Entropy Analysis of Heart Rate Variability in Different Sleep Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
- For 270-s epoch length:
- For 300-s epoch length:
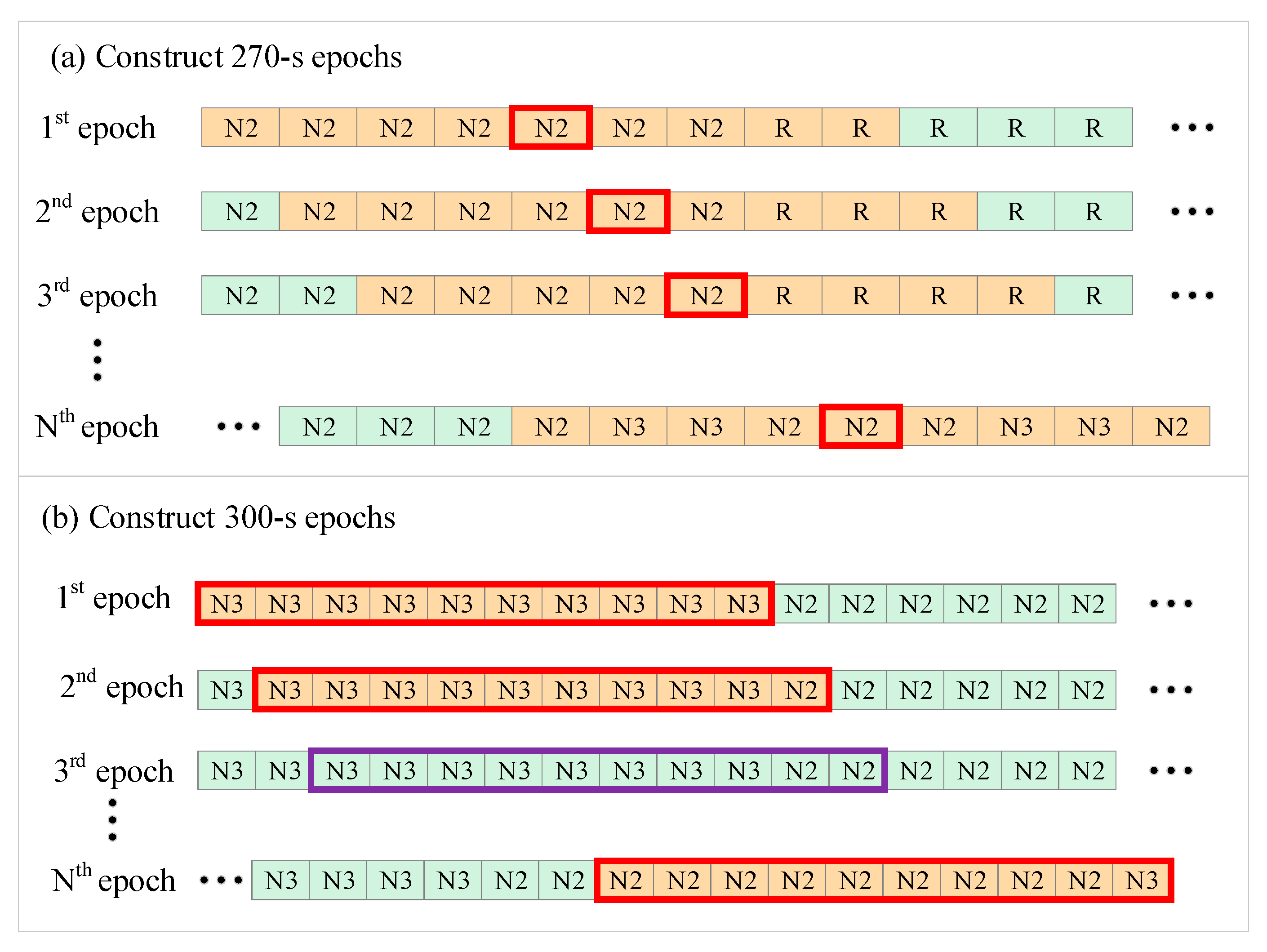
2.2. Entropy Analysis of HRV Time Series
2.3. Linear Measures of HRV Time Series
2.4. Statistical Analysis
2.5. Sleep Staging
- (1)
- a three-class classification task to differentiate among W, NREM, and REM;
- (2)
- a four-class classification task to differentiate among W, light sleep (LS, combined N1 and N2), deep sleep (DS, or N3), and REM;
- (3)
- a five-class classification task to differentiate among W, N1, N2, N3, and REM. Different models were trained for each classification task and then tested.
