Biodiversity and Constrained Information Dynamics in Ecosystems: A Framework for Living Systems
Abstract
1. Introduction
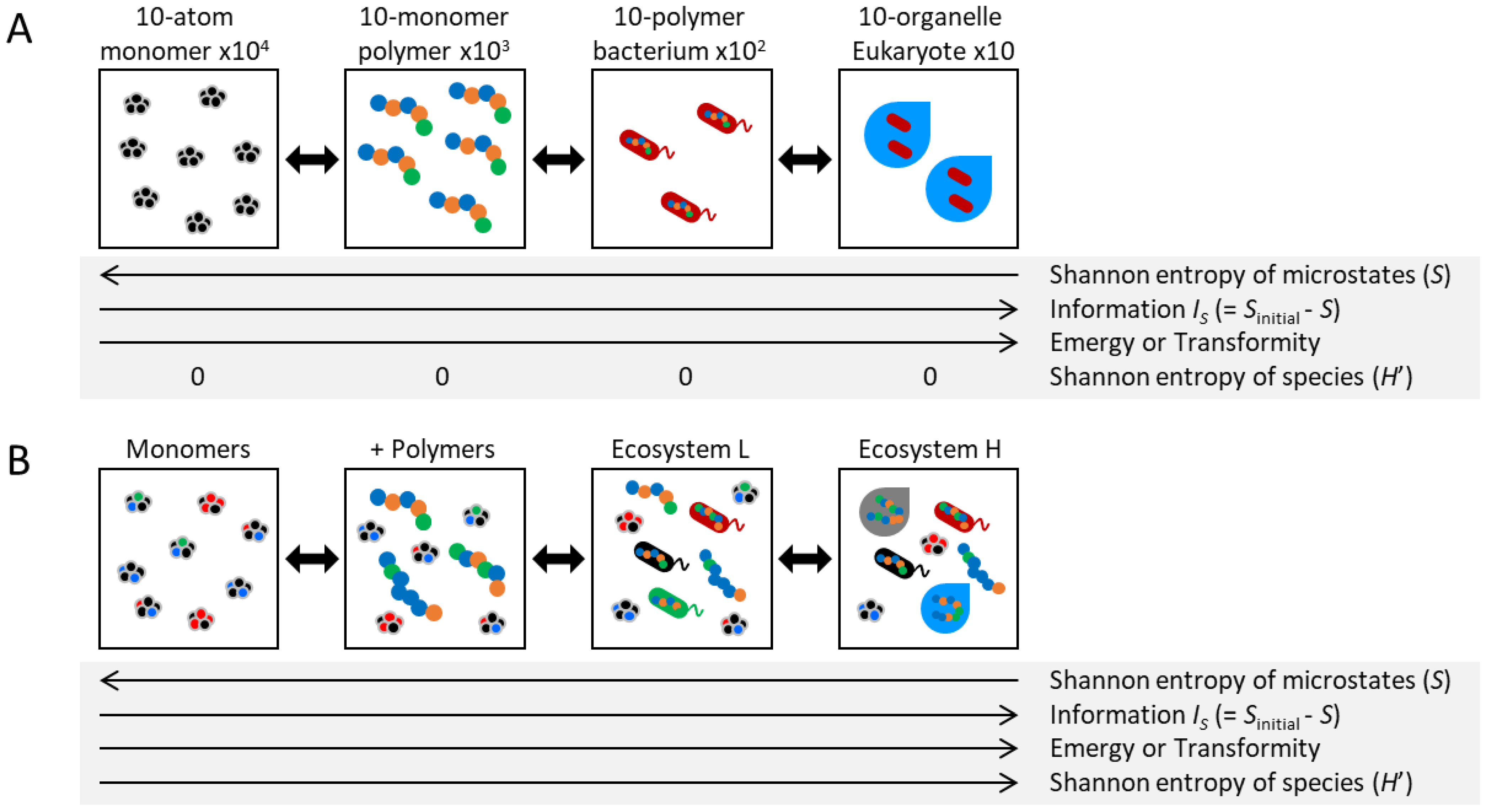
1.1. Ecosystem Framework and Macroscopic Parameters
1.2. Information Dynamics in Living Systems: Macroscopic and Microscopic Perspectives
1.3. Diversity and Information Dynamics in Ecosystems: Necessity of Adaptability
1.4. Mechanism for Information Increase and Identifying Information Carriers in Ecosystems
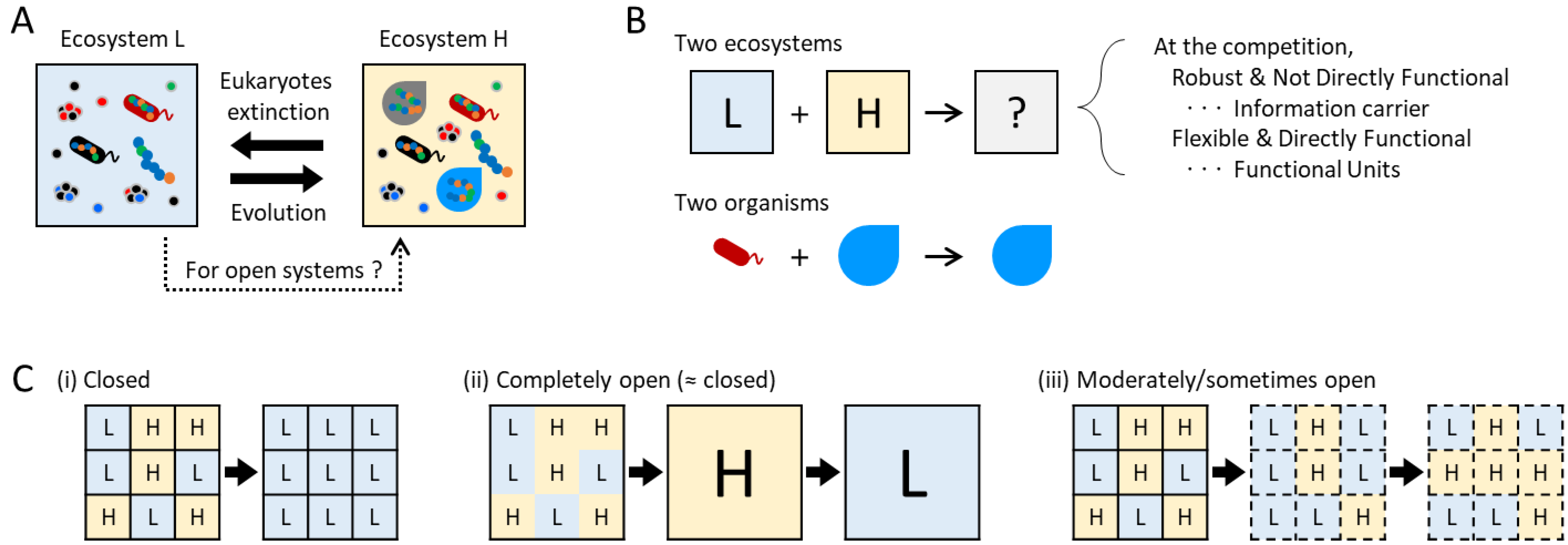
1.5. Freedom and Constraints, Homeostasis and Homeorhesis
1.6. Using Experimental Ecosystems as a Phenomenological Approach
1.7. Experiments in This Study
2. Materials and Methods
2.1. Microorganisms
2.2. Microcosm Experiments
2.3. Measurements
3. Results and Discussion
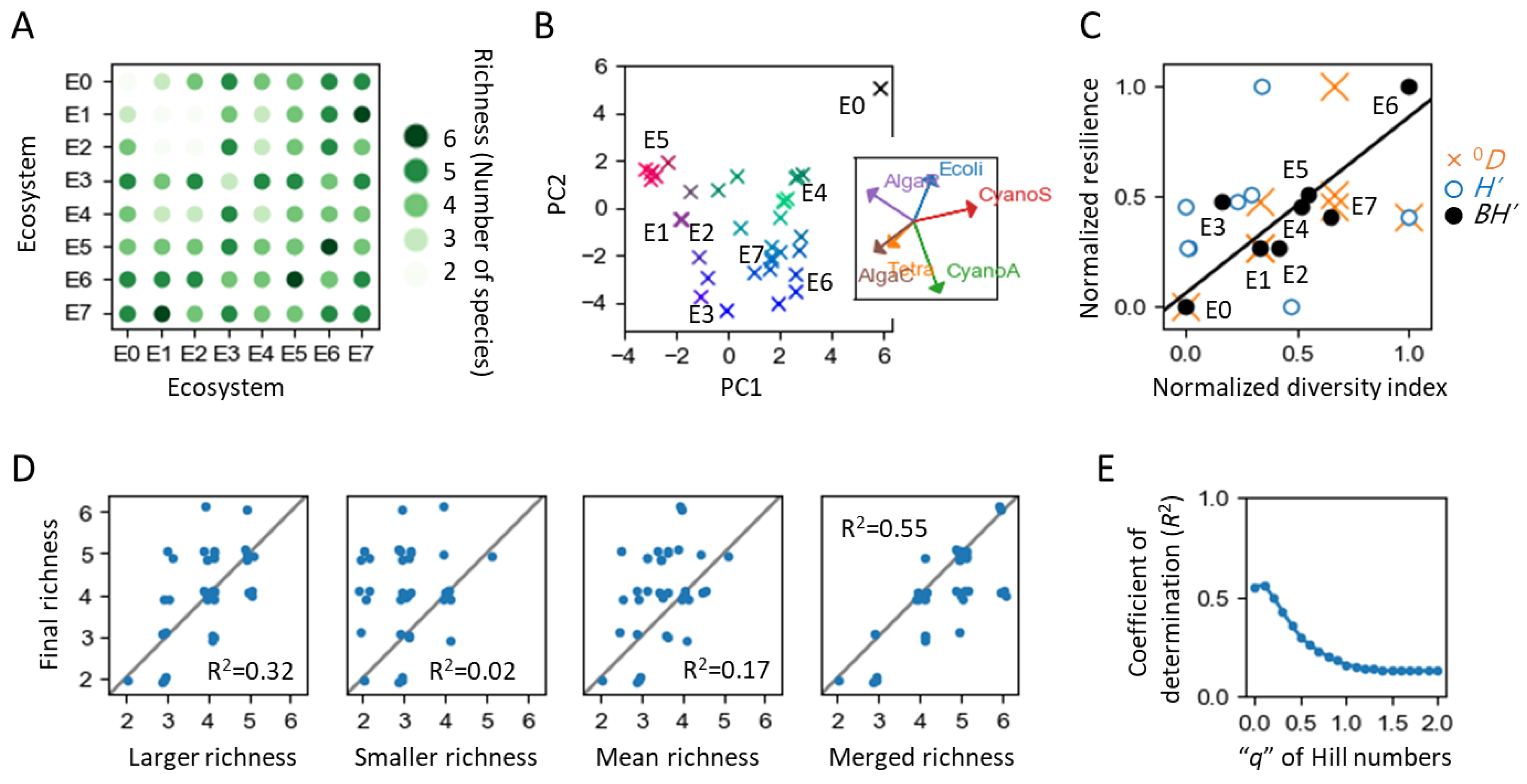
3.1. Ecosystems Used as Initial State
3.2. Ecosystem Coalescence Experiments for Investigating Competitive Stability and Information Carrier
3.3. Ecosystem Constraints for Investigating the Dominant Mode Hypothesis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conrad, M. Adaptability: The Significance of Variability from Molecule to Ecosystem; Springer: Berlin/Heidelberg, Germany, 1983. [Google Scholar]
- England, J.L. Dissipative adaptation in driven self-assembly. Nat. Nanotechnol. 2015, 10, 919–923. [Google Scholar] [CrossRef] [PubMed]
- Odum, E.P.; Barrett, G.W. Fundamentals of Ecology, 5th ed.; Thomson Brooks/Cole: Belmont, CA, USA, 2005. [Google Scholar]
- May, R.M. Will a Large Complex System Be Stable. Nature 1972, 238, 413–414. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, M. Foraging adaptation and the relationship between food-web complexity and stability. Science 2003, 299, 1388–1391. [Google Scholar] [CrossRef]
- Wilmers, C.C.; Sinha, S.; Brede, M. Examining the effects of species richness on community stability: An assembly model approach. Oikos 2002, 99, 363–367. [Google Scholar] [CrossRef]
- Nature. Why interdisciplinary research matters. Nature 2015, 525, 305. [Google Scholar] [CrossRef] [PubMed]
- Cover, T.M.; Thomas, J.A. Elements of information theory second edition solutions to problems. Internet Access 2006, 19–20. [Google Scholar]
- Seifert, U. Stochastic thermodynamics, fluctuation theorems and molecular machines. Rep. Prog. Phys. 2012, 75, 126001. [Google Scholar] [CrossRef]
