1. Introduction
A breakthrough in information theory happened in 2010 when Williams and Beer [
1] published a method called
partial information decomposition, which provided a framework under which the mutual information, shared between the inputs and output of a probabilistic system, could be decomposed into components that measure different aspects of the information: the unique information that each input conveys about the output; the shared information that all inputs possess regarding the output; the information that the inputs in combination have about the output. They also defined a measure of redundancy, Imin, together with a method for obtaining a partial information decomposition (PID), which is also commonly referred to as
Imin. Several authors criticised the definition of the redundancy component in the Imin PID [
2,
3,
4,
5], thus spawning several new methods for computing a partial information decomposition. Harder and colleagues [
2] defined a new measure of redundancy based on information projections, with a PID denoted here as
Iproj, and they introduced the distinction between source redundancy and mechanistic redundancy. Griffiths and Koch [
4] developed a measure of synergy, while independently, Bertschinger and colleagues focused on defining a measure of unique information and defined an optimisation approach to estimate each of the four PID components; it turned out that both approaches resulted in the same decomposition, commonly called
Ibroja. Taking a pointwise approach, Ince [
5] considered measuring redundancy at the level of each individual realisation of the probabilistic system by considering a common change in surprisal, thus creating a measure of redundancy for the system, and PID
Iccs. James and colleagues defined a measure of unique information by using a lattice of maximum entropy distributions based on dependency constraints, with PID,
Idep. Finn and Lizier [
6] introduced a very detailed pointwise approach, with PID,
Ipm. Niu and Quinn [
7] produced a decomposition,
Iig, based on information geometry. Makkeh and colleagues [
8,
9] defined a measure of shared information and PID,
Isx. Most recently, Kolchinsky [
10] used a general approach to define a measure of redundancy based on Blackwell ordering. The corresponding PID has been named
Iprec.The Imin, Iproj, Ibroja and Idep PIDs are guaranteed to have nonnegative components. In [
7], it is claimed that Iig is a nonnegative PID, but examples of systems producing a negative estimate of redundancy were discovered in 2020. Nevertheless, this PID has given sensible results in many systems. No guarantee of non-negativity was given for the Iprec PID, and an example of negative synergy has been found. The other three methods are defined in a pointwise manner by considering information measures at the level of individual realisations and defining partial information components at this level. Pointwise PIDs can produce negative components and they are described as providing
misinformation in such cases [
11].
An important feature of PID is that it enables the shared information and synergistic information in a system to be estimated separately. This provides an advance on earlier research in which the interaction information was used to estimate synergy [
12,
13], but could be negative, and the three-way mutual information [
14] or coinformation [
15], which also could be negative, was used as an objective function in neural networks with two distinct sets of inputs from receptive and contextual fields, respectively.
Partial information decomposition has been applied to data in neuroscience, neuroimaging, neural networks and cellular automata; see, e.g., [