3. Results
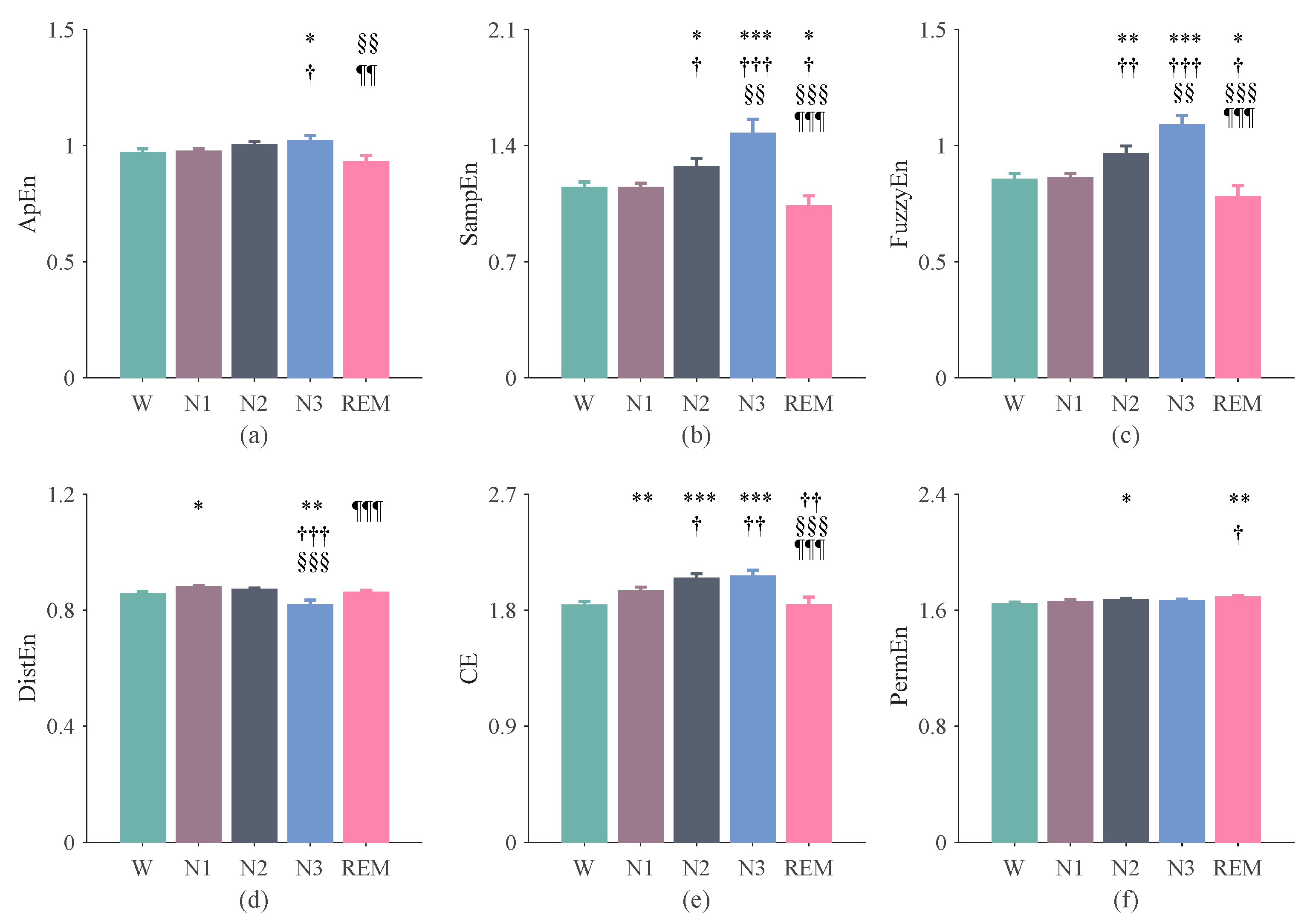
3.1. The Results of Entropy Indices Using 270-s Epochs
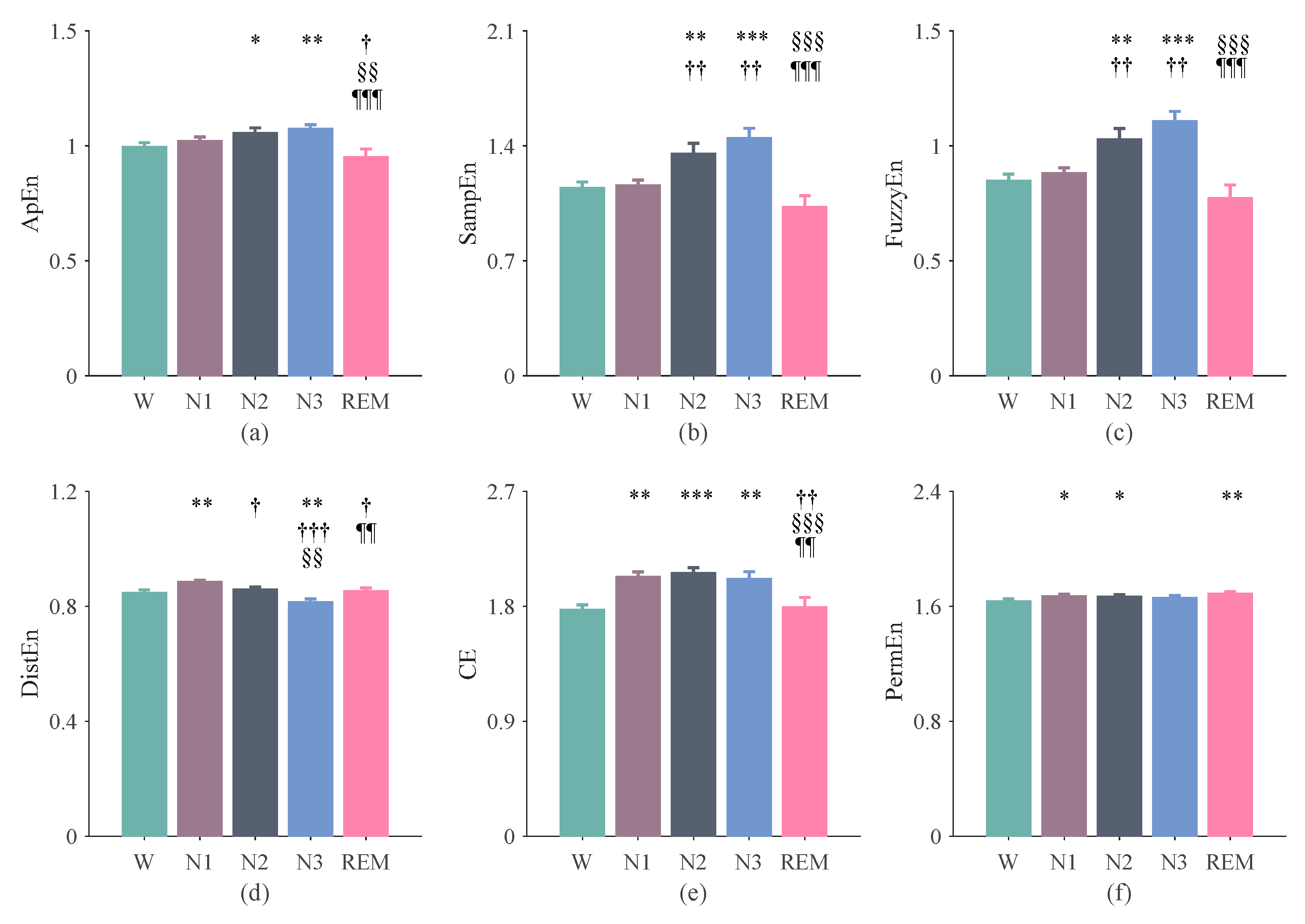
3.2. The Results of Entropy Indices Using 300-s Epochs
3.3. Sleep Staging
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Somers, V.K.; Dyken, M.; Mark, A.L.; Abboud, F. Sympathetic-Nerve Activity during Sleep in Normal Subjects. N. Engl. J. Med. 1993, 328, 303–307. [Google Scholar] [CrossRef]
- Cabiddu, R.; Cerutti, S.; Viardot, G.; Werner, S.; Bianchi, A.M. Modulation of the Sympatho-Vagal Balance during Sleep: Frequency Domain Study of Heart Rate Variability and Respiration. Front. Physiol. 2012, 3, 45. [Google Scholar] [CrossRef] [Green Version]
- de Zambotti, M.; Trinder, J.; Silvani, A.; Colrain, I.M.; Baker, F.C. Dynamic coupling between the central and autonomic nervous systems during sleep: A review. Neurosci. Biobehav. Rev. 2018, 90, 84–103. [Google Scholar] [CrossRef]
- Mancia, G. Autonomic Modulation of the Cardiovascular System during Sleep. N. Engl. J. Med. 1993, 328, 347–349. [Google Scholar] [CrossRef]
- Iber, C.; Ancoli-Israel, S.; Chesson, A.L.; Quan, S. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications; American Academy of Sleep Medicine: Westchester, IL, USA, 2007. [Google Scholar]
- Garbarino, S.; Lanteri, P.; Feeling, N.R.; Jarczok, M.N.; Quintana, D.S.; Koenig, J.; Sannita, W.G. Circadian Rhythms, Sleep, and the Autonomic Nervous System. Psychophysiology 2020, 34, 1–9. [Google Scholar] [CrossRef]
- Toscani, L.; Gangemi, P.F.; Parigi, A.; Silipo, R.; Ragghianti, P.; Sirabella, E.; Morelli, M.; Bagnoli, L.; Vergassola, R.; Zaccara, G. Human heart rate variability and sleep stages. Neurol. Sci. 1996, 17, 437–439. [Google Scholar] [CrossRef]
- Sztajzel, J. Heart Rate Variability: A Noninvasive Electrocardiographic Method to Measure the Autonomic Nervous System. Swiss Med. Wkly. 2004, 134, 514–522. [Google Scholar]
- Žemaitytė, D.; Varoneckas, G.; Sokolov, E. Heart Rhythm Control during Sleep. Psychophysiology 1984, 21, 279–289. [Google Scholar] [CrossRef]
- Versace, F.; Mozzato, M.; Tona, G.D.M.; Cavallero, C.; Stegagno, L. Heart rate variability during sleep as a function of the sleep cycle. Biol. Psychol. 2003, 63, 149–162. [Google Scholar] [CrossRef]