- Sagawa, T. Second law, entropy production, and reversibility in thermodynamics of information. In Energy Limits in Computation: A Review of Landauer’s Principle, Theory and Experiments; Springer: Cham, Switzerland, 2019; pp. 101–139. [Google Scholar]
- Lairez, D. Thermodynamical versus logical irreversibility: A concrete objection to Landauer’s principle. arXiv 2023, arXiv:2301.07026. [Google Scholar] [CrossRef]
- Baez, J.C.; Pollard, B.S. Relative entropy in biological systems. Entropy 2016, 18, 46. [Google Scholar] [CrossRef]
- Harper, M.; Fryer, D.E. Stability of evolutionary dynamics on time scales. arXiv 2012, arXiv:1210.5539. [Google Scholar]
- Hau, J.L.; Bakshi, B.R. Promise and problems of emergy analysis. Ecol. Model. 2004, 178, 215–225. [Google Scholar] [CrossRef]
- Wang, Q.; Xiao, H.; Ma, Q.; Yuan, X.; Zuo, J.; Zhang, J.; Wang, S.; Wang, M. Review of Emergy Analysis and Life Cycle Assessment: Coupling Development Perspective. Sustainability 2020, 12, 367. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.-H.; Jost, L. Phylogenetic diversity measures and their decomposition: A framework based on Hill numbers. Biodivers. Conserv. Phylogenetic Syst. 2016, 14, 141–172. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Leinster, T. Entropy and Diversity: The Axiomatic Approach; Cambridge University Press: Cambridge, UK; London, UK, 2021. [Google Scholar]
- Chao, A.; Chiu, C.-H.; Jost, L. Unifying Species Diversity, Phylogenetic Diversity, Functional Diversity, and Related Similarity and Differentiation Measures Through Hill Numbers. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 297–324. [Google Scholar] [CrossRef]
- Chao, A.; Jost, L. Estimating diversity and entropy profiles via discovery rates of new species. Methods Ecol. Evol. 2015, 6, 873–882. [Google Scholar] [CrossRef]
- Ito, S.; Sagawa, T. Maxwell’s demon in biochemical signal transduction with feedback loop. Nat. Commun. 2015, 6, 7498. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Kobayashi, T.J. Connection between the bacterial chemotactic network and optimal filtering. Phys. Rev. Lett. 2021, 126, 128102. [Google Scholar] [CrossRef]
- England, J.L. Statistical physics of self-replication. J. Chem. Phys. 2013, 139, 121923. [Google Scholar] [CrossRef] [PubMed]
- Bonduriansky, R.; Day, T. Extended Heredity: A New Understanding of Inheritance and Evolution; Princeton University Press: Princeton, NJ, USA, 2020. [Google Scholar]