16,
17,
18,
19,
20,
21,
22,
23]. A major selling point of the Isx PID is that the components are differentiable, unlike other PIDs. This makes it possible to build neural networks with a particular neural goal involving PID components as the objective function [
24,
25]. For an overview of PID, see [
26], and for an excellent tutorial, see [
27].
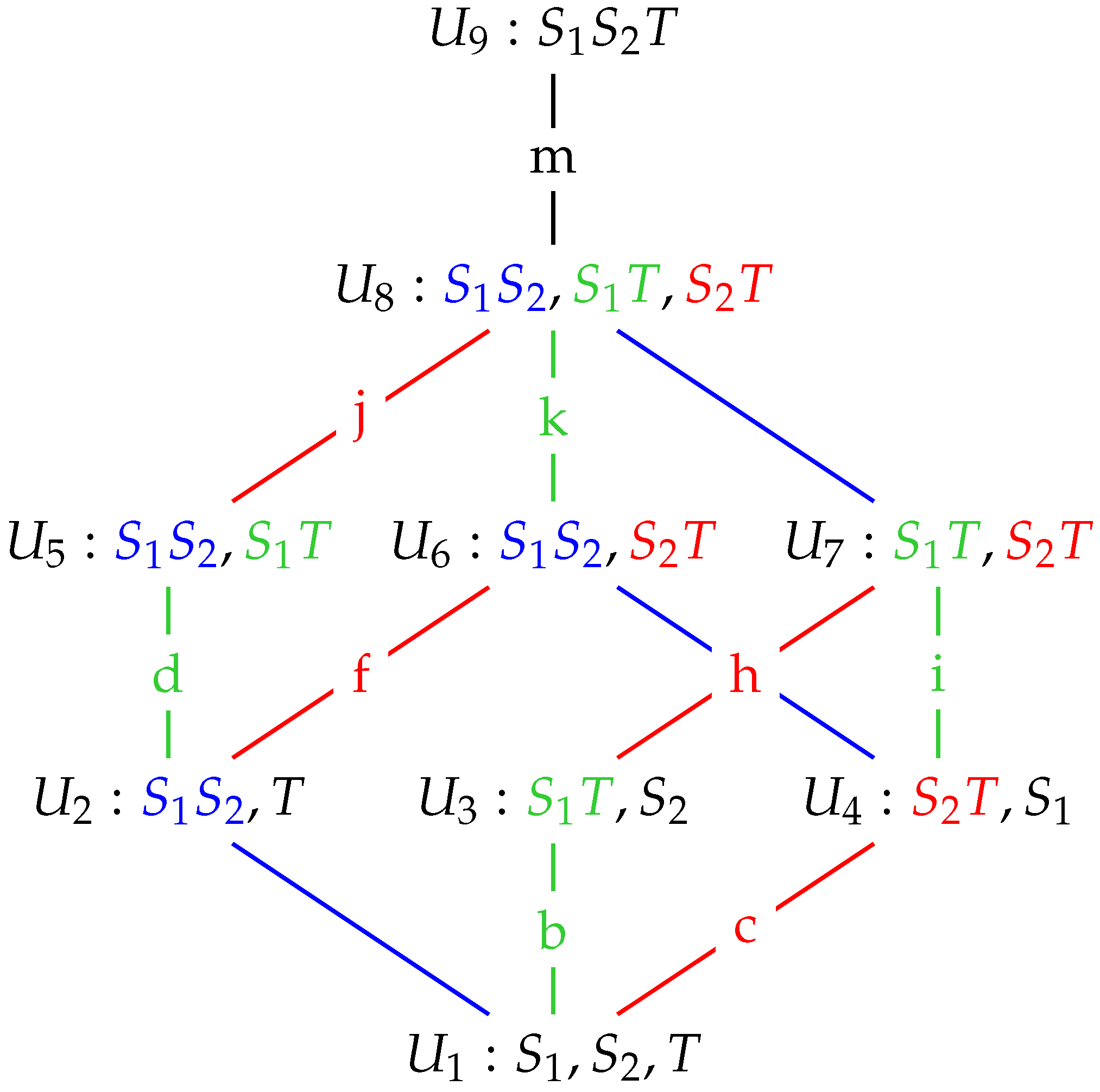
We will provide a systematic comparison of the different methods by applying the Ibroja, Idep, Iccs, Ipm and Isx PIDs to data recorded in two different studies. Further detail and illustration of these PID methods are provided in Appendices
Appendix A,
Appendix B,
Appendix C,
Appendix D,
Appendix E,
Appendix F,
Appendix G and
Appendix H. In
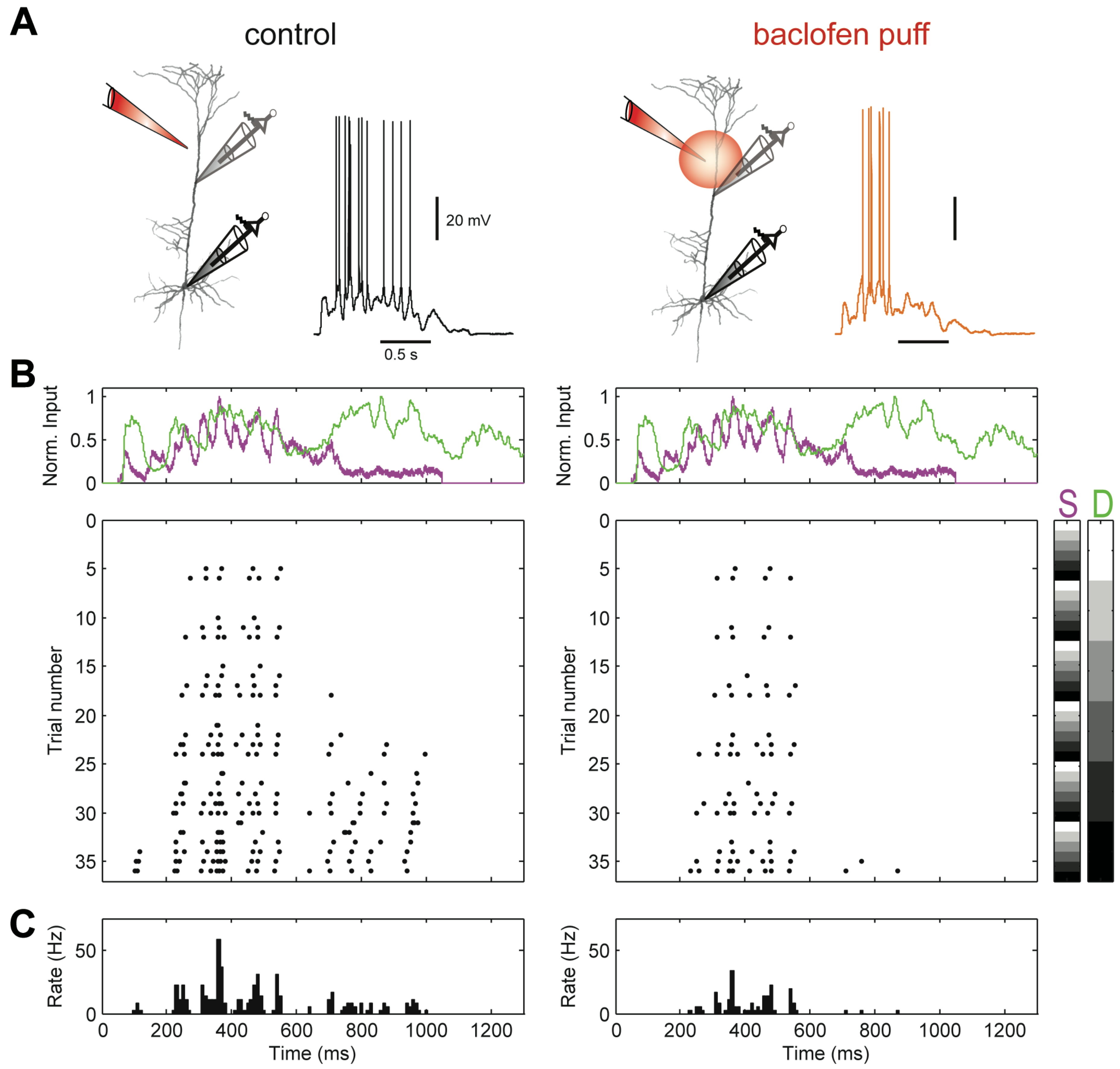
Appendix I, further comparisons are provided involving the Imin, Iproj, Iig and Iprec PIDs. First, we present comparisons of detailed analyses of physiological data recorded from cortical layer 5b (L5b) pyramidal neurons in a study of GABA
receptor-mediated regulation of dendro-somatic synergy [
28]. The influence of GABA
receptor-mediated inhibition of the apical dendrites evoked by local application of the GABA
receptor agonist baclofen will be studied by making within-neuron paired comparisons of PID components. Secondly, we will also shed light on unique information asymmetries as revealed by the PID analyses, as well as discussing the evidence for apical amplification in the presence and absence of GABA
receptor-mediated inhibition of apical dendrites.
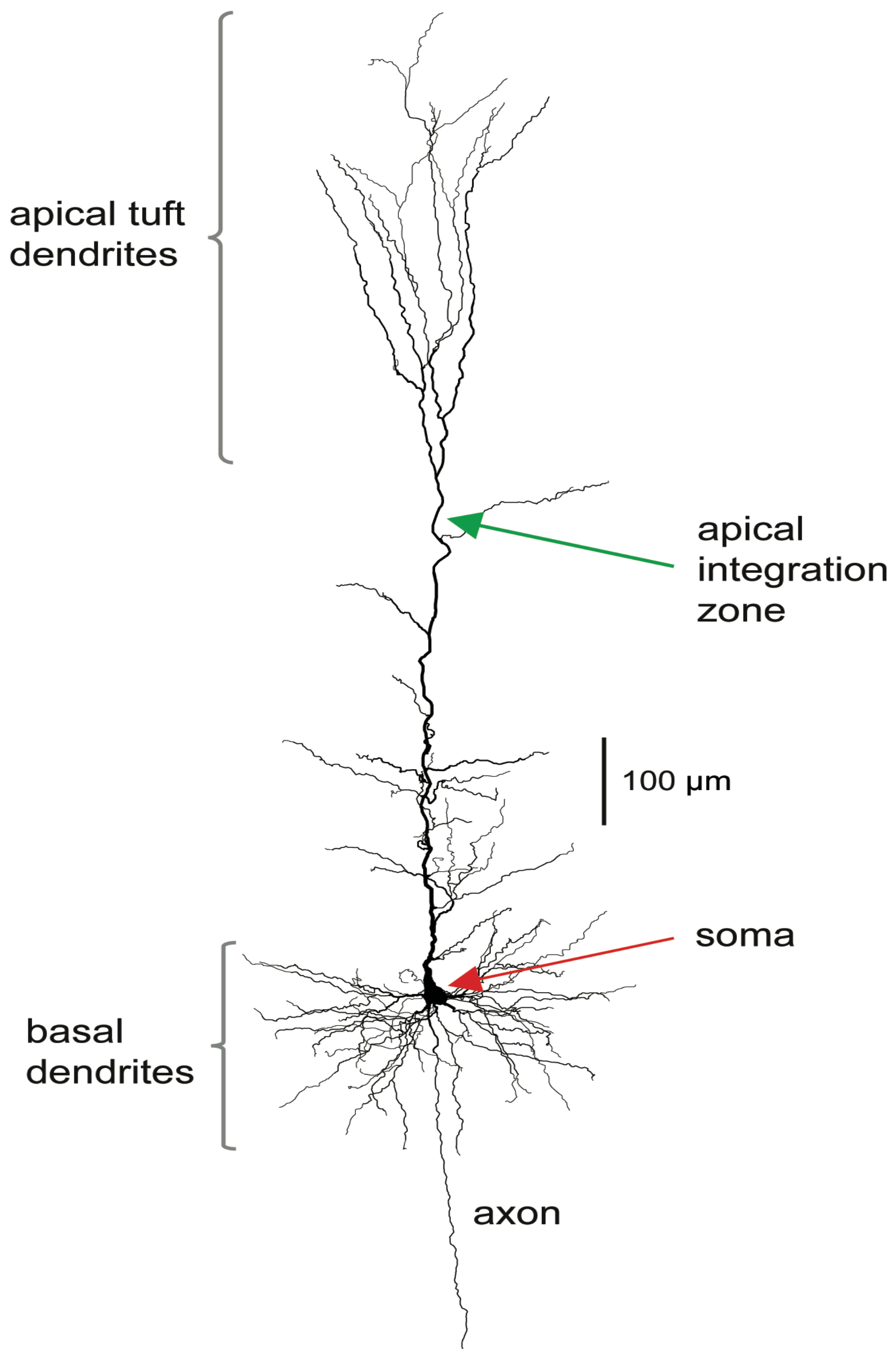
The stereotypical morphology of pyramidal neurons suggests that they have at least two functionally distinct sets of fine dendrites, the basal dendrites that feed directly into the cell body, or soma, from where output action potential(AP)s are generated, and the dendrites of the apical tuft, which are far more distant from the soma and connected to it by the apical trunk. Inputs to the branches of the apical tuft arise from diverse sources that specify various aspects of the context within which the feedforward input to the basal dendrites is processed [
29,
30]. These apical inputs are summed at an integration zone near the top of the apical trunk (see
Figure 1), which, when sufficiently activated, generates Ca
-dependent regenerative potentials in the apical trunk, thus providing a cellular mechanism by which these pyramidal cells can respond more strongly to activation of their basal dendrites when that coincides with activation of their apical dendrites [
31]. Though these experiments require an exceptionally high level of technical expertise, there are now many anatomical and physiological studies indicating that some classes of pyramidal cell can operate as context-sensitive two-point processors, rather than as integrate-and-fire point processors [
31,
32,
33]. Thick-tufted L5b pyramidal cells are the class of pyramidal cell in which operational modes approximating context-sensitive two-point processing have most clearly been demonstrated, but it may apply to some other classes of pyramidal cell also, though not to all [
34]. Mechanistically, these operations are supported by dendritic nonlinear integration of synaptic inputs, including dendritic Ca
spikes and the voltage-dependence of the transfer resistance from dendrite to soma [
28,
35]. These advances in our knowledge of the division of labor between apical and basal dendrites now give the interaction between apical and basal dendritic compartments a prominent role within the broader field of dendritic computation [
36].
