Ungulate Detection and Species Classification from Camera Trap Images Using RetinaNet and Faster R-CNN
Abstract
:1. Introduction
- We compiled new ungulates in the wild dataset that was collected over four years.
- We compared the performance of different backbones in a three-module architecture on the new dataset. Thus, we created a baseline accuracy for animal localization and classification tasks on the new dataset.
- We investigated the effect of data augmentation on the performance.
2. Related Work
3. Methodology
3.1. Detection Neural Networks
- 1.
- Backbone network—DNN consisting of convolutional layers which are used for the feature extraction from the input image. Usually, backbone networks which are pre-trained on a natural image dataset, such as ImageNet, are used. Common networks used as the backbone are ResNet50 [21], VGG16 [22], Inception-ResNetV2 [23] and DarkNet-19 [24].
- 2.
- Neck—DNN module on top of the backbone network. The neck network takes and processes inputs from the different layers of the backbone, harnessing advantages of data pattern distribution over different feature map scales by using FPN (Feature Pyramid Network) [25].
- 3.
- Head—A feed-forward neural network which performs the classification or regression task. The detector could have multiple heads for performing different classification and regression tasks.
3.2. One-Stage Detectors
3.3. Two-Stage Detectors
4. Database: Preparation and Pre-Processing
4.1. Dataset
4.2. Training Data
4.3. Pre-Processing
5. Experimental Results and Discussion
5.1. Experiments
- 1.
- Faster RCNN–ResNet50 network and RetinaNet were trained for 34,850 iterations (10 epochs) on the training dataset with a batch size of 4, learning rate of 0.0001 and Adam optimizer for the weight update. Other batch sizes and optimizers were tested, but those mentioned above were selected because they produced seemingly good convergence for the first training epoch.
- 2.
- To assess the effectiveness of the learning strategies, the RetinaNet results from the first experiment were compared with the control cases featuring RetinaNet without pre-trained weights and RetinaNet results for the corresponding number of iterations on the training set without augmentation.
5.2. Experiment 1
5.3. Experiment 2
5.4. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valente, A.M.; Acevedo, P.; Figueiredo, A.M.; Fonseca, C.; Torres, R.T. Overabundant wild ungulate populations in Europe: Management with consideration of socio-ecological consequences. Mammal Rev. 2020, 50, 353–366. [Google Scholar] [CrossRef]
- Carpio, A.J.; Apollonio, M.; Acevedo, P. Wild ungulate overabundance in Europe: Contexts, causes, monitoring and management recommendations. Mammal Rev. 2021, 51, 95–108. [Google Scholar] [CrossRef]
- Langbein, J.; Putman, R.; Pokorny, B. Traffic collisions involving deer and other ungulates in Europe and available measures for mitigation. In Ungulate Management in Europe: Problems and Practices; Cambridge University Press: Cambridge, UK, 2010; pp. 215–259. [Google Scholar]
- Rovero, F.; Zimmermann, F.; Berzi, D.; Meek, P. “Which camera trap type and how many do I need?” A review of camera features and study designs for a range of wildlife research applications. Hystrix 2013, 24, 148–156. [Google Scholar] [CrossRef]
- Massei, G.; Coats, J.; Lambert, M.S.; Pietravalle, S.; Gill, R.; Cowan, D. Camera traps and activity signs to estimate wild boar density and derive abundance indices. Pest Manag. Sci. 2018, 74, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.E.; Spitzer, R.; Allen, A.M.; Hofmeester, T.R.; Ericsson, G.; Widemo, F.; Singh, N.J.; Cromsigt, J.P. Pictures or pellets? Comparing camera trapping and dung counts as methods for estimating population densities of ungulates. Remote Sens. Ecol. Conserv. 2018, 4, 173–183. [Google Scholar] [CrossRef]
- Molloy, S.W. A Practical Guide to Using Camera Traps for Wildlife Monitoring in Natural Resource Management Projects; SWCC [Camera Trapping Guide]; Edith Cowan University: Perth, Australia, 2018. [Google Scholar]
- Norouzzadeh, M.S.; Nguyen, A.; Kosmala, M.; Swanson, A.; Palmer, M.S.; Packer, C.; Clune, J. Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proc. Natl. Acad. Sci. USA 2018, 115, E5716–E5725. [Google Scholar] [CrossRef] [Green Version]
