2 Diabetes. J. Diabetes Res. 2017, 2017, 6549242. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.P.; Purushothaman, K.R.; Levy, N.S.; Purushothaman, M.; Strauss, M.; Asleh, R.; Marsh, S.; Cohen, O.; Moestrup, S.K.; Moller, H.J.; et al. Downregulation of the hemoglobin scavenger receptor in individuals with diabetes and the Hp 2-2 genotype: Implications for the response to intraplaque hemorrhage and plaque vulnerability. Circ. Res. 2007, 101, 106–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Liu, L.; Zhang, X.; Fan, X.; Krishnamurthy, P.; Verma, S.; Tongers, J.; Misener, S.; Ashcherkin, N.; Sun, H.; et al. IL-10 provides cardioprotection in diabetic myocardial infarction via upregulation of Heme clearance pathways. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Snyder, M.P. Metformin Affects Heme Function as a Possible Mechanism of Action. G3 Genes Genomes Genet. 2018, 9, 513–522. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-W.; He, S.-J.; Feng, X.; Cheng, J.; Luo, Y.-T.; Tian, L.; Huang, Q. Metformin: A review of its potential indications. Drug Des. Dev. Ther. 2017, 11, 2421–2429. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yang, Z.; Xu, H.; Zhang, P.; Gao, Z.; Li, H. Insulin enhances the peroxidase activity of heme by forming heme-insulin complex: Relevance to type 2 diabetes mellitus. Int. J. Biol. Macromol. 2017, 102, 1009–1015. [Google Scholar] [CrossRef]
- Mukherjee, S.; Dey, S.G. Heme bound amylin: Spectroscopic characterization, reactivity, and relevance to type 2 diabetes. Inorg. Chem. 2013, 52, 5226–5235. [Google Scholar] [CrossRef]
- Saitoh, S.; Okano, S.; Nohara, H.; Nakano, H.; Shirasawa, N.; Naito, A.; Yamamoto, M.; Kelly, V.P.; Takahashi, K.; Tanaka, T.; et al. 5-aminolevulinic acid (ALA) deficiency causes impaired glucose tolerance and insulin resistance coincident with an attenuation of mitochondrial function in aged mice. PLoS ONE 2018, 13, e0189593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handschin, C.; Lin, J.; Rhee, J.; Peyer, A.K.; Chin, S.; Wu, P.H.; Meyer, U.A.; Spiegelman, B.M. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell 2005, 122, 505–515. [Google Scholar] [CrossRef] [Green Version]
- Higashikawa, F.; Noda, M.; Awaya, T.; Tanaka, T.; Sugiyama, M. 5-aminolevulinic acid, a precursor of heme, reduces both fasting and postprandial glucose levels in mildly hyperglycemic subjects. Nutrition 2013, 29, 1030–1036. [Google Scholar] [CrossRef]