- Scott, M.J.; Randolph, P.H.; Leier, C.V. Reproducibility of systolic and diastolic time intervals in normal humans: An important issue in clinical cardiovascular pharmacology. J. Cardiovasc. Pharmacol. 1989, 13, 125–130. [Google Scholar]
- Elsenbruch, S.; Harnish, M.J.; Orr, W.C. Heart Rate Variability during Waking and Sleep in Healthy Males and Females. Sleep 1999, 22, 1067–1071. [Google Scholar] [CrossRef] [Green Version]
- Stein, P.K.; Pu, Y. Heart rate variability, sleep and sleep disorders. Sleep Med. Rev. 2012, 16, 47–66. [Google Scholar] [CrossRef]
- Busek, P.; Vanková, J.; Opavsky, J.; Salinger, J.; Nevsimalova, S. Spectral analysis of the heart rate variability in sleep. Physiol. Res. 2004, 54, 369–376. [Google Scholar]
- Bashan, A.; Bartsch, R.; Kantelhardt, J.W.; Havlin, S.; Ivanov, P.C. Network physiology reveals relations between network topology and physiological function. Nat. Commun. 2012, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Lavanga, M.; Bollen, B.; Jansen, K.; Ortibus, E.; Naulaers, G.; Van Huffel, S.; Caicedo, A. A Bradycardia-Based Stress Calculator for the Neonatal Intensive Care Unit: A Multisystem Approach. Front. Physiol. 2020, 11, 741. [Google Scholar] [CrossRef]
- Voss, A.; Schroeder, R.; Vallverdu, M.; Cygankiewicz, I.; Vazquez, R.; De Luna, A.B.; Caminal, P. Linear and nonlinear heart rate variability risk stratification in heart failure patients. In Proceedings of the 2008 Computers in Cardiology, Bologna, Italy, 14–17 September 2008; pp. 557–560. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Li, K.; Zheng, D.; Liu, C.; Hou, Y. Assessing the complexity of short-term heartbeat interval series by distribution entropy. Med. Biol. Eng. Comput. 2014, 53, 77–87. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [Green Version]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol.-Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.-B.; He, W.-X.; Liu, H. Measuring time series regularity using nonlinear similarity-based sample entropy. Phys. Lett. A 2008, 372, 7140–7146. [Google Scholar] [CrossRef]
- Shi, B.; Wang, L.; Yan, C.; Chen, D.; Liu, M.; Li, P. Nonlinear heart rate variability biomarkers for gastric cancer severity: A pilot study. Sci. Rep. 2019, 9, 13833. [Google Scholar] [CrossRef] [Green Version]
