Subcellular Localizations of Catalase and Exogenously Added Fatty Acid in Chlamydomonas reinhardtii
Abstract
:1. Introduction
- (1)
- Is there a catalase isoform localized in C. reinhardtii peroxisomes?
- (2)
- Where are exogenously added FAs incorporated into C. reinhardtii?
2. Materials and Methods
2.1. Algal Strains and Culture Conditions
2.2. Fatty Acid Preparations
2.3. Fluorescent Protein Constructs and Algal Transformation
2.4. Fluorescence Microscopy Analysis
3. Results
3.1. A Catalase Isoform Is Localized in Peroxisomes
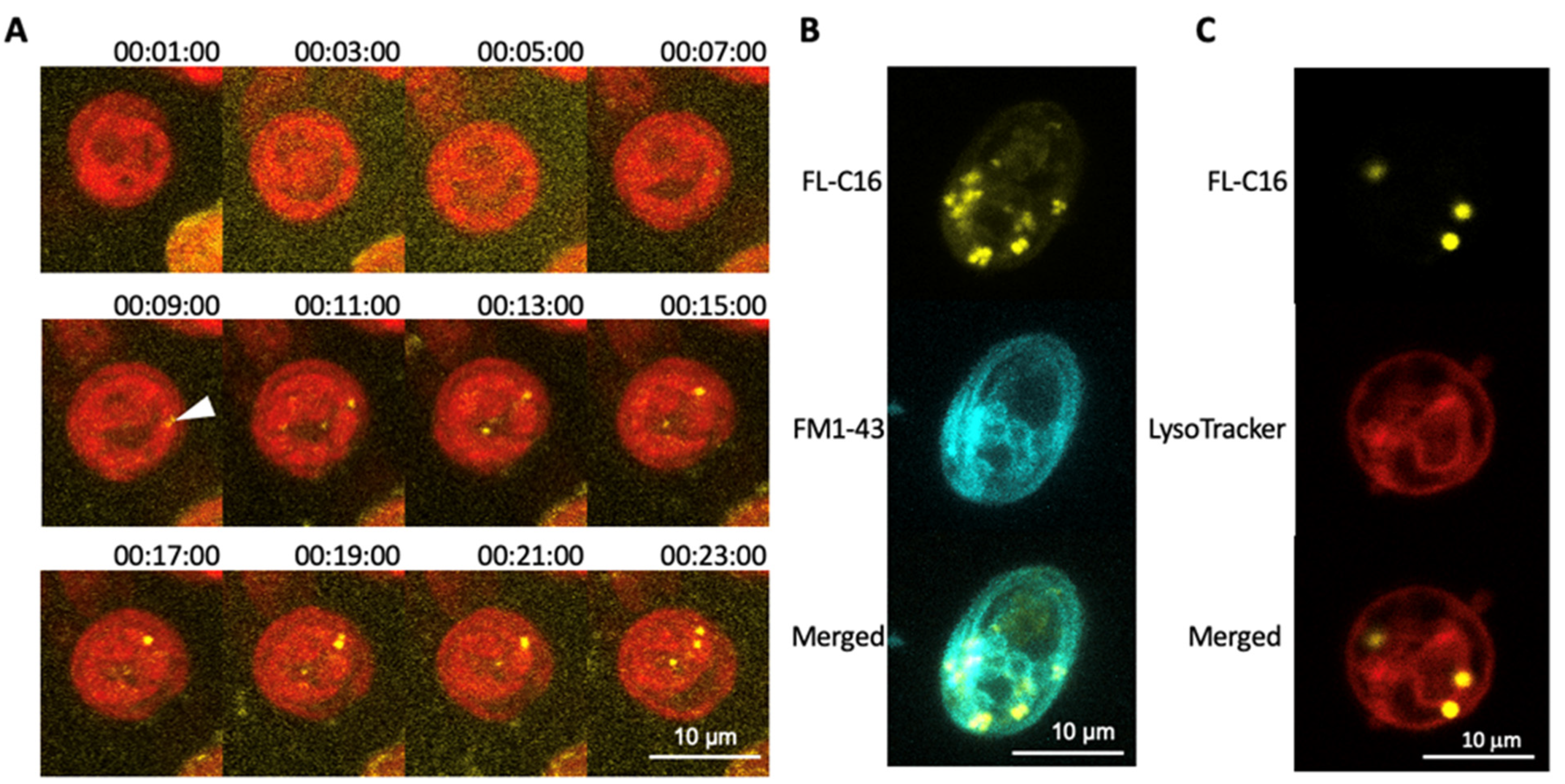
3.2. Exogenously Added Fatty Acids Are Incorporated into Cells within 10 Min via a Non-Endocytic Mechanism and Are Not Directed to Lysosomes
3.3. FAIMs Are Not Peroxisomes or Sub-Compartments in Mitochondria
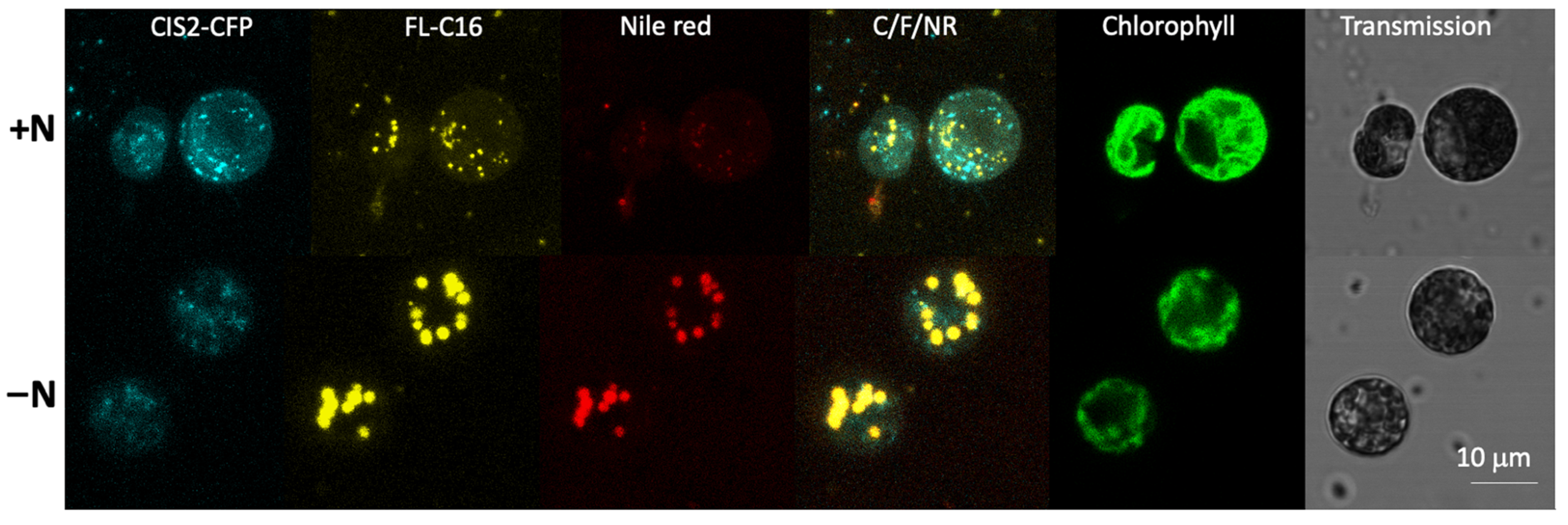
3.4. Exogenously Added FA Are Localized in Lipid Droplets
4. Discussion
4.1. Peroxisomal Localization of CAT1 Supports the Idea That the β-Oxidation Occurs in the Peroxisome
4.2. Separation of FAIMs and Peroxisomes Supports Membrane Turnover Observations
4.3. Biogenesis of FAIMs Remains a Question
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harris, E. Chlamydomonas as a Model Organism. Annu. Rev. Plant Physiol. 2001, 52, 363–406. [Google Scholar] [CrossRef] [Green Version]
- Rochaix, J.-D. Chlamydomonas Reinhardtii As The Photosynthetic Yeast. Annu. Rev. 1995, 29, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Liang, Y.; Légeret, B.; Beyly-Adriano, A.; Blangy, S.; Haslam, R.P.; Napier, J.A.; Beisson, F.; Peltier, G.; Li-Beisson, Y. Chlamydomonas carries out fatty acid β-oxidation in ancestral peroxisomes using a bona fide acyl-CoA oxidase. Plant J. 2017, 90, 358–371. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Sommerfeld, M.; Jarvis, E.; Ghirardi, M.; Posewitz, M.; Seibert, M.; Darzins, A. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008, 54, 621–639. [Google Scholar] [CrossRef]
- Mussgnug, J.H. Genetic tools and techniques for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2015, 99, 5407–5418. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Marechal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Benning, C. Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 2013, 24, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.G.; Beevers, H. Mitochondria and glyoxysomes from castor bean endosperm. Enzyme constitutents and catalytic capacity. J. Biol. Chem. 1969, 244, 3507–3513. [Google Scholar] [CrossRef]
- Lauersen, K.J.; Willamme, R.; Coosemans, N.; Joris, M.; Kruse, O.; Remacle, C. Peroxisomal microbodies are at the crossroads of acetate assimilation in the green microalga Chlamydomonas reinhardtii. Algal Res. 2016, 16, 266–274. [Google Scholar] [CrossRef]