The second study [
37] considers data on spike counts obtained using an amended version of the Hay compartmental model [
38]. Spike counts are available for many different combinations of basal and apical inputs. While PIDs can be computed for the entire dataset, an interesting diversity of balance between basal drive and apical drive is revealed by applying the PID methods to subsets of the data defined by various combinations of basal and apical inputs. For one set of subsets, this reveals a bifurcation in unique information asymmetry and a difference among the methods in how this is expressed. A different analysis of subsets allows a full discussion of the extent to which evidence of cooperative context-sensitivity is revealed by the nature of the PID components.
Many empirical findings have been interpreted as indicating that cooperative context-sensitivity is common throughout perceptual and higher cognitive regions of the mammalian neocortex. For example, consider the effect of a flanking context on the ability to detect a short faint line. A surrounding context is neither necessary nor sufficient for that task, but many psychophysical and physiological studies show that context can have large effects, as reviewed, for example, by Lamme [
39,
40,
41]. In [
42,
43], theoretical studies on the effects of context (then called ‘contextual modulation’) were explored. The ideal properties of cooperative context-sensitivity are described in
Section 2.
The goal of this work is two-fold. First, for the datasets considered, we wish to compare the results obtained by employing the different PID methods on probability distributions defined using real and simulated data. Secondly, we intend to use the various PIDs to make inferences about the functioning of the pyramidal cells under investigation.
4. Conclusions and Discussion
4.1. Rat Somatosensory Cortical L5b Pyramidal Neuron Recording Data
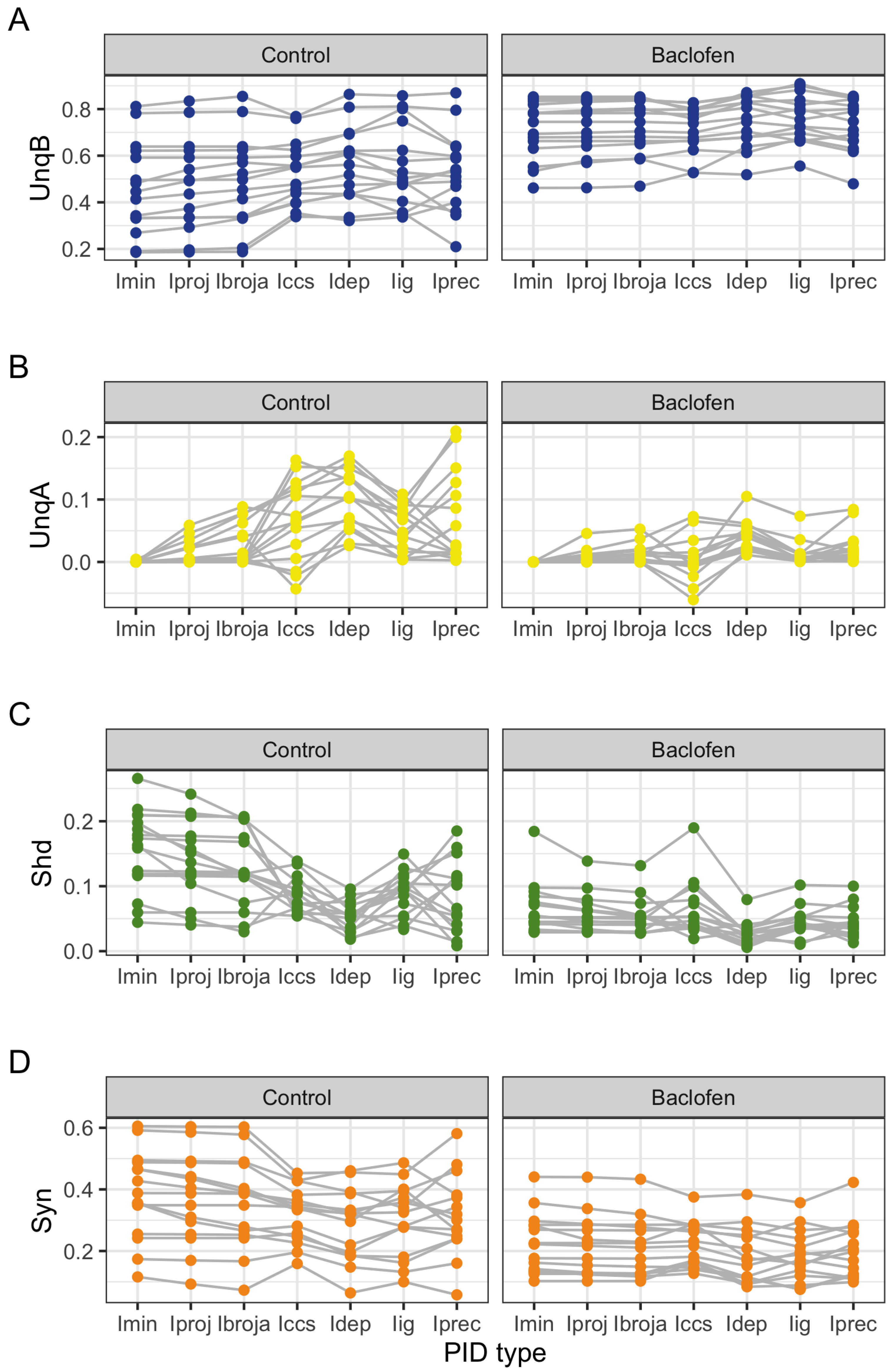
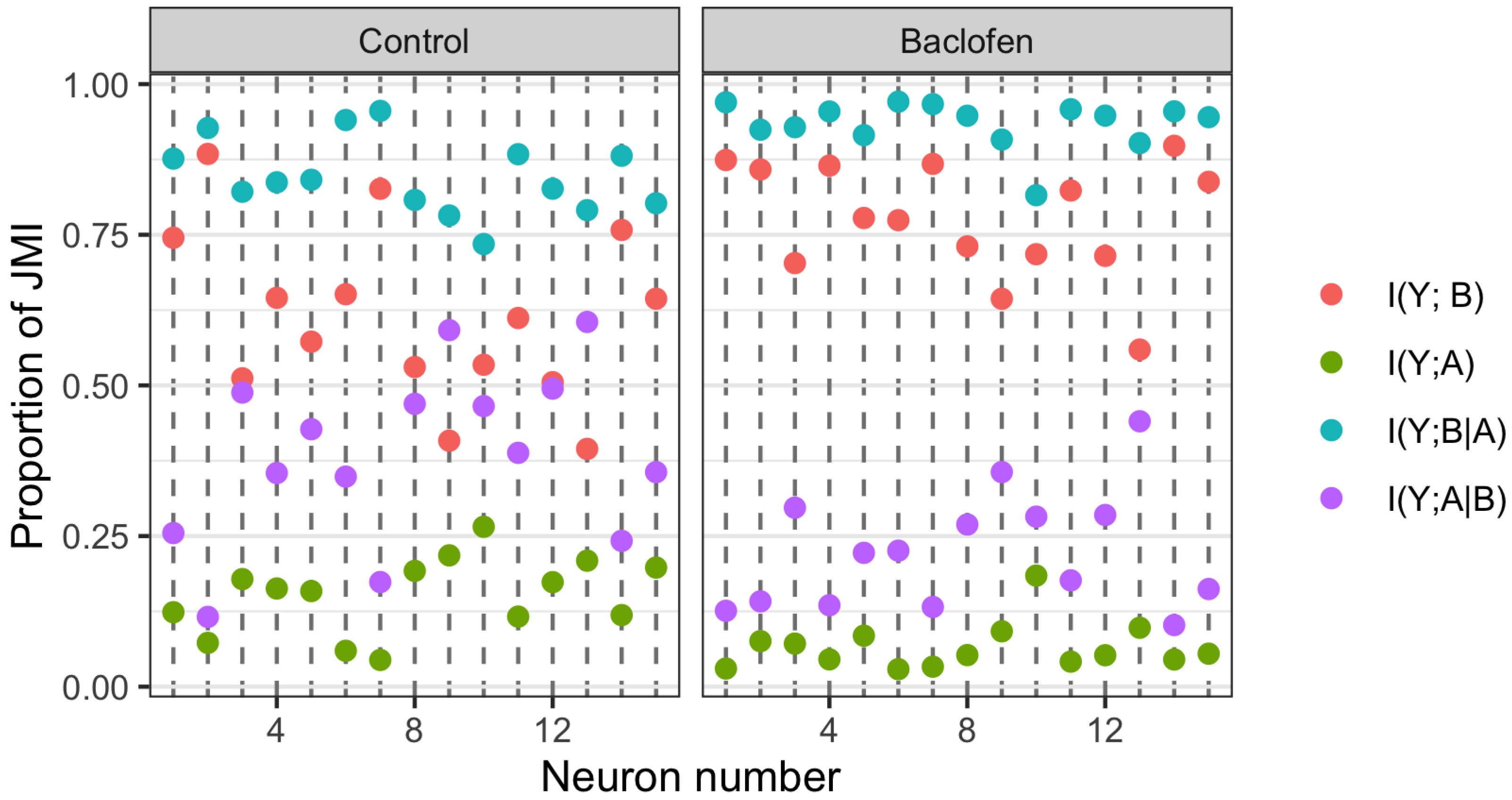
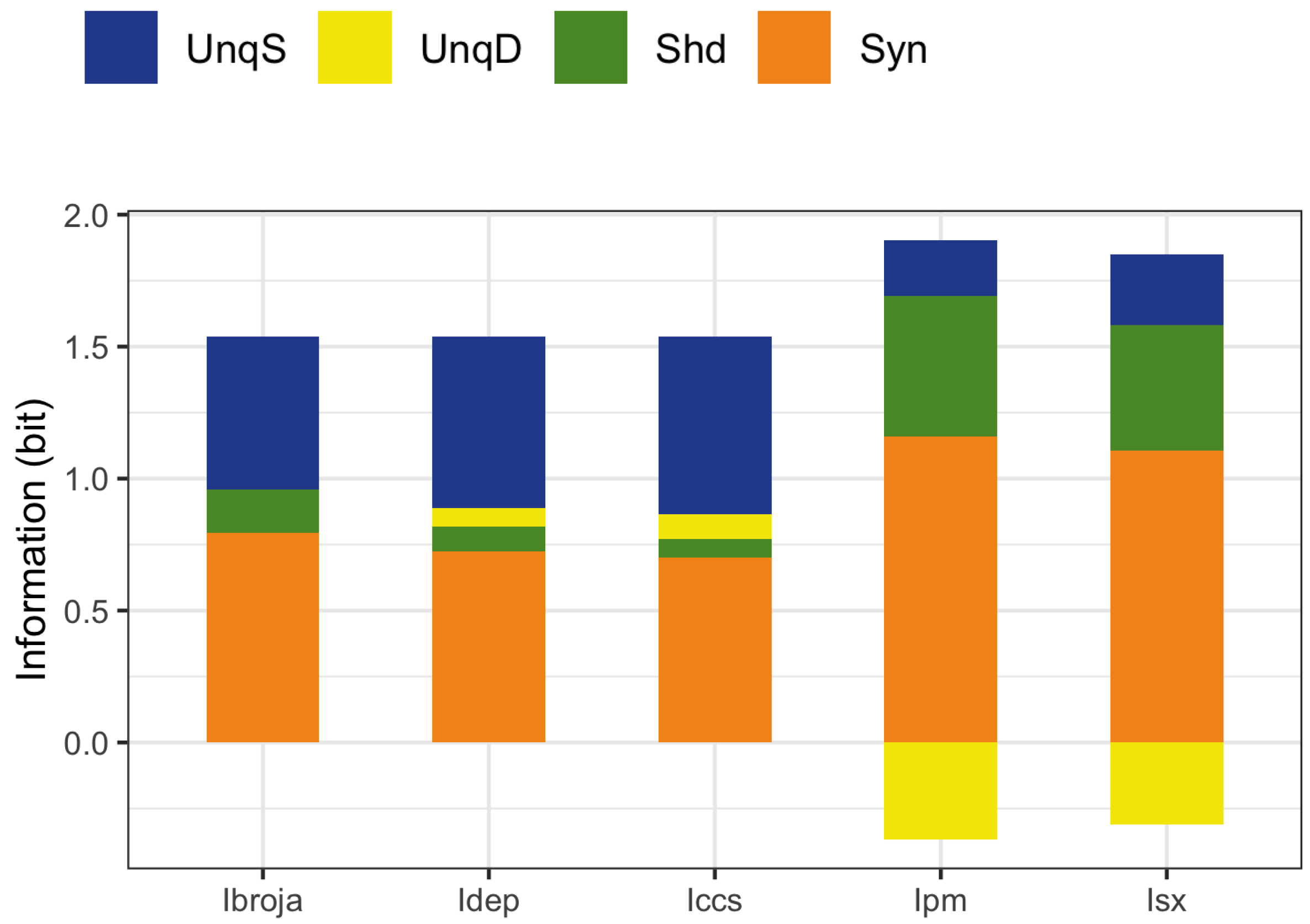
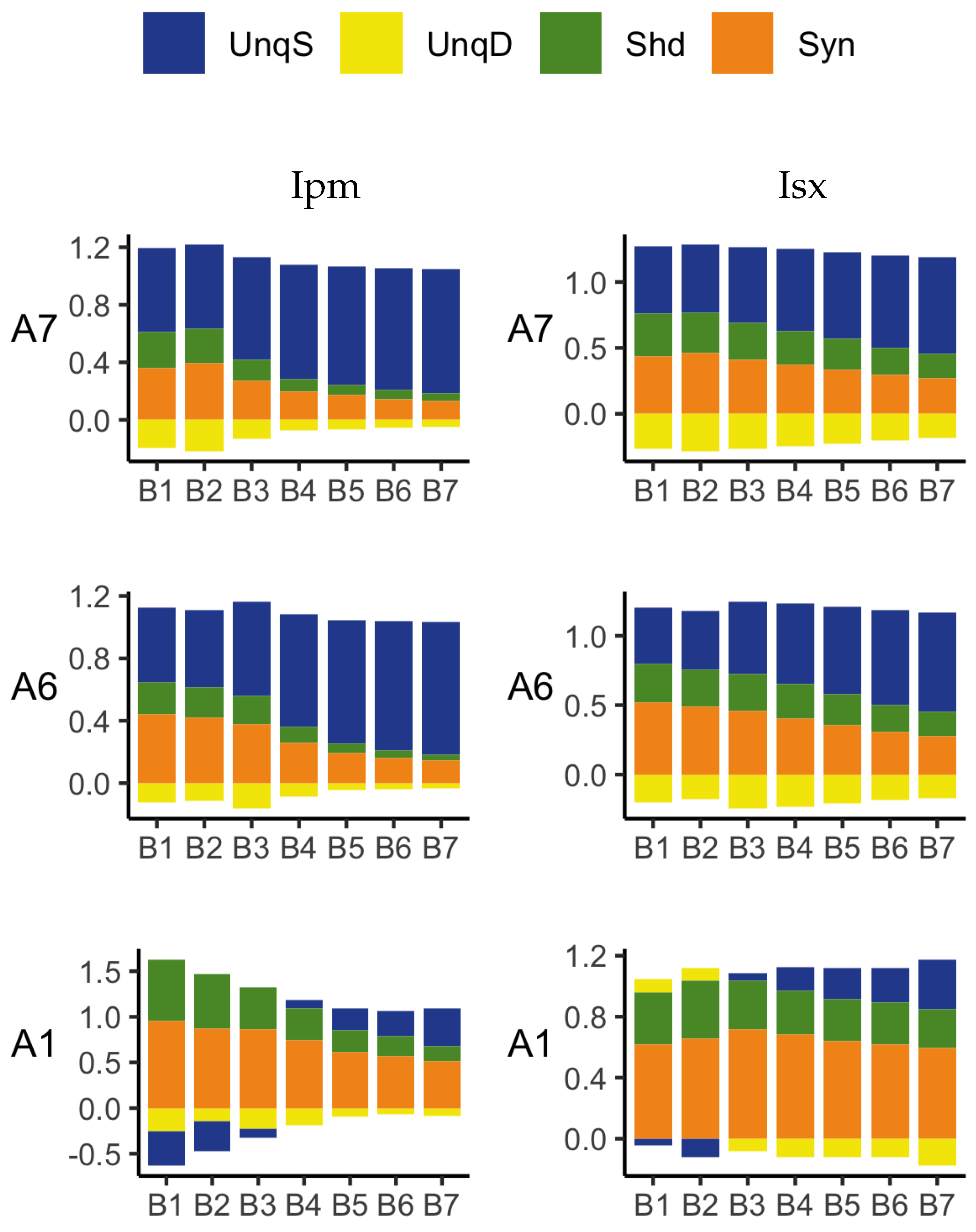
The PID analyses reveal that the Ibroja, Idep and Iccs methods produce broadly similar PIDs for the 15 neurons under each experimental condition, whereas the Ipm and Isx methods produce components that have very different values than those given by the Ibroja, Idep and Iccs methods. In particular, the Ipm and Isx methods produce much larger estimates of shared information and synergy. The Ipm method even produces some values for synergy that are larger than the joint mutual information, which seems nonsensical.