- Rodriguez, B.L.; Curb, J.D.; Davis, J.; Shintani, T.; Perez, M.H.; Apau-Ludlum, N.; Johnson, C.; Harrigan, R.C. Use of the dietary supplement 5-aminiolevulinic acid (5-ALA) and its relationship with glucose levels and hemoglobin A1C among individuals with prediabetes. Clin. Transl. Sci. 2012, 5, 314–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
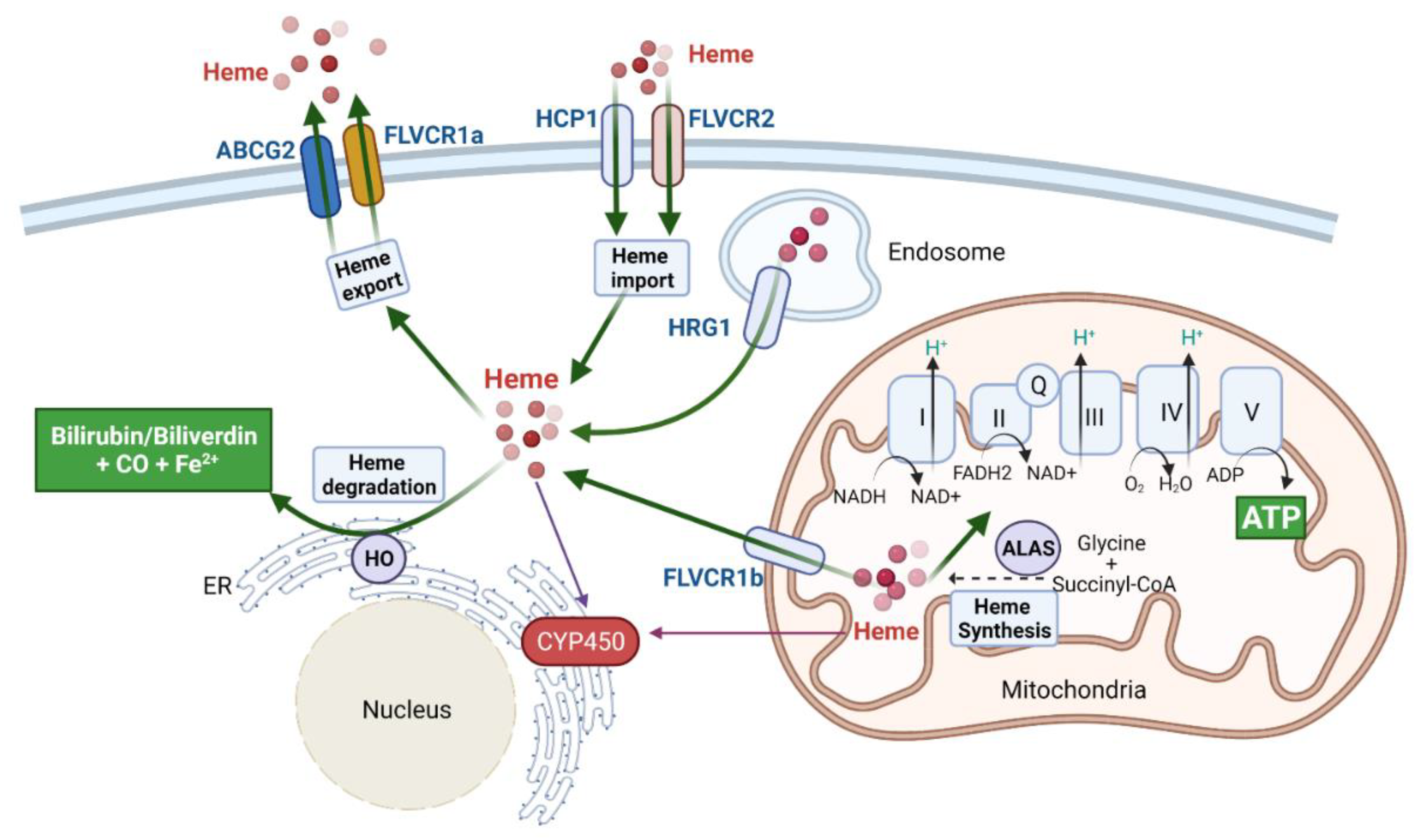


| Protein Name | Binding Residues | Binding Affinity | Heme Function | Heme in Proteins Involved in Pathogenesis of Disease | Reference |
|---|---|---|---|---|---|
| BACH1 | CP motifs in HRM3-6 | Kd = 1.37 × 10−5 M (C-terminal region) | Promote the dissociation of BACH1 from DNA; promote nuclear export and ubiquitination of BACH1 | Heme destabilizes BACH1, leading to the inhibition of different types of cancer | [136,143,144,145] |
| PGRMC1 | Tyr113, Tyr107, Lys163, and Tyr164 | Kd = 50 nM | Mediate PGRMC1 dimerization | Mediates PGRMC1 regulated EGFR and cytochromes P450 activity in colon cancer cells and hepatoma cells | [138,146,147] |
| P53 | Cys275/Cys277 | Kd = ~1.2 µM | Mediate P53 destabilization | Heme–P53 may mediate colon carcinoma cell suppression based on iron-deprivation | [139] |
| CBS | Cys52/His65 Cys15/His22 | Kd = 2.18 ± 0.64 µM (Cys15/His22) | Promote CBS folding and assembling | Lack of in vivo study. Limiting heme inhibits CBS activity without abolishing the enzymatic activity in vitro. | [140,148,149] |
| sGC | His105 in β1 subunit | Contains heme as a cofactor | Cause conformational change to initiate the first step in sGC activation | A cofactor that is required for essential sGC activity | [150,151,152] |
| NOS | NOS1/nNOS: Cys419 NOS2/iNOS: Cys184 NOS3/eNOS: Cys200 | Contains heme as a cofactor | Cofactor to NOS which catalyzes NO synthesis | Heme is required to maintain basic enzyme function including NOS homodimerization and catalysis of NO synthesis | [153,154,155] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Ashrafi, A.; Modareszadeh, P.; Deese, A.R.; Chacon Castro, M.D.C.; Alemi, P.S.; Zhang, L. An Analysis of the Multifaceted Roles of Heme in the Pathogenesis of Cancer and Related Diseases. Cancers 2021, 13, 4142. https://doi.org/10.3390/cancers13164142
Wang T, Ashrafi A, Modareszadeh P, Deese AR, Chacon Castro MDC, Alemi PS, Zhang L. An Analysis of the Multifaceted Roles of Heme in the Pathogenesis of Cancer and Related Diseases. Cancers. 2021; 13(16):4142. https://doi.org/10.3390/cancers13164142
Chicago/Turabian StyleWang, Tianyuan, Adnin Ashrafi, Pouya Modareszadeh, Alexander R. Deese, Maria Del Carmen Chacon Castro, Parinaz Sadat Alemi, and Li Zhang. 2021. "An Analysis of the Multifaceted Roles of Heme in the Pathogenesis of Cancer and Related Diseases" Cancers 13, no. 16: 4142. https://doi.org/10.3390/cancers13164142