- Bandt, C.; Pompe, B. Permutation Entropy: A Natural Complexity Measure for Time Series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef]
- Vigo, D.E.; Dominguez, J.; Guinjoan, S.M.; Scaramal, M.; Ruffa, E.; Solernó, J.; Siri, L.N.; Cardinali, D.P. Nonlinear analysis of heart rate variability within independent frequency components during the sleep–wake cycle. Auton. Neurosci. 2010, 154, 84–88. [Google Scholar] [CrossRef]
- Xiao, M.; Yan, H.; Yang, X.; Li, Y.; Zhu, R. Multiscale Entropy Based Analysis of HRV during Sleep. In Proceedings of the 2012 5th International Conference on BioMedical Engineering and Informatics, Chongqing, China, 16–18 October 2012; pp. 558–562. [Google Scholar]
- Ebrahimi, F.; Setarehdan, S.-K.; Ayala-Moyeda, J.; Nazeran, H. Automatic sleep staging using empirical mode decomposition, discrete wavelet transform, time-domain, and nonlinear dynamics features of heart rate variability signals. Comput. Methods Programs Biomed. 2013, 112, 47–57. [Google Scholar] [CrossRef]
- Yoon, H.; Hwang, S.H.; Choi, J.-W.; Lee, Y.J.; Jeong, D.-U.; Park, K.S. REM sleep estimation based on autonomic dynamics using R–R intervals. Physiol. Meas. 2017, 38, 631–651. [Google Scholar] [CrossRef]
- Mitsukura, Y.; Fukunaga, K.; Yasui, M.; Mimura, M. Sleep stage detection using only heart rate. Health Inform. J. 2019, 26, 376–387. [Google Scholar] [CrossRef]
- Fonseca, P.; Van Gilst, M.M.; Radha, M.; Ross, M.; Moreau, A.; Cerny, A.; Anderer, P.; Long, X.; Van Dijk, J.P.; Overeem, S. Automatic sleep staging using heart rate variability, body movements, and recurrent neural networks in a sleep disordered population. Sleep 2020, 43, zsaa048. [Google Scholar] [CrossRef]
- Radha, M.; Fonseca, P.; Moreau, A.; Ross, M.; Cerny, A.; Anderer, P.; Long, X.; Aarts, R.M. Sleep stage classification from heart-rate variability using long short-term memory neural networks. Sci. Rep. 2019, 9, 14149. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, Q.; Liu, C.; Shashikumar, S.P.; Nemati, S.; Clifford, G.D. Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol. Meas. 2018, 39, 124005. [Google Scholar] [CrossRef]