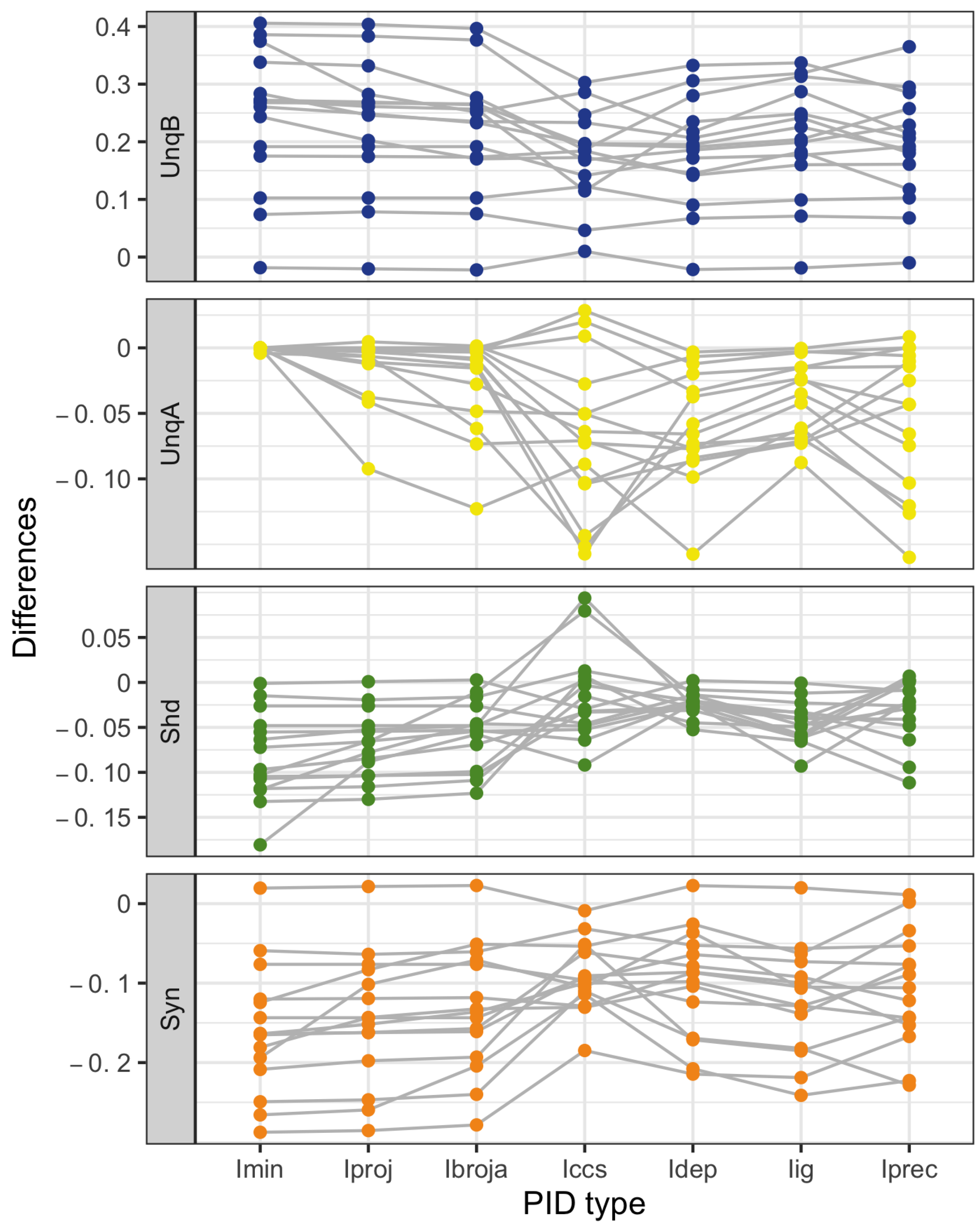
When the relative values of the PID components are considered—as with the within-neuron differences in the PID components used in the investigation of the effect of baclofen—these differences can be considered on the same scale for all five PID methods, and the results are generally more similar, although not for the shared information component. Statistical testing shows that for the synergy component, five independent researchers, each using one of the five methods, would arrive at the same formal statistical conclusion. Were they to consider the shared information, however, the researchers using the Ipm or Isx methods would reach a different formal statistical conclusion than obtained by those using the Ibroja, Idep and Iccs methods.
While the values of the unique information asymmetry are the same for all five methods, the asymmetry is expressed in different ways. The Ibroja, Idep and Iccs methods all exhibit strong basal drive and there is evidence of apical amplification for several neurons. Examination of the within-neuron differences and statistical testing conducted on the asymmetry values provide support for the conclusion in [
28] regarding the effect of GABA
R-mediated inhibition. Neither conclusion applies to the Ipm and Isx methods.