- Pracharoenwattana, I.; Smith, S.M. When is a peroxisome not a peroxisome? Trends Plant Sci. 2008, 13, 522–525. [Google Scholar] [CrossRef]
- Rengel, R.; Smith, R.T.; Haslam, R.P.; Sayanova, O.; Vila, M.; León, R. Overexpression of acetyl-CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii. Algal Res. 2018, 31, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Stern, D.; Witman, G.B. The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use; Academic Press: Waltham, MA, USA, 2008; Volume 1. [Google Scholar]
- Kato, J.; Yamahara, T.; Tanaka, K.; Takio, S.; Satoh, T. Characterization of catalase from green algae Chlamydomonas reinhardtii. J. Plant Physiol. 1997, 151, 262–268. [Google Scholar] [CrossRef]
- Spalding, M.H. Chapter 8–The CO2-Concentrating Mechanism and Carbon Assimilation. In The Chlamydomonas Sourcebook, 2nd ed.; Harris, E.H., Stern, D.B., Witman, G.B., Eds.; Academic Press: London, UK, 2009; pp. 257–301. [Google Scholar]
- Nakamura, Y.; Kanakagiri, S.; Van, K.; He, W.; Spalding, M.H. Disruption of the glycolate dehydrogenase gene in the high-CO2-requiring mutant HCR89 of Chlamydomonas reinhardtii. Can. J. Bot. 2005, 83, 820–833. [Google Scholar] [CrossRef]
- Hayashi, Y.; Sato, N.; Shinozaki, A.; Watanabe, M. Increase in peroxisome number and the gene expression of putative glyoxysomal enzymes in Chlamydomonas cells supplemented with acetate. J. Plant Res. 2014, 128, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Young, D.Y.; Shachar-Hill, Y. Large fluxes of fatty acids from membranes to triacylglycerol and back during N-deprivation and recovery in Chlamydomonas. Plant Physiol. 2020, 185, 796–814. [Google Scholar] [CrossRef]
- McCammon, M.T.; Veenhuis, M.; Trapp, S.B.; Goodman, J.M. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J. Bacteriol. 1990, 172, 5816–5827. [Google Scholar] [CrossRef] [Green Version]
- Faergeman, N.J.; DiRusso, C.C.; Elberger, A.; Knudsen, J.; Black, P.N. Disruption of the Saccharomyces cerevisiae homologue to the murine fatty acid transport protein impairs uptake and growth on long-chain fatty acids. J. Biol. Chem. 1997, 272, 8531–8538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faergeman, N.; Black, P.N.; Zhao, X.D.; Knudsen, J.; DiRusso, C.C. The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular Utilization. J. Biol. Chem. 2001, 276, 37051–37059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Z.; DiRusso, C.C.; Ctrnacta, V.; Black, P.N. Fatty acid transport in Saccharomyces cerevisiae. Directed mutagenesis of FAT1 distinguishes the biochemical activities associated with Fat1p. J. Biol. Chem. 2002, 277, 31062–31071. [Google Scholar] [CrossRef] [Green Version]