- Walch, O.; Huang, Y.; Forger, D.; Goldstein, C. Sleep stage prediction with raw acceleration and photoplethysmography heart rate data derived from a consumer wearable device. Sleep 2019, 42, zsz180. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef] [Green Version]
- Ichimaru, Y.; Moody, G. Development of the polysomnographic database on CD-ROM. Psychiatry Clin. Neurosci. 1999, 53, 175–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNames, J.; Thong, T.; Aboy, M. Impulse Rejection Filter for Artifact Removal in Spectral Analysis of Biomedical Signals. In Proceedings of the 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Francisco, CA, USA, 1–5 September 2004; Volume 1, pp. 145–148. [Google Scholar]
- Shi, B.; Motin, M.; Wang, X.; Karmakar, C.; Li, P. Bivariate Entropy Analysis of Electrocardiographic RR–QT Time Series. Entropy 2020, 22, 1439. [Google Scholar] [CrossRef] [PubMed]
- Pincus, S. Approximate entropy (ApEn) as a complexity measure. Chaos Interdiscip. J. Nonlinear Sci. 1995, 5, 110–117. [Google Scholar] [CrossRef]
- Li, P. EZ Entropy: A software application for the entropy analysis of physiological time-series. Biomed. Eng. Online 2019, 18, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zhuang, J.; Yu, W.; Wang, Z. Measuring complexity using FuzzyEn, ApEn, and SampEn. Med. Eng. Phys. 2009, 31, 61–68. [Google Scholar] [CrossRef]
- Porta, A.; Baselli, G.; Liberati, D.; Montano, N.; Cogliati, C.; Gnecchi-Ruscone, T.; Malliani, A.; Cerutti, S. Measuring regularity by means of a corrected conditional entropy in sympathetic outflow. Biol. Cybern. 1998, 78, 71–78. [Google Scholar] [CrossRef]
- Shi, B.; Zhang, Y.; Yuan, C.; Wang, S.; Li, P. Entropy Analysis of Short-Term Heartbeat Interval Time Series during Regular Walking. Entropy 2017, 19, 568. [Google Scholar] [CrossRef]
- Makowiec, D.; Kaczkowska, A.; Wejer, D.; Żarczyńska-Buchowiecka, M.; Struzik, Z.R. Entropic Measures of Complexity of Short-Term Dynamics of Nocturnal Heartbeats in an Aging Population. Entropy 2015, 17, 1253–1272. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, F.; Ginsberg, J.P. An Overview of Heart Rate Variability Metrics and Norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, Q.; Xin, Y.; Lu, X. Obstructive Sleep Apnea Detection Using Sleep Architecture. In Proceedings of the 2020 IEEE International Conference on Mechatronics and Automation (ICMA), Beijing, China, 13–16 October 2020; pp. 255–260. [Google Scholar]