As to the question of which method(s) to rely on, it seems wise, for probability distributions of the type considered in this study, to employ the Ibroja, Idep and Iccs methods since they give broadly similar results, rather than the Ipm or Isx method.
4.2. Simulated Mouse L5b Neuron Model Data
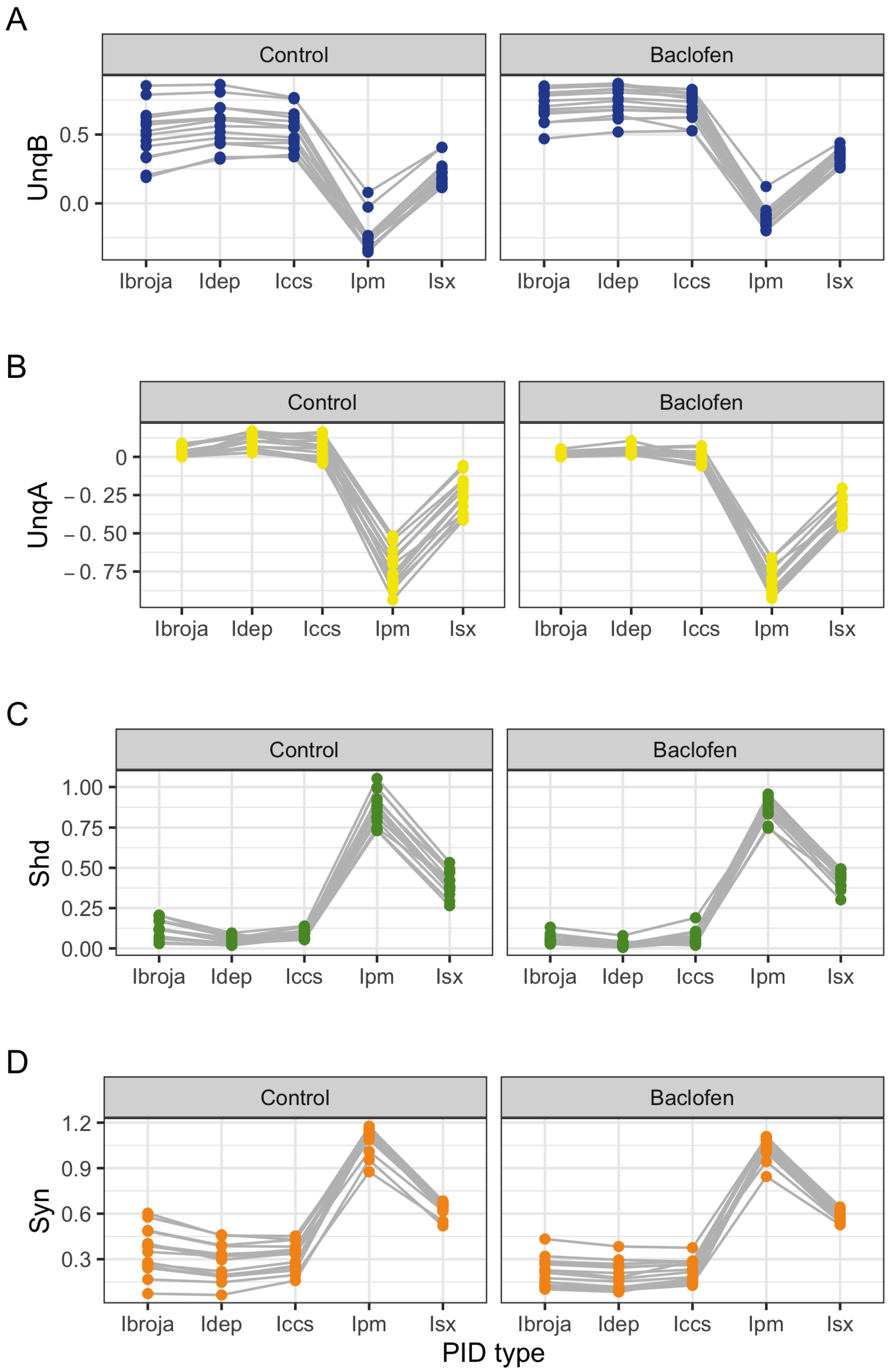
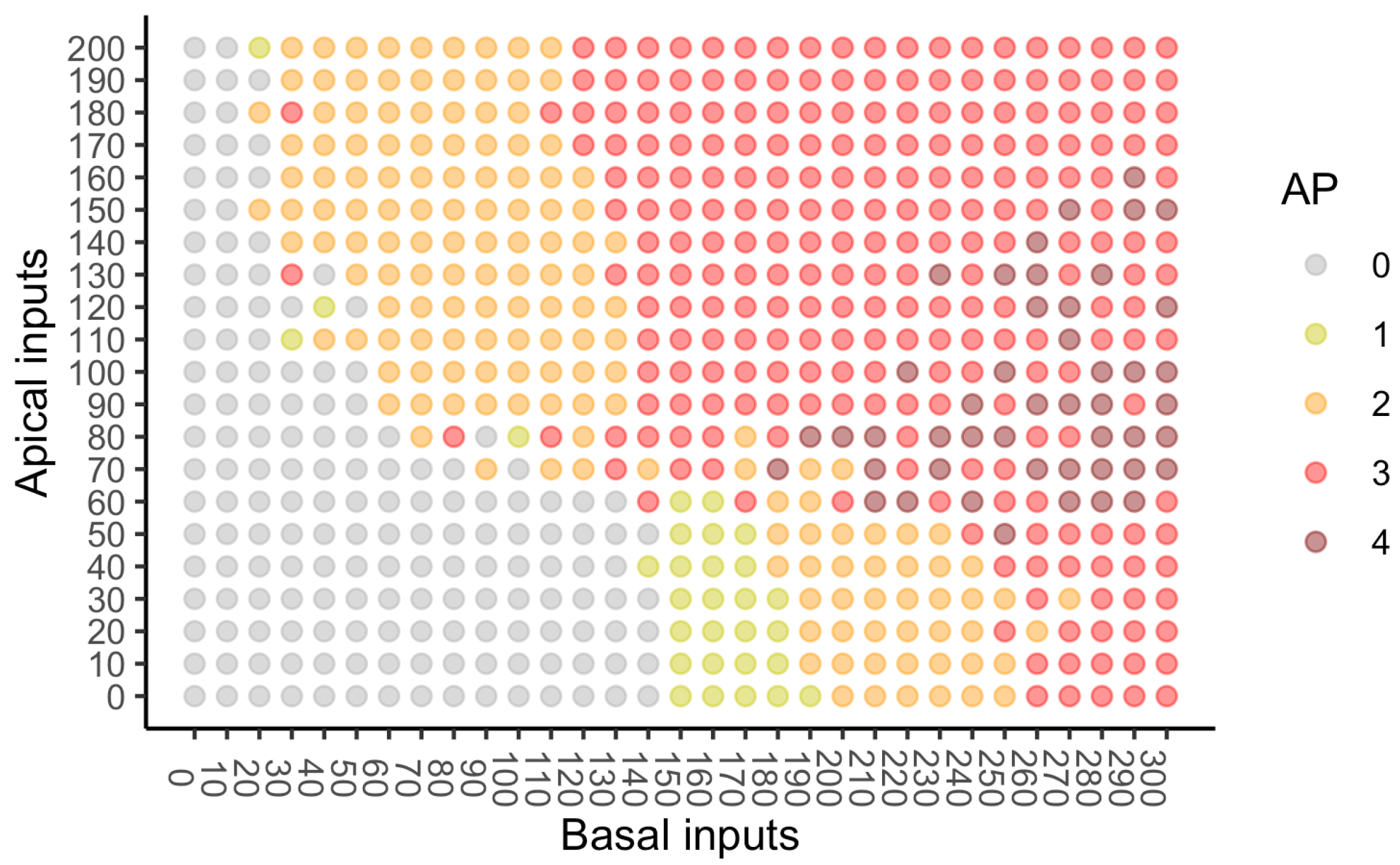
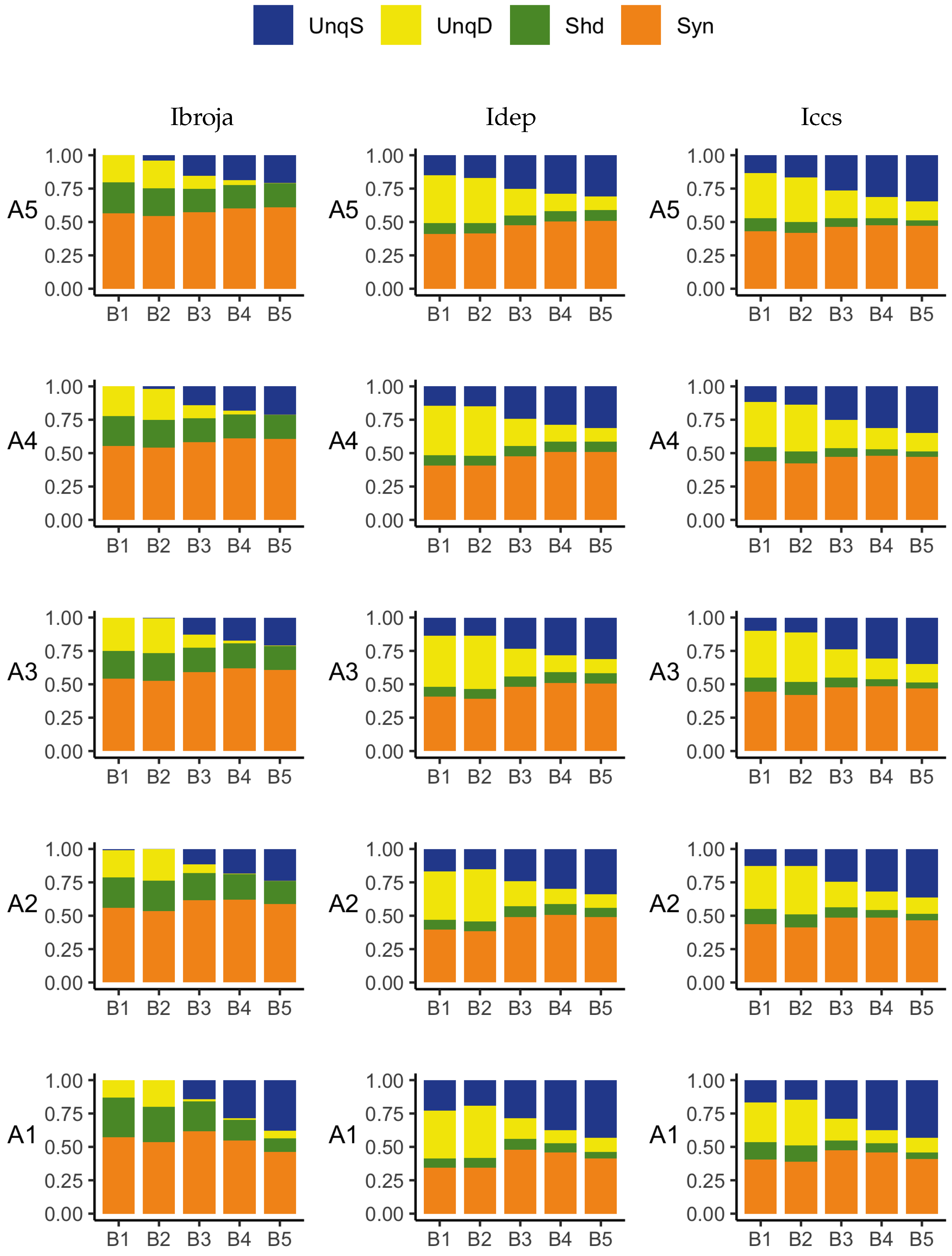