- Jia, B.; Song, Y.; Wu, M.; Lin, B.; Xiao, K.; Hu, Z.; Huang, Y. Characterization of long-chain acyl-CoA synthetases which stimulate secretion of fatty acids in green algae Chlamydomonas reinhardtii. Biotechnol. Biofuels 2016, 9, 184. [Google Scholar] [CrossRef] [Green Version]
- Kato, N.; Dong, T.; Bailey, M.; Lum, T.; Ingram, D. Triacylglycerol mobilization is suppressed by brefeldin A in Chlamydomonas reinhardtii. Plant Cell Physiol. 2013, 54, 1585–1599. [Google Scholar] [CrossRef] [Green Version]
- Giroud, C.; Eichenberger, W. Lipids of Chlamydomonas Reinhardtii: Incorporation of 14C-Acetate, Palmitate and Oleate into Different Lipids and Evidence for Lipid-Linked Desaturation of Fatty Acids. In Biological Role of Plant Lipids; Biacs, P.A., Gruiz, K., Kremmer, T., Eds.; Springer: Boston, MA, USA, 1989; pp. 61–64. [Google Scholar]
- Neupert, J.; Karcher, D.; Bock, R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009, 57, 1140–1150. [Google Scholar] [CrossRef]
- Gorman, D.S.; Levine, R.P. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 1965, 54, 1665–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauersen, K.J.; Kruse, O.; Mussgnug, J.H. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 2015, 99, 3491–3503. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef] [Green Version]
- Reumann, S. Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 2004, 135, 783–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konno, T.; Pinho Melo, E.; Lopes, C.; Mehmeti, I.; Lenzen, S.; Ron, D.; Avezov, E. ERO1-independent production of H2O2 within the endoplasmic reticulum fuels Prdx4-mediated oxidative protein folding. J. Cell Biol. 2015, 211, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.D.; Saaranen, M.J.; Karala, A.R.; Lappi, A.K.; Wang, L.; Raykhel, I.B.; Alanen, H.I.; Salo, K.E.; Wang, C.C.; Ruddock, L.W. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol 2011, 406, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0, Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Tardif, M.; Atteia, A.; Specht, M.; Cogne, G.; Rolland, N.; Brugiere, S.; Hippler, M.; Ferro, M.; Bruley, C.; Peltier, G.; et al. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012, 29, 3625–3639. [Google Scholar] [CrossRef] [Green Version]
- Pelham, H.R. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 1990, 15, 483–486. [Google Scholar] [CrossRef]
- Thumser, A.E.; Storch, J. Characterization of a BODIPY-labeled fluorescent fatty acid analogue. Binding to fatty acid-binding proteins, intracellular localization, and metabolism. Mol. Cell Biochem. 2007, 299, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kolahi, K.; Louey, S.; Varlamov, O.; Thornburg, K. Real-Time Tracking of BODIPY-C12 Long-Chain Fatty Acid in Human Term Placenta Reveals Unique Lipid Dynamics in Cytotrophoblast Cells. PLoS ONE 2016, 11, e0153522. [Google Scholar]