- Swanson, A.; Kosmala, M.; Lintott, C.; Simpson, R.; Smith, A.; Packer, C. Snapshot Serengeti, high-frequency annotated camera trap images of 40 mammalian species in an African savanna. Sci. Data 2015, 2, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Whytock, R.; Świeżewski, J.; Zwerts, J.A.; Bara-Słupski, T.; Pambo, A.F.K.; Rogala, M.; Boekee, K.; Brittain, S.; Cardoso, A.W.; Henschel, P.; et al. High performance machine learning models can fully automate labeling of camera trap images for ecological analyses. bioRxiv 2020. [Google Scholar] [CrossRef]
- Avots, E.; Jermakovs, K.; Bachmann, M.; Päeske, L.; Ozcinar, C.; Anbarjafari, G. Ensemble approach for detection of depression using EEG features. Entropy 2022, 24, 211. [Google Scholar] [CrossRef]
- Kamińska, D.; Aktas, K.; Rizhinashvili, D.; Kuklyanov, D.; Sham, A.H.; Escalera, S.; Nasrollahi, K.; Moeslund, T.B.; Anbarjafari, G. Two-Stage Recognition and beyond for Compound Facial Emotion Recognition. Electronics 2021, 10, 2847. [Google Scholar] [CrossRef]
- Tabak, M.A.; Norouzzadeh, M.S.; Wolfson, D.W.; Sweeney, S.J.; VerCauteren, K.C.; Snow, N.P.; Halseth, J.M.; Di Salvo, P.A.; Lewis, J.S.; White, M.D.; et al. Machine learning to classify animal species in camera trap images: Applications in ecology. Methods Ecol. Evol. 2019, 10, 585–590. [Google Scholar] [CrossRef] [Green Version]
- Carl, C.; Schönfeld, F.; Profft, I.; Klamm, A.; Landgraf, D. Automated detection of European wild mammal species in camera trap images with an existing and pre-trained computer vision model. Eur. J. Wildl. Res. 2020, 66, 62. [Google Scholar] [CrossRef]
- Choinski, M.; Rogowski, M.; Tynecki, P.; Kuijper, D.P.; Churski, M.; Bubnicki, J.W. A first step towards automated species recognition from camera trap images of mammals using AI in a European temperate forest. arXiv 2021, arXiv:2103.11052. [Google Scholar]
- Ieracitano, C.; Mammone, N.; Versaci, M.; Varone, G.; Ali, A.R.; Armentano, A.; Calabrese, G.; Ferrarelli, A.; Turano, L.; Tebala, C.; et al. A Fuzzy-enhanced Deep Learning Approach for Early Detection of Covid-19 Pneumonia from Portable Chest X-ray Images. Neurocomputing 2022, 481, 202–215. [Google Scholar] [CrossRef]
- Obeso, A.M.; Benois-Pineau, J.; Vázquez, M.S.G.; Acosta, A.Á.R. Visual vs. internal attention mechanisms in deep neural networks for image classification and object detection. Pattern Recognit. 2022, 123, 108411. [Google Scholar] [CrossRef]
- Aktas, K.; Demirel, M.; Moor, M.; Olesk, J.; Ozcinar, C.; Anbarjafari, G. Spatiotemporal based table tennis stroke-type assessment. Signal Image Video Process. 2021, 15, 1593–1600. [Google Scholar] [CrossRef]
- Norouzzadeh, M.S.; Morris, D.; Beery, S.; Joshi, N.; Jojic, N.; Clune, J. A deep active learning system for species identification and counting in camera trap images. arXiv 2019, arXiv:cs.LG/1910.09716. [Google Scholar] [CrossRef]
- Zhang, Z.; He, Z.; Cao, G.; Cao, W. Animal Detection From Highly Cluttered Natural Scenes Using Spatiotemporal Object Region Proposals and Patch Verification. IEEE Trans. Multimed. 2016, 18, 2079–2092. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:cs.CV/1409.1556. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. arXiv 2016, arXiv:cs.CV/1612.08242. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. arXiv 2017, arXiv:cs.CV/1612.03144. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:cs.CV/2004.10934. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; pp. 21–37. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. arXiv 2018, arXiv:cs.CV/1708.02002. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.B.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. arXiv 2015, arXiv:1506.01497. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, V.; Hebert, M.; Sminchisescu, C.; Weiss, Y. (Eds.) Computer Vision—ECCV 2018: 15th European Conference, Munich, Germany, 8–14 September 2018, Proceedings, Part XVI; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11220. [Google Scholar] [CrossRef]
- The Nature Conservancy (2021): Channel Islands Camera Traps 1.0. The Nature Conservancy. Dataset. Available online: https://lila.science/datasets (accessed on 10 February 2022).