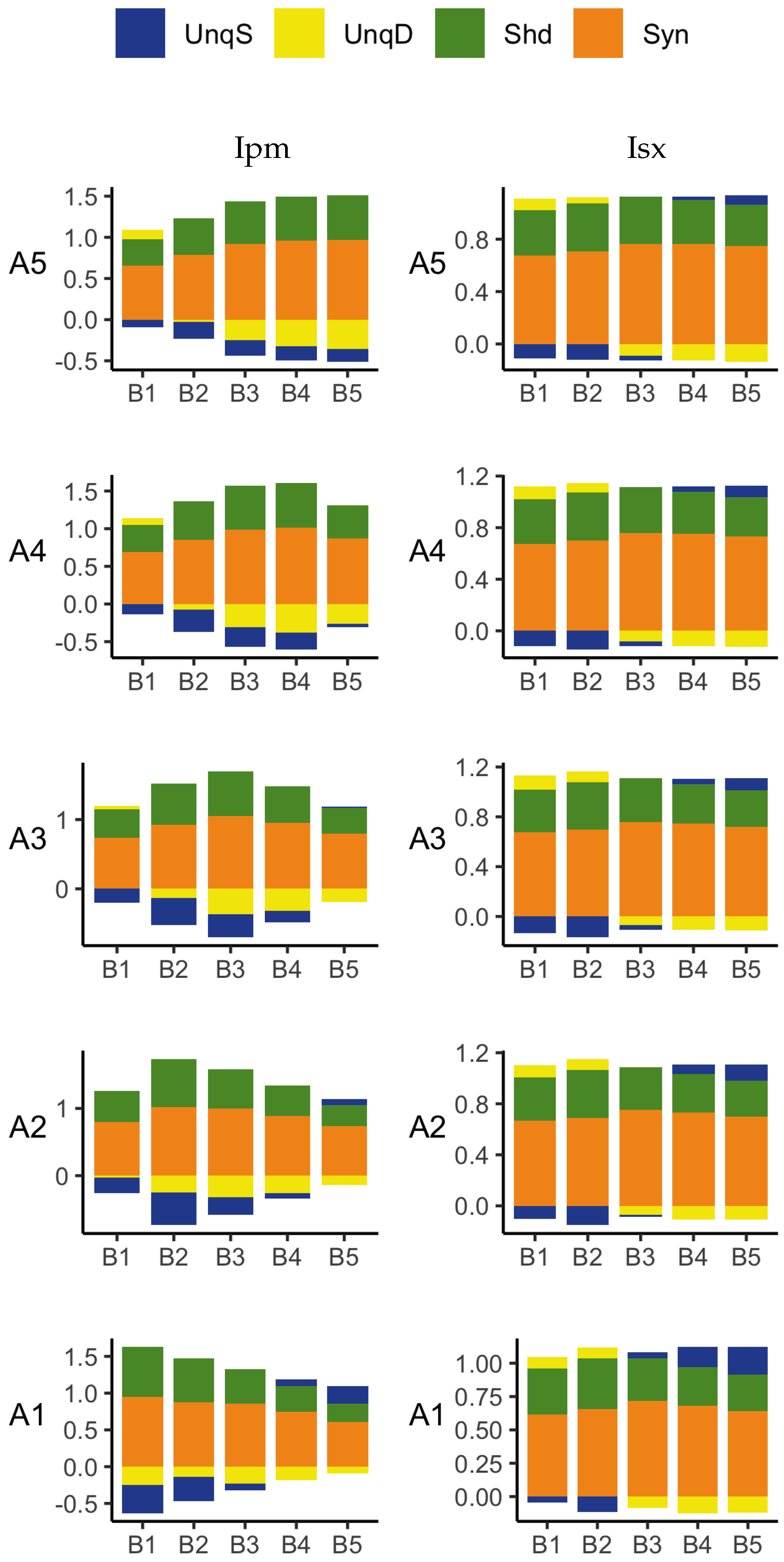
The PID analyses of the full dataset again reveal differences among the five methods, with the Ibroja, Idep and Iccs decompositions again being broadly similar. The Ipm and Isx methods transmit higher percentages of the information as synergy and shared information and an appreciable percentage as unique apical misinformation that is larger in magnitude than the transmitted unique basal information.