- Xavier, M.J.; Roman, S.D.; Aitken, R.J.; Nixon, B. Transgenerational inheritance: How impacts to the epigenetic and genetic information of parents affect offspring health. Hum. Reprod. Update 2019, 25, 519–541. [Google Scholar] [CrossRef]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef]
- Takeuchi, N.; Kaneko, K. The origin of the central dogma through conflicting multilevel selection. Proc. R. Soc. B 2019, 286, 20191359. [Google Scholar] [CrossRef]
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef]
- Kobayashi, I.; Sasa, S.-I. Characterizing the asymmetry in hardness between synthesis and destruction of heteropolymers. Phys. Rev. Lett. 2022, 128, 247801. [Google Scholar] [CrossRef]
- Matsubara, Y.J.; Kaneko, K. Kinetic selection of template polymer with complex sequences. Phys. Rev. Lett. 2018, 121, 118101. [Google Scholar] [CrossRef] [PubMed]
- Odum, E.P. The Strategy of Ecosystem Development: An understanding of ecological succession provides a basis for resolving man’s conflict with nature. Science 1969, 164, 262–270. [Google Scholar] [CrossRef]
- Yamagishi, J.F.; Saito, N.; Kaneko, K. Adaptation of metabolite leakiness leads to symbiotic chemical exchange and to a resilient microbial ecosystem. PLoS Comp. Biol. 2021, 17, e1009143. [Google Scholar] [CrossRef]
- Elton, C.S. The Ecology of Invasions by Animals and Plants; Springer: New York, NY, USA, 1958. [Google Scholar]
- Darwin, C. The Origin of Species by Means of Natural Selection, 6th ed.; The Modern Library: New York, NY, USA, 1872. [Google Scholar]
- Hosoda, K.; Suzuki, S.; Yamauchi, Y.; Shiroguchi, Y.; Kashiwagi, A.; Ono, N.; Mori, K.; Yomo, T. Cooperative adaptation to establishment of a synthetic bacterial mutualism. PLoS ONE 2011, 6, e17105. [Google Scholar] [CrossRef] [PubMed]
- Braun, E. The unforeseen challenge: From genotype-to-phenotype in cell populations. Rep. Prog. Phys. 2015, 78, 036602. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Sherwin, W.B. Entropy, or Information, Unifies Ecology and Evolution and Beyond. Entropy 2018, 20, 727. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Hamada, T.; Shimokawa, T.; Tanifuji, M.; Yanagida, T. Stochastic process underlying emergent recognition of visual objects hidden in degraded images. PLoS ONE 2014, 9, e115658. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Seno, S.; Murata, T. Simulating Reaction Time for Eureka Effect in Visual Object Recognition Using Artificial Neural Network. IIAI Lett. Inform. Interdiscip. Res. 2023, 3. [Google Scholar] [CrossRef]