- Zhao, X.; Rong, P.; Sun, G.; Zhang, B. Automatic Sleep Staging Based on XGBOOST Physiological Signals. In Proceedings of the 11th International Conference on Modelling, Identification and Control (ICMIC2019), Tianjin, China, 13–15 July 2019; pp. 1095–1106. [Google Scholar]
- Shahriari, B.; Swersky, K.; Wang, Z.; Adams, R.P.; De Freitas, N. Taking the Human Out of the Loop: A Review of Bayesian Optimization. Proc. IEEE 2015, 104, 148–175. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, S.M.; Erion, G.; Chen, H.; De Grave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Al-Angari, H.M.; Sahakian, A.V. Use of Sample Entropy Approach to Study Heart Rate Variability in Obstructive Sleep Apnea Syndrome. IEEE Trans. Biomed. Eng. 2007, 54, 1900–1904. [Google Scholar] [CrossRef]
- Yan, C.; Liu, C.; Yao, L.; Wang, X.; Wang, J.; Li, P. Short-Term Effect of Percutaneous Coronary Intervention on Heart Rate Variability in Patients with Coronary Artery Disease. Entropy 2021, 23, 540. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.-K. Multiscale Entropy Analysis of Complex Physiologic Time Series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [Green Version]
- De Wel, O.; Lavanga, M.; Dorado, A.C.; Jansen, K.; Dereymaeker, A.; Naulaers, G.; Van Huffel, S. Complexity Analysis of Neonatal EEG Using Multiscale Entropy: Applications in Brain Maturation and Sleep Stage Classification. Entropy 2017, 19, 516. [Google Scholar] [CrossRef] [Green Version]
- Lavanga, M.; Bollen, B.; Caicedo, A.; Dereymaeker, A.; Jansen, K.; Ortibus, E.; Van Huffel, S.; Naulaers, G. The effect of early procedural pain in preterm infants on the maturation of electroencephalogram and heart rate variability. Pain 2020, 162, 1556–1566. [Google Scholar] [CrossRef]
- Li, P.; Karmakar, C.; Yan, C.; Palaniswami, M.; Liu, C. Classification of 5-S Epileptic EEG Recordings Using Distribution Entropy and Sample Entropy. Front. Physiol. 2016, 7, 136. [Google Scholar] [CrossRef] [Green Version]