- Rigal, A.; Doyle, S.M.; Robert, S. Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells. Methods Mol. Biol. 2015, 1242, 93–103. [Google Scholar]
- Jaishy, B.; Abel, E.D. Lipids, lysosomes, and autophagy. J. Lipid Res. 2016, 57, 1619–1635. [Google Scholar] [CrossRef] [Green Version]
- DeVorkin, L.; Gorski, S.M. LysoTracker staining to aid in monitoring autophagy in Drosophila. Cold Spring Harb. Protoc. 2014, 2014, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Sengupta, N.; Morgan, J.A. Metabolic flux analysis of heterotrophic growth in Chlamydomonas reinhardtii. PLoS ONE 2017, 12, e0177292. [Google Scholar] [CrossRef] [Green Version]
- Li-Beisson, Y.; Beisson, F.; Riekhof, W. Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 2015, 82, 504–522. [Google Scholar] [CrossRef] [PubMed]
- Durante, L.; Hubner, W.; Lauersen, K.J.; Remacle, C. Characterization of the GPR1/FUN34/YaaH protein family in the green microalga Chlamydomonas suggests their role as intracellular membrane acetate channels. Plant Direct 2019, 3, e00148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siaut, M.; Cuine, S.; Cagnon, C.; Fessler, B.; Nguyen, M.; Carrier, P.; Beyly, A.; Beisson, F.; Triantaphylides, C.; Li-Beisson, Y.; et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: Characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Drummen, G.P.; van Liebergen, L.C.; Op den Kamp, J.A.; Post, J.A. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic. Biol. Med. 2002, 33, 473–490. [Google Scholar] [CrossRef]
- Rambold, A.S.; Cohen, S.; Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 2015, 32, 678–692. [Google Scholar] [CrossRef] [Green Version]
- Chu, M.; Sampath, H.; Cahana, D.Y.; Kahl, C.A.; Somwar, R.; Cornea, A.; Roberts, C.T., Jr.; Varlamov, O. Spatiotemporal dynamics of triglyceride storage in unilocular adipocytes. Mol. Biol. Cell. 2014, 25, 4096–4105. [Google Scholar] [CrossRef]
- Somwar, R.; Roberts, C.T.; Varlamov, O. Live-cell imaging demonstrates rapid cargo exchange between lipid droplets in adipocytes. FEBS Lett. 2011, 585, 1946–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulermo, R.; Gamboa-Melendez, H.; Ledesma-Amaro, R.; Thevenieau, F.; Nicaud, J.M. Unraveling fatty acid transport and activation mechanisms in Yarrowia lipolytica. Biochim. Biophys. Acta 2015, 1851, 1202–1217. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Moscatelli, J.; Smith, E.M.; Banerjee, C.; Puchner, E.M. Single-molecule localization microscopy and tracking with red-shifted states of conventional BODIPY conjugates in living cells. Nat. Commun. 2019, 10, 3400. [Google Scholar] [CrossRef] [Green Version]