- Hosoda, K.; Nishida, K.; Seno, S.; Mashita, T.; Kashioka, H.; Ohzawa, I. It’s DONE: Direct ONE-shot learning with quantile weight imprinting. arXiv 2022, arXiv:2204.13361. [Google Scholar] [CrossRef]
- Power, D.A.; Watson, R.A.; Szathmáry, E.; Mills, R.; Powers, S.T.; Doncaster, C.P.; Czapp, B. What can ecosystems learn? Expanding evolutionary ecology with learning theory. Biol. Direct 2015, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Friston, K.J. The free-energy principle: A unified brain theory? Nat. Rev. Neurosci. 2010, 11, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Ulanowicz, R.E. The balance between adaptability and adaptation. Biosystems 2002, 64, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. The Strategy of the Genes. A Discussion of Some Aspects of Theoretical Biology; George Allen & Unwin, Ltd.: Crow’s nest, Australia, 1957. [Google Scholar]
- Cannon, W.B. The Wisdom of the Body; The Norton Library: Norton, MA, USA, 1932. [Google Scholar]
- Furusawa, C.; Kaneko, K. Formation of dominant mode by evolution in biological systems. Phys. Rev. E 2018, 97, 042410. [Google Scholar] [CrossRef]
- Marconi, U.M.B.; Puglisi, A.; Rondoni, L.; Vulpiani, A. Fluctuation–dissipation: Response theory in statistical physics. Phys. Rep. 2008, 461, 111–195. [Google Scholar] [CrossRef]
- Kaneko, K. Phenotypic plasticity and robustness: Evolutionary stability theory, gene expression dynamics model, and laboratory experiments. In Evolutionary Systems Biology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 249–278. [Google Scholar]
- Maeda, T.; Iwasawa, J.; Kotani, H.; Sakata, N.; Kawada, M.; Horinouchi, T.; Sakai, A.; Tanabe, K.; Furusawa, C. High-throughput laboratory evolution reveals evolutionary constraints in Escherichia coli. Nat. Commun. 2020, 11, 5970. [Google Scholar] [CrossRef]
- Kaneko, K.; Furusawa, C.; Yomo, T. Universal relationship in gene-expression changes for cells in steady-growth state. Phys. Rev. X 2015, 5, 011014. [Google Scholar] [CrossRef]
- Furusawa, C.; Kaneko, K. Global relationships in fluctuation and response in adaptive evolution. J. R. Soc. Interface 2015, 12, 20150482. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Barzel, B.; Barabási, A.-L. Universal resilience patterns in complex networks. Nature 2016, 530, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.Y.; D’Odorico, P.; Suweis, S. Dimensionality reduction of complex dynamical systems. Iscience 2021, 24, 101912. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Slotine, J.-J.; Barabási, A.-L. Controllability of complex networks. Nature 2011, 473, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.-J.; Schuppert, A. Few inputs can reprogram biological networks. Nature 2011, 478, E4. [Google Scholar] [CrossRef] [PubMed]