- Lavanga, M.; Heremans, E.; Moeyersons, J.; Bollen, B.; Jansen, K.; Ortibus, E.; Naulaers, G.; Van Huffel, S.; Caicedo, A. Maturation of the Autonomic Nervous System in Premature Infants: Estimating Development Based on Heart-Rate Variability Analysis. Front. Physiol. 2021, 11, 1709. [Google Scholar] [CrossRef]
- Togo, F.; Yamamoto, Y. Decreased fractal component of human heart rate variability during non-REM sleep. Am. J. Physiol. Circ. Physiol. 2001, 280, H17–H21. [Google Scholar] [CrossRef]
- Bunde, A.; Havlin, S.; Kantelhardt, J.W.; Penzel, T.; Peter, J.-H.; Voigt, K. Correlated and Uncorrelated Regions in Heart-Rate Fluctuations during Sleep. Phys. Rev. Lett. 2000, 85, 3736–3739. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Teng, J.; Qi, X.; Wei, S.; Liu, C. Comparison between heart rate variability and pulse rate variability during different sleep stages for sleep apnea patients. Technol. Health Care 2017, 25, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Xiong, J.; Cao, Z.; Wang, X.; Li, J.; Liu, C. Decreased sample entropy during sleep-to-wake transition in sleep apnea patients. Physiol. Meas. 2021, 42, 044001. [Google Scholar] [CrossRef] [PubMed]
- Surantha, N.; Lesmana, T.F.; Isa, S.M. Sleep stage classification using extreme learning machine and particle swarm optimization for healthcare big data. J. Big Data 2021, 8, 1–17. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Setarehdan, S.-K.; Nazeran, H. Automatic sleep staging by simultaneous analysis of ECG and respiratory signals in long epochs. Biomed. Signal Process. Control 2015, 18, 69–79. [Google Scholar] [CrossRef]
- Wei, R.; Zhang, X.; Wang, J.; Dang, X. The research of sleep staging based on single-lead electrocardiogram and deep neural network. Biomed. Eng. Lett. 2017, 8, 87–93. [Google Scholar] [CrossRef]
- Tanida, K.; Shibata, M.; Heitkemper, M.M. Sleep Stage Assessment Using Power Spectral Indices of Heart Rate Variability with a Simple Algorithm. Biol. Res. Nurs. 2012, 15, 264–272. [Google Scholar] [CrossRef]
- Yücelbaş, Ş.; Yücelbaş, C.; Tezel, G.; Özşen, S.; Yosunkaya, Ş. Automatic sleep staging based on SVD, VMD, HHT and morphological features of single-lead ECG signal. Expert Syst. Appl. 2018, 102, 193–206. [Google Scholar] [CrossRef]
- Liu, C.; Li, K.; Zhao, L.; Liu, F.; Zheng, D.; Liu, C.; Liu, S. Analysis of heart rate variability using fuzzy measure entropy. Comput. Biol. Med. 2013, 43, 100–108. [Google Scholar] [CrossRef]