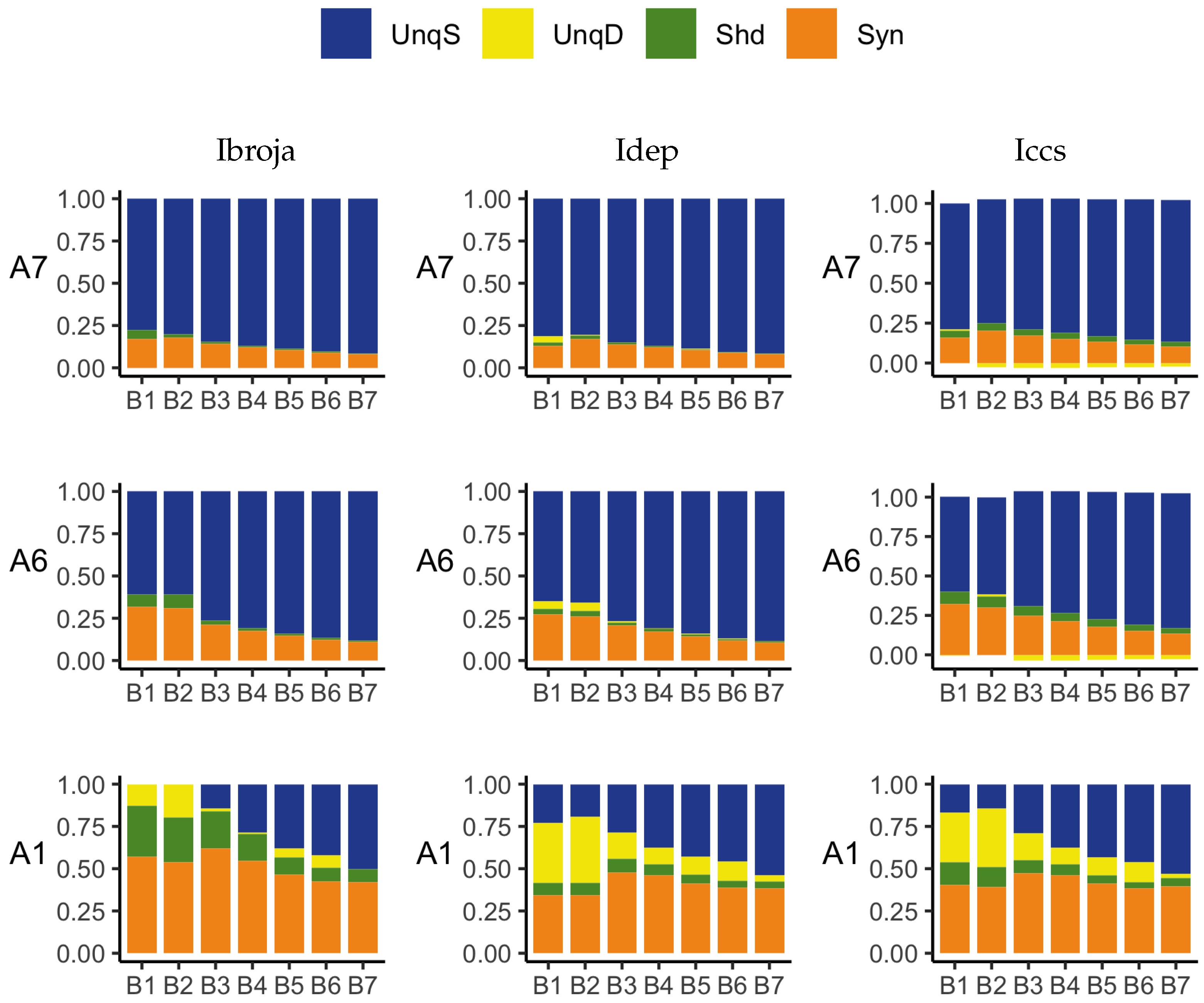
A richer picture emerges when various subsets of the data are analysed. When the basal and apical inputs are treated on an equal footing, and various combinations of strengths of basal and apical inputs considered, we find that there is a bifurcation in unique information asymmetry for all PIDs. While the values of the asymmetries are fixed by classical measures of mutual information, the nature of the asymmetries is only revealed by the PIDs. For Ibroja, Idep and Iccs, we find that as the strength of the basal input increases, to the extent that it is sufficient to drive AP output, there is a switch from apical drive to basal drive, and that this occurs at the same strength of basal input for every strength of apical input. We also find that this bifurcation happens when we consider combinations where the basal and apical strengths are equal. The Ipm and Isx PIDs express the asymmetry in terms of combinations of basal and apical misinformation or a combination of a unique information with a unique misinformation.
In a second exploration of subsets, increasing basal strengths were considered for three fixed apical strengths. With regard to cooperative context-sensitivity, we find that all five PIDs provide at least some support for the ideal properties. The Ibroja PID satisfies the properties to the fullest extent, with Idep and Iccs close behind. Ipm and Isx provide partial support.
A challenge in interpreting the results of any PID analysis is that the underlying reality of any system under investigation is not known; it seems there is no way to know what the true levels of the PID components actually are. It is possible to define fairly simple probability models in which there is a clear expectation that synergy, unique information, shared information, or a combination of these components, should be present, and such distributions are often used when evaluating a new PID or performing comparisons among several existing ones. For some of these standard distributions, the existing methods can agree, but on others they do not. It is, therefore, useful to consider several different PIDs and to place more emphasis on those findings where the different PIDs produce the expected results on simple probability distributions and where they produce similar results for the system under investigation. The findings obtained in the analyses considered herein with Ibroja, Iccs and Idep are very plausible since they generally accord with expectations based on the current understanding of L5b pyramidal cells.
4.3. Biological Implications
Twentieth century psychology and systems neuroscience were built on the assumption that neurons, in general, operate as point processors that nearly linearly sum all their synaptic inputs and signal the extent to which that net sum exceeds a threshold. Direct physiological studies of communication between the apical integration zone and the soma in layer 5 pyramidal cells of the neocortex show that assumption to be false [
31,
35,
55,
56,
57]. Synaptic activation of the apical integration zone that has a limited effect on axonal AP output by itself can greatly increase the response AP output induced by more proximal synaptic inputs occurring at about the same time. This study provides the first systematic comparison of the most widely used information decomposition methods on physiological data from L5b pyramidal cells, which are known to have particularly prominent dendritic non-linearities. These analyses strongly support two important interpretations of physiological data in previous reports, despite certain limitations of the used data sets. A technical limitation of the first study is that direct current injection was used as an experimental approximation of synaptic inputs. In the second study, the model neuron is expected to provide limited accuracy in the precise AP number evoked by synaptic inputs due to the intrinsic difficulties in appropriately modeling the fast underlying conductances [
37]. Despite these different constraints, our analyses converge at the conclusion that apical dendritic inputs may mainly contribute to synergy, i.e., have a modulatory role, rather than driving output information. The reason for this is that, in the investigated pyramidal neurons, apical dendritic inputs are bound to recruit an amplifying Ca
spike mechanism in the apical dendrite associated with bursts of several APs if they were to activate somatic APs directly. Therefore, apical dendritic inputs cannot provide the graded impact on AP output that basal dendritic inputs do. We can conclude that under these circumstances, the role of apical dendritic inputs is largely restricted to amplifying output rather than driving output information. Second, the results directly show that this amplification is reduced by inhibitory input to the apical zone, which implies that amplification is tightly regulated by apical inhibition. Based on recent physiological studies [
58,
59,
60], other neuromodulatory systems are expected to play similar regulatory roles. Together, these observations support the idea that apical amplification may be an important mechanism for contextual modulation and conscious perception.