- Sadtler, P.T.; Quick, K.M.; Golub, M.D.; Chase, S.M.; Ryu, S.I.; Tyler-Kabara, E.C.; Yu, B.M.; Batista, A.P. Neural constraints on learning. Nature 2014, 512, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Abbaspourazad, H.; Choudhury, M.; Wong, Y.T.; Pesaran, B.; Shanechi, M.M. Multiscale low-dimensional motor cortical state dynamics predict naturalistic reach-and-grasp behavior. Nat. Commun. 2021, 12, 607. [Google Scholar] [CrossRef]
- Csete, M.; Doyle, J. Bow ties, metabolism and disease. Trends Biotechnol. 2004, 22, 446–450. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- San Miguel, M. Frontiers in Complex Systems. Front. Complex Syst. 2023, 1, 1080801. [Google Scholar] [CrossRef]
- Donohue, I.; Hillebrand, H.; Montoya, J.M.; Petchey, O.L.; Pimm, S.L.; Fowler, M.S.; Healy, K.; Jackson, A.L.; Lurgi, M.; McClean, D. Navigating the complexity of ecological stability. Ecol. Lett. 2016, 19, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Struhl, K. From E. coli to elephants. Nature 2002, 417, 22–23. [Google Scholar] [CrossRef]
- Taub, F.B. A Biological Model of a Freshwater Community—A Gnotobiotic Ecosystem. Limnol. Oceanogr. 1969, 14, 136–142. [Google Scholar] [CrossRef]
- Beyers, R.J.; Odum, H.T. Ecological Microcosms; Springer: New York, NY, USA, 1993. [Google Scholar]
- Tilman, D.; Wedin, D.; Knops, J. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature 1996, 379, 718–720. [Google Scholar] [CrossRef]
- Isbell, F.; Calcagno, V.; Hector, A.; Connolly, J.; Harpole, W.S.; Reich, P.B.; Scherer-Lorenzen, M.; Schmid, B.; Tilman, D.; van Ruijven, J.; et al. High plant diversity is needed to maintain ecosystem services. Nature 2011, 477, 199–202. [Google Scholar] [CrossRef]
- Naeem, S.; Thompson, L.J.; Lawler, S.P.; Lawton, J.H.; Woodfin, R.M. Declining biodiversity can alter the performance of ecosystems. Nature 1994, 368, 734–737. [Google Scholar] [CrossRef]
- Naeem, S.; Li, S.B. Consumer species richness and autotrophic biomass. Ecology 1998, 79, 2603–2615. [Google Scholar] [CrossRef]
- Benton, T.G.; Solan, M.; Travis, J.M.J.; Sait, S.M. Microcosm experiments can inform global ecological problems. Trends Ecol. Evol. 2007, 22, 516–521. [Google Scholar] [CrossRef]