- Ji, L.; Li, P.; Li, K.; Wang, X.; Liu, C. Analysis of short-term heart rate and diastolic period variability using a refined fuzzy entropy method. Biomed. Eng. Online 2015, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
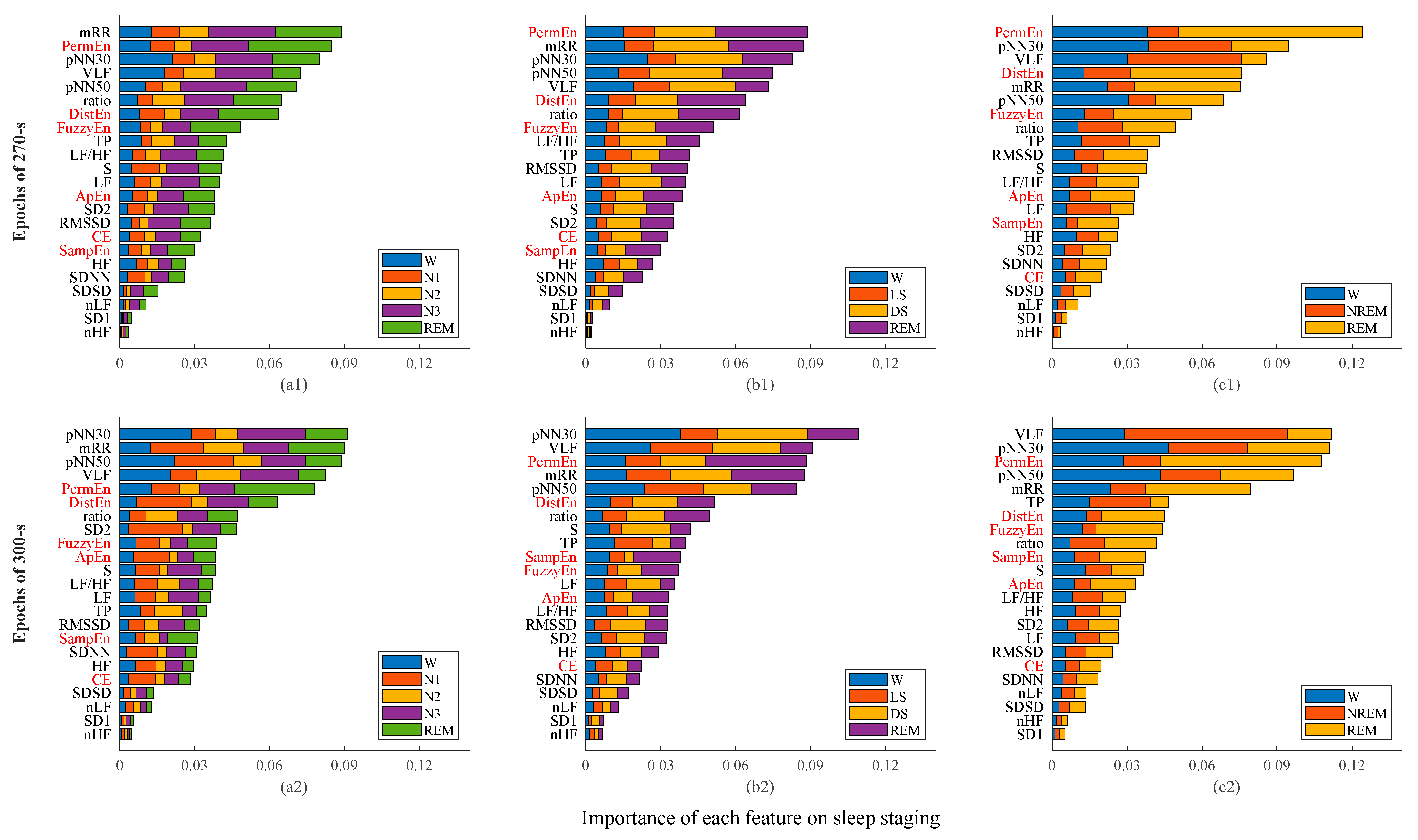
| Classes | Acc (%) | κ (a.u.) | |||
|---|---|---|---|---|---|
| Only Linear HRV Features | Linear HRV Features + Entropy Features | Only Linear HRV Features | Linear HRV Features + Entropy Features | ||
| 270-s | 5 | 41.2 ± 6.6 | 42.1 ± 7.4 | 0.17 ± 0.08 | 0.17 ± 0.10 |
| 4 | 56.1 ± 9.1 | 59.1 ± 8.9 | 0.22 ± 0.15 | 0.25 ± 0.16 | |
| 3 | 59.1 ± 8.2 | 60.8 ± 9.5 | 0.23 ± 0.15 | 0.27 ± 0.17 | |
| 300-s | 5 | 53.9 ± 13.5 * | 54.3 ± 14.3 * | 0.29 ± 0.17 * | 0.29 ± 0.19 * |
| 4 | 61.4 ± 12.1 | 63.1 ± 13.3 | 0.35 ± 0.20 * | 0.36 ± 0.24 * | |
| 3 | 65.5 ± 9.9 | 67.5 ± 11.6 * | 0.37 ± 0.19 * | 0.40 ± 0.21 * | |
| Work | Classes | Acc (%) |
|---|---|---|
| Yasue et al. [28] | 5 | 66 |
| Our work | 5 | 54.3 |
| Li et al. [31] | 4 | 75.4/65.9 a |
| Mustafa et al. [30] | 4 | 77 |
| Surantha et al. [59] | 4 | 71.52 |
| Ebrahimi et al. [60] | 4 | 89.32 |
| Tanida et al. [62] | 4 | 56 |
| Our work | 4 | 63.1 |
| Wei et al. [61] | 3 | 77 |
| Li et al. [31] | 3 | 81.6/75.3 a |
| Yücelbaş et al. [63] | 3 | 76.79 |
| Our work | 3 | 67.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, C.; Li, P.; Yang, M.; Li, Y.; Li, J.; Zhang, H.; Liu, C. Entropy Analysis of Heart Rate Variability in Different Sleep Stages. Entropy 2022, 24, 379. https://doi.org/10.3390/e24030379
Yan C, Li P, Yang M, Li Y, Li J, Zhang H, Liu C. Entropy Analysis of Heart Rate Variability in Different Sleep Stages. Entropy. 2022; 24(3):379. https://doi.org/10.3390/e24030379
Chicago/Turabian StyleYan, Chang, Peng Li, Meicheng Yang, Yang Li, Jianqing Li, Hongxing Zhang, and Chengyu Liu. 2022. "Entropy Analysis of Heart Rate Variability in Different Sleep Stages" Entropy 24, no. 3: 379. https://doi.org/10.3390/e24030379
APA StyleYan, C., Li, P., Yang, M., Li, Y., Li, J., Zhang, H., & Liu, C. (2022). Entropy Analysis of Heart Rate Variability in Different Sleep Stages. Entropy, 24(3), 379. https://doi.org/10.3390/e24030379