- Joshi, A.S.; Nebenfuehr, B.; Choudhary, V.; Satpute-Krishnan, P.; Levine, T.P.; Golden, A.; Prinz, W.A. Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 2018, 9, 2940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.S.; Martin, O.C.; Pagano, R.E. Changes in the spectral properties of a plasma membrane lipid analog during the first seconds of endocytosis in living cells. Biophys. J. 1997, 72, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Black, P.N.; DiRusso, C.C. A live-cell high-throughput screening assay for identification of fatty acid uptake inhibitors. Anal. Biochem. 2005, 336, 11–19. [Google Scholar] [CrossRef]
- Carten, J.D.; Bradford, M.K.; Farber, S.A. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 2011, 360, 276–285. [Google Scholar] [CrossRef] [Green Version]


| Target | Oligonucleotide 5′–3′ |
|---|---|
| CAT1-NFw | AATTTCATATGGACCCCGCCAAGATCCGCCCCAGCAGCGCCTACAACACCCCCTACTG |
| CAT1-NRw | AAATTGGATCCCACGGGGGCGCCGCTGTTGGTGGTCCAGTAGGGGGTGTTGTAGGCGCTGC |
| CAT1-CFw | AATTTGATATCGTGGGCTACTGGAGCCAGGCCGACCCCCAGCTGGGCGCCCGCATCGCCGC |
| CAT1-CRv | AAATTGAATTCTTACAGGCAGCCGCGGCCCTGCAGCTTGGCGGCGATGCGGGCGCCCAGCTGGG |
| CAT2-NFw | AATTTCATATGCGCGACAAGGCCCTGATCACCCTGCTGCTGGCCGCCAGCGCCGCCTT |
| CAT2-NRv | AAATTGGATCCGCCGAAGGGGCACTTGCAGGTGGCGAAGGCGGCGCTGGCGGCCAGCAGCA |
| CAT2-CFw | AATTTGATATCGCCGCCGTGGTGCAGCTGGTGCTGGAGGCCAGCTACGGCGCCGCCGCCCG |
| CAT2-CRv | AAATTGAATTCTTACAGCTCCACGAACTCCTCCTCCACGCGGGCGGCGGCGCCGTAGCTGGCCT |
| Set 1 | Excitation (nm) | Emission (nm) |
| CFP | 458 | 463–490 |
| FLC-16 | 500 | 505–530 |
| MitoTracker | 580 | 585–620 * |
| Chlorophylls | 458 | 697–800 |
| Set 2 | Excitation (nm) | Emission (nm) |
| CFP | 458 | 463–490 |
| FLC-16 | 500 | 505–530 |
| Nile Red | 540 | 545–600 * |
| Chlorophylls | 458 | 697–800 |
| Set 3 | Excitation (nm) | Emission (nm) |
| FL-C16 | 502 | 507–540 |
| FM1-43 | 470 | 565–655 |
| Set 4 | Excitation (nm) | Emission (nm) |
| FL-C16 | 500 | 505–532 |
| LysoTracker | 576 | 583–643C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, N.; Nelson, G.; Lauersen, K.J. Subcellular Localizations of Catalase and Exogenously Added Fatty Acid in Chlamydomonas reinhardtii. Cells 2021, 10, 1940. https://doi.org/10.3390/cells10081940
Kato N, Nelson G, Lauersen KJ. Subcellular Localizations of Catalase and Exogenously Added Fatty Acid in Chlamydomonas reinhardtii. Cells. 2021; 10(8):1940. https://doi.org/10.3390/cells10081940
Chicago/Turabian StyleKato, Naohiro, Gabela Nelson, and Kyle J. Lauersen. 2021. "Subcellular Localizations of Catalase and Exogenously Added Fatty Acid in Chlamydomonas reinhardtii" Cells 10, no. 8: 1940. https://doi.org/10.3390/cells10081940