- Hosoda, K.; Seno, S.; Murakami, N.; Matsuda, H.; Osada, Y.; Kamiura, R.; Kondoh, M. Synthetic model ecosystem of 12 cryopreservable microbial species allowing for a noninvasive approach. Biosystems 2023, 235, 105087. [Google Scholar] [CrossRef]
- Rillig, M.C.; Antonovics, J.; Caruso, T.; Lehmann, A.; Powell, J.R.; Veresoglou, S.D.; Verbruggen, E. Interchange of entire communities: Microbial community coalescence. Trends Ecol. Evol. 2015, 30, 470–476. [Google Scholar] [CrossRef]
- Diaz-Colunga, J.; Lu, N.; Sanchez-Gorostiaga, A.; Chang, C.-Y.; Cai, H.S.; Goldford, J.E.; Tikhonov, M.; Sánchez, Á. Top-down and bottom-up cohesiveness in microbial community coalescence. Proc. Natl. Acad. Sci. USA 2022, 119, e2111261119. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.S.; Frentz, Z.; Leibler, S. Homeorhesis and ecological succession quantified in synthetic microbial ecosystems. Proc. Natl. Acad. Sci. USA 2019, 116, 14852–14861. [Google Scholar] [CrossRef] [PubMed]
- Hekstra, D.R.; Leibler, S. Contingency and Statistical Laws in Replicate Microbial Closed Ecosystems. Cell 2012, 149, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Jacques, S.M.; Pires, A.P.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 2016, 1, 0015. [Google Scholar] [CrossRef] [PubMed]
- Goldford, J.E.; Lu, N.; Bajić, D.; Estrela, S.; Tikhonov, M.; Sanchez-Gorostiaga, A.; Segrè, D.; Mehta, P.; Sanchez, A. Emergent simplicity in microbial community assembly. Science 2018, 361, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Tsuda, S.; Kadowaki, K.; Nakamura, Y.; Nakano, T.; Ishii, K. Population-reaction model and microbial experimental ecosystems for understanding hierarchical dynamics of ecosystems. Biosystems 2016, 140, 28–34. [Google Scholar] [CrossRef]
- Momeni, B.; Chen, C.-C.; Hillesland, K.L.; Waite, A.; Shou, W. Using artificial systems to explore the ecology and evolution of symbioses. Cell. Mol. Life Sci. 2011, 68, 1353–1368. [Google Scholar] [CrossRef]
- Tenaillon, O.; Barrick, J.E.; Ribeck, N.; Deatherage, D.E.; Blanchard, J.L.; Dasgupta, A.; Wu, G.C.; Wielgoss, S.; Cruveiller, S.; Medigue, C.; et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 2016, 536, 165–170. [Google Scholar] [CrossRef]
- Blount, Z.D.; Lenski, R.E.; Losos, J.B. Contingency and determinism in evolution: Replaying life’s tape. Science 2018, 362, eaam5979. [Google Scholar] [CrossRef]
- Nakajima, T.; Sano, A.; Matsuoka, H. Auto-/heterotrophic endosymbiosis evolves in a mature stage of ecosystem development in a microcosm composed of an alga, a bacterium and a ciliate. Biosystems 2009, 96, 127–135. [Google Scholar] [CrossRef]
- Germond, A.; Kunihiro, T.; Inouhe, M.; Nakajima, T. Physiological changes of a green alga (Micractinium sp.) involved in an early-stage of association with Tetrahymena thermophila during 5-year microcosm culture. Biosystems 2013, 114, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Fujikawa, Y.; Matsubara, T.; Karita, M.; Sano, A. Differentiation of a free-living alga into forms with ecto- and endosymbiotic associations with heterotrophic organisms in a 5-year microcosm culture. Biosystems 2015, 131, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T. Symbiogenesis is driven through hierarchical reorganization of an ecosystem under closed or semi-closed conditions. Biosystems 2021, 205, 104427. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.M. Simple conditions for growth of unicellular blue-green algae on plates1, 2. J. Phycol. 1968, 4, 1–4. [Google Scholar] [CrossRef]
- Bertani, G. Studies on Lysogenesis.1. The Mode of Phage Liberation by Lysogenic Escherichia-coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Hosoda, K.; Habuchi, M.; Suzuki, S.; Miyazaki, M.; Takikawa, G.; Sakurai, T.; Kashiwagi, A.; Sueyoshi, M.; Matsumoto, Y.; Kiuchi, A. Adaptation of a cyanobacterium to a biochemically rich environment in experimental evolution as an initial step toward a chloroplast-like state. PLoS ONE 2014, 9, e98337. [Google Scholar] [CrossRef]
- Azuma, Y.; Tsuru, S.; Habuchi, M.; Takami, R.; Takano, S.; Yamamoto, K.; Hosoda, K. Synthetic symbiosis between a cyanobacterium and a ciliate toward novel chloroplast-like endosymbiosis. Sci. Rep. 2023, 13, 6104. [Google Scholar] [CrossRef] [PubMed]
- Wilhm, J.L. Use of Biomass Units in Shannon’s Formula. Ecology 1968, 49, 153–156. [Google Scholar] [CrossRef]
- Hossain, M.; Amakawa, T.; Sekiguchi, H. Density, biomass and community structure of megabenthos in Ise Bay, central Japan. Fish. Sci. 1996, 62, 350–360. [Google Scholar] [CrossRef][Green Version]
- Zhuang, S. Species richness, biomass and diversity of macroalgal assemblages in tidepools of different sizes. Mar. Ecol. Prog. Ser. 2006, 309, 67–73. [Google Scholar] [CrossRef][Green Version]
- Menalled, U.D.; Adeux, G.; Cordeau, S.; Smith, R.G.; Mirsky, S.B.; Ryan, M.R. Cereal rye mulch biomass and crop density affect weed suppression and community assembly in no-till planted soybean. Ecosphere 2022, 13, e4147. [Google Scholar] [CrossRef]
- Tan, X.P.; Shan, Y.Q.; Wang, X.; Liu, R.P.; Yao, Y.L. Comparison of the predictive ability of spectral indices for commonly used species diversity indices and Hill numbers in wetlands. Ecol. Indic. 2022, 142, 109233. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef] [PubMed]
- Payton, I.J.; Fenner, M.; Lee, W.G. Keystone Species: The Concept and Its Relevance for Conservation Management in New Zealand; Department of Conservation: Wellington, New Zealand, 2002.
- Chao, A.; Chiu, C.H.; Villéger, S.; Sun, I.F.; Thorn, S.; Lin, Y.C.; Chiang, J.M.; Sherwin, W.B. An attribute-diversity approach to functional diversity, functional beta diversity, and related (dis) similarity measures. Ecol. Monogr. 2019, 89, e01343. [Google Scholar] [CrossRef]



| Species | Abbreviation | Classification | Functional Group | Shape, Approx. Vol. | |
|---|---|---|---|---|---|
| #0 | Escherichia coli | Ecoli | Proteobacteria | Decomposer | Rod, 1 μm3 |
| #1 | Tetrahymena thermophila | Tetra | Ciliophora | Consumer | Oval, 104 μm3 |
| #2 | Anabaenopsis circularis | CyanoA | Cyanobacteria | Producer | Filamentous, 103 μm3 |
| #3 | Synechocystis sp. 6803 | CyanoS | Cyanobacteria | Producer | Spherical, 10 μm3 |
| #4 | Raphidocelis subcapitata | AlgaR | Chlorophyta | Producer | Crescent, 102 μm3 |
| #5 | Chlorella vulgaris | AlgaC | Chlorophyta | Producer | Spherical, 102 μm3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosoda, K.; Seno, S.; Kamiura, R.; Murakami, N.; Kondoh, M. Biodiversity and Constrained Information Dynamics in Ecosystems: A Framework for Living Systems. Entropy 2023, 25, 1624. https://doi.org/10.3390/e25121624
Hosoda K, Seno S, Kamiura R, Murakami N, Kondoh M. Biodiversity and Constrained Information Dynamics in Ecosystems: A Framework for Living Systems. Entropy. 2023; 25(12):1624. https://doi.org/10.3390/e25121624
Chicago/Turabian StyleHosoda, Kazufumi, Shigeto Seno, Rikuto Kamiura, Naomi Murakami, and Michio Kondoh. 2023. "Biodiversity and Constrained Information Dynamics in Ecosystems: A Framework for Living Systems" Entropy 25, no. 12: 1624. https://doi.org/10.3390/e25121624
APA StyleHosoda, K., Seno, S., Kamiura, R., Murakami, N., & Kondoh, M. (2023). Biodiversity and Constrained Information Dynamics in Ecosystems: A Framework for Living Systems. Entropy, 25(12), 1624. https://doi.org/10.3390/e25121624