- Yousif, H.; Kays, R.; He, Z. Dynamic Programming Selection of Object Proposals for Sequence-Level Animal Species Classification in the Wild. IEEE Trans. Circuits Syst. Video Technol. 2019. [Google Scholar]
- Anton, V.; Hartley, S.; Geldenhuis, A.; Wittmer, H.U. Monitoring the mammalian fauna of urban areas using remote cameras and citizen science. J. Urban Ecol. 2018, 4, juy002. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2020, arXiv:cs.LG/1905.11946. [Google Scholar]
- Montavon, G.; Orr, G.; Müller, K.R. Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2012; Volume 7700. [Google Scholar]
- Oļševskis, E.; Schulz, K.; Staubach, C.; Seržants, M.; Lamberga, K.; Pūle, D.; Ozoliņš, J.; Conraths, F.; Sauter-Louis, C. African swine fever in Latvian wild boar—A step closer to elimination. Transbound. Emerg. Dis. 2020, 67, 2615–2629. [Google Scholar] [CrossRef]
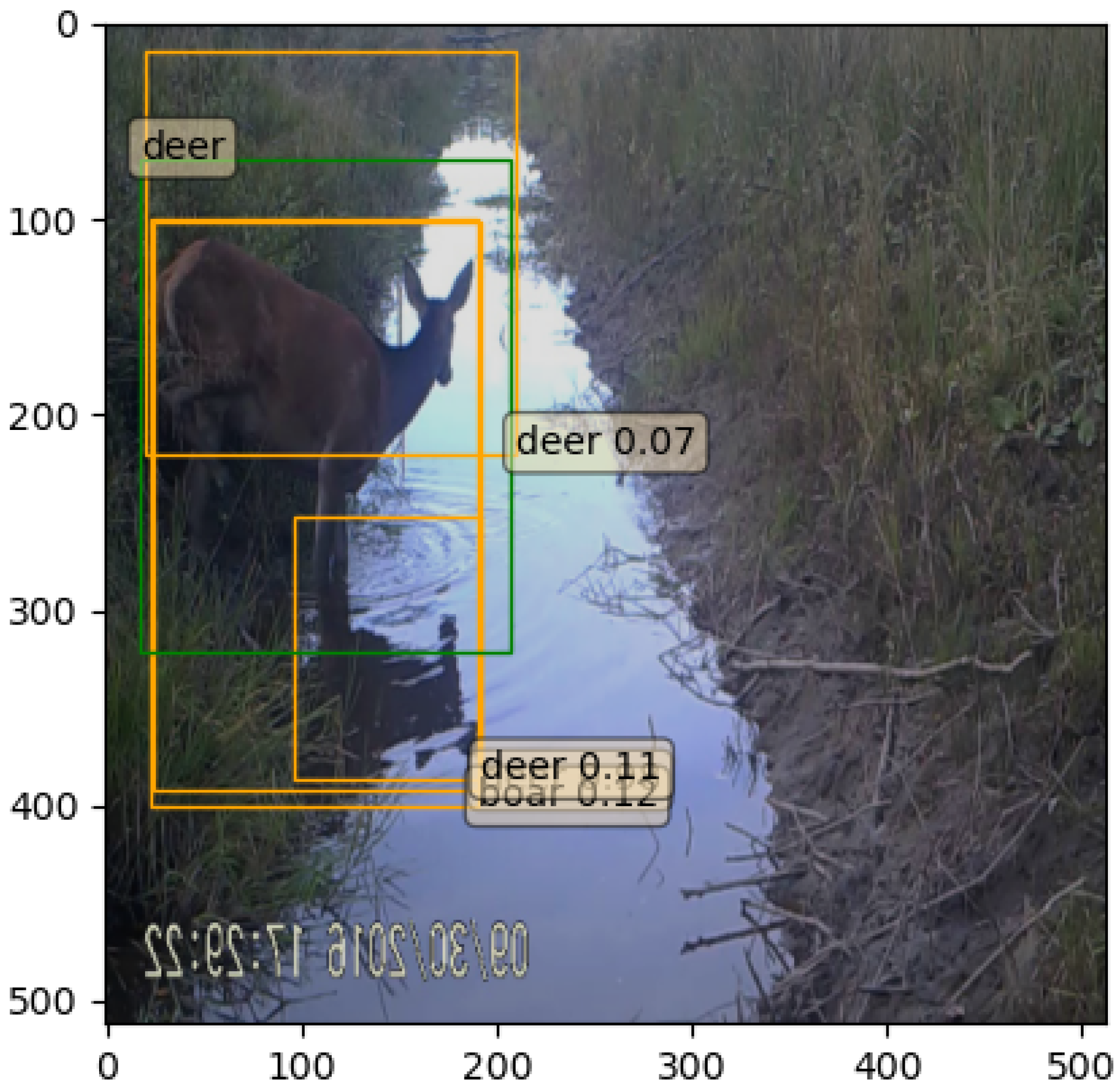
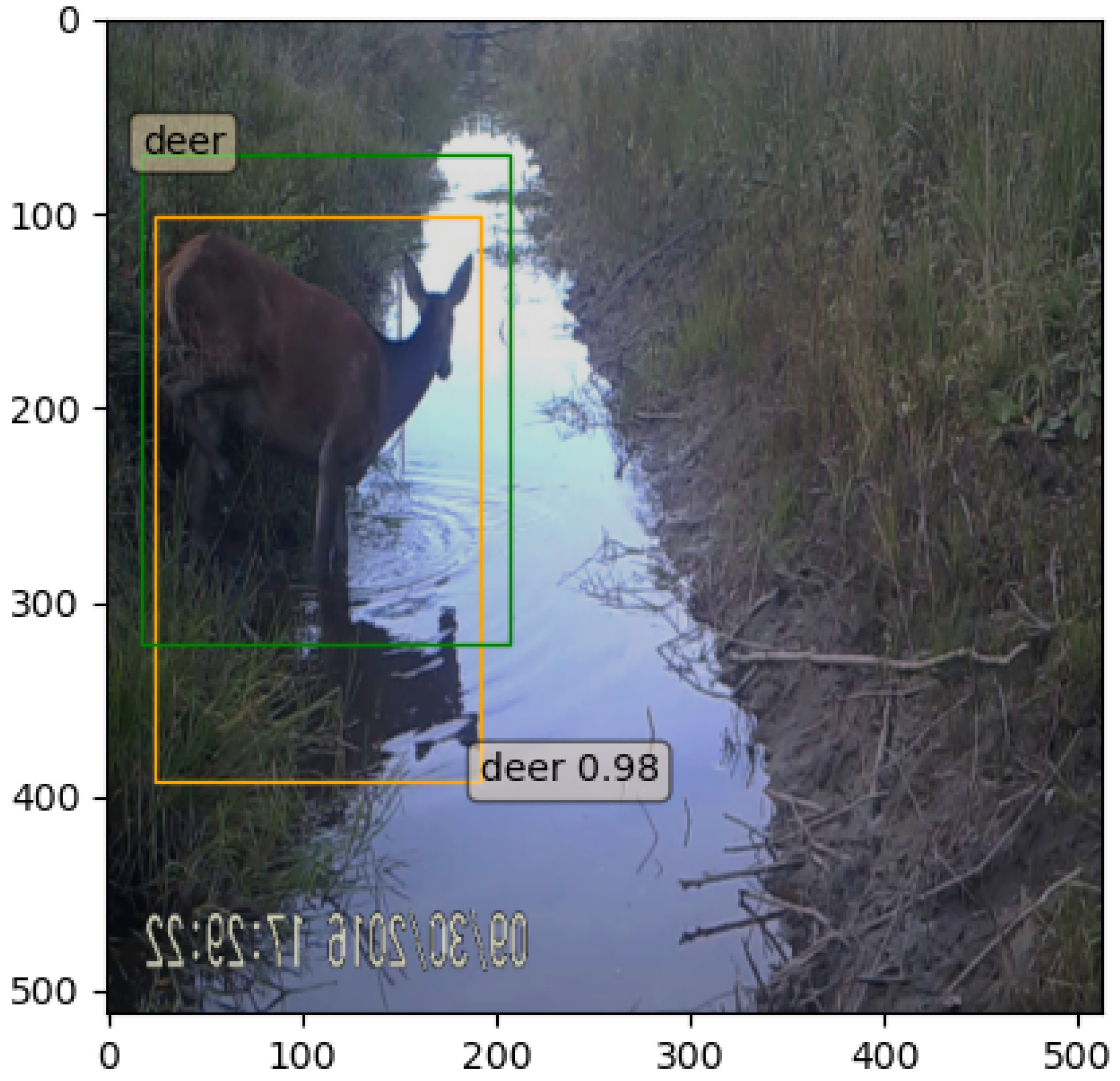

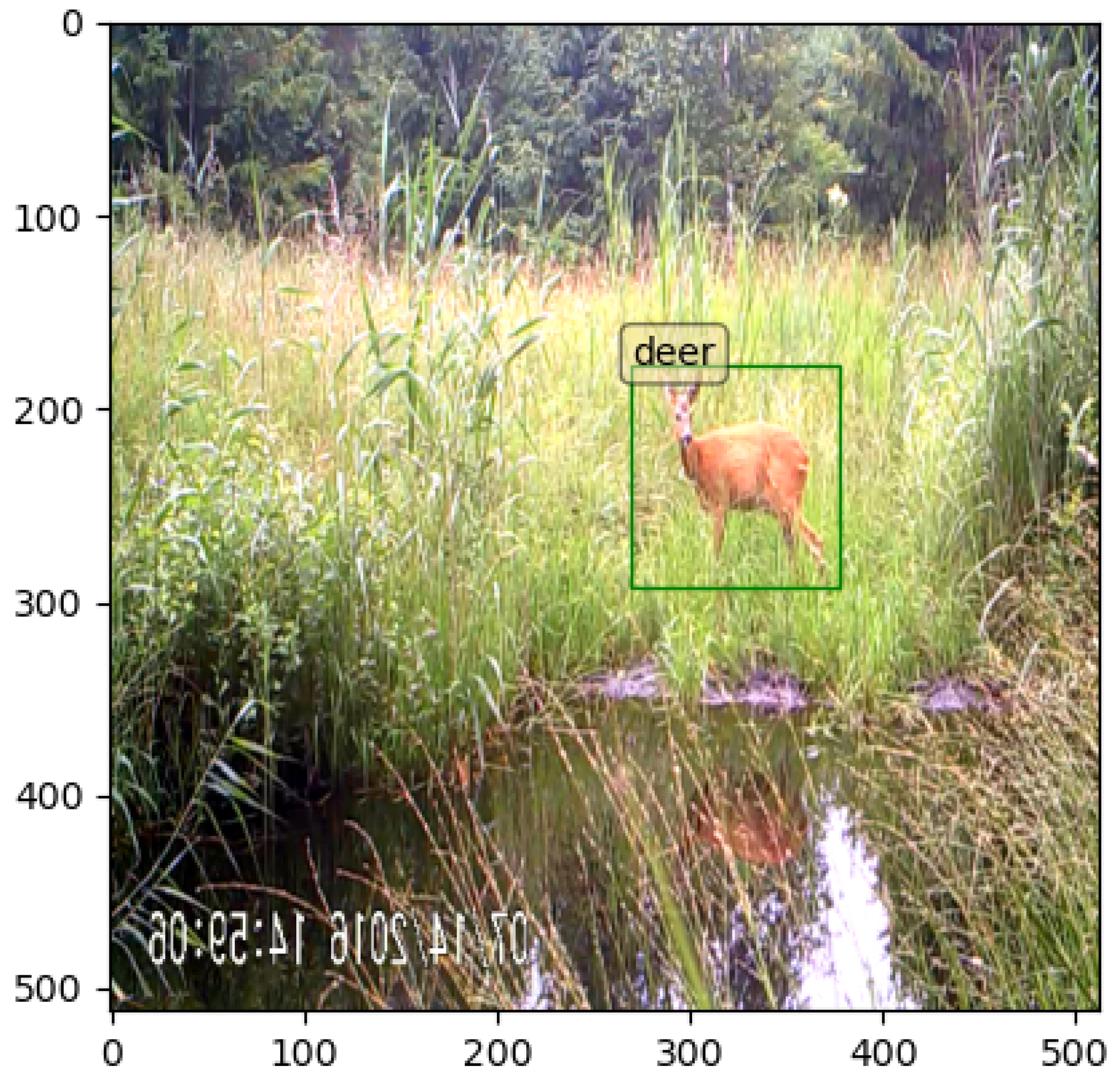


| Species or Group Name | Scientific Name | Number of Annotations |
|---|---|---|
| Deer | Cervidae | 516 |
| Wild boar | Sus Scrofa | 526 |
| Other species | 86 | |
| Total count | 1128 | |
| Species or Group Name | Scientific Name | Number of Annotations | Number of Augmented Samples | Total Number of Annotations |
|---|---|---|---|---|
| Deer | Cervidae | 6970 | 0 | 6970 |
| Wild boar | Sus Scrofa | 2642 | 4328 | 6970 |
| Total count | 9612 | 4328 | 13,940 |
| Model | Metrics | Number of Iterations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3485 | 6970 | 10,455 | 13,940 | 17,425 | 20,910 | 24,395 | 27,880 | 31,365 | 34,850 | ||
| Faster R-CNN | mAP @0.5:0.05:0.95 | 0.1832 | 0.2697 | 0.2238 | 0.1913 | 0.1848 | 0.2618 | 0.2551 | 0.2449 | 0.2241 | 0.2582 |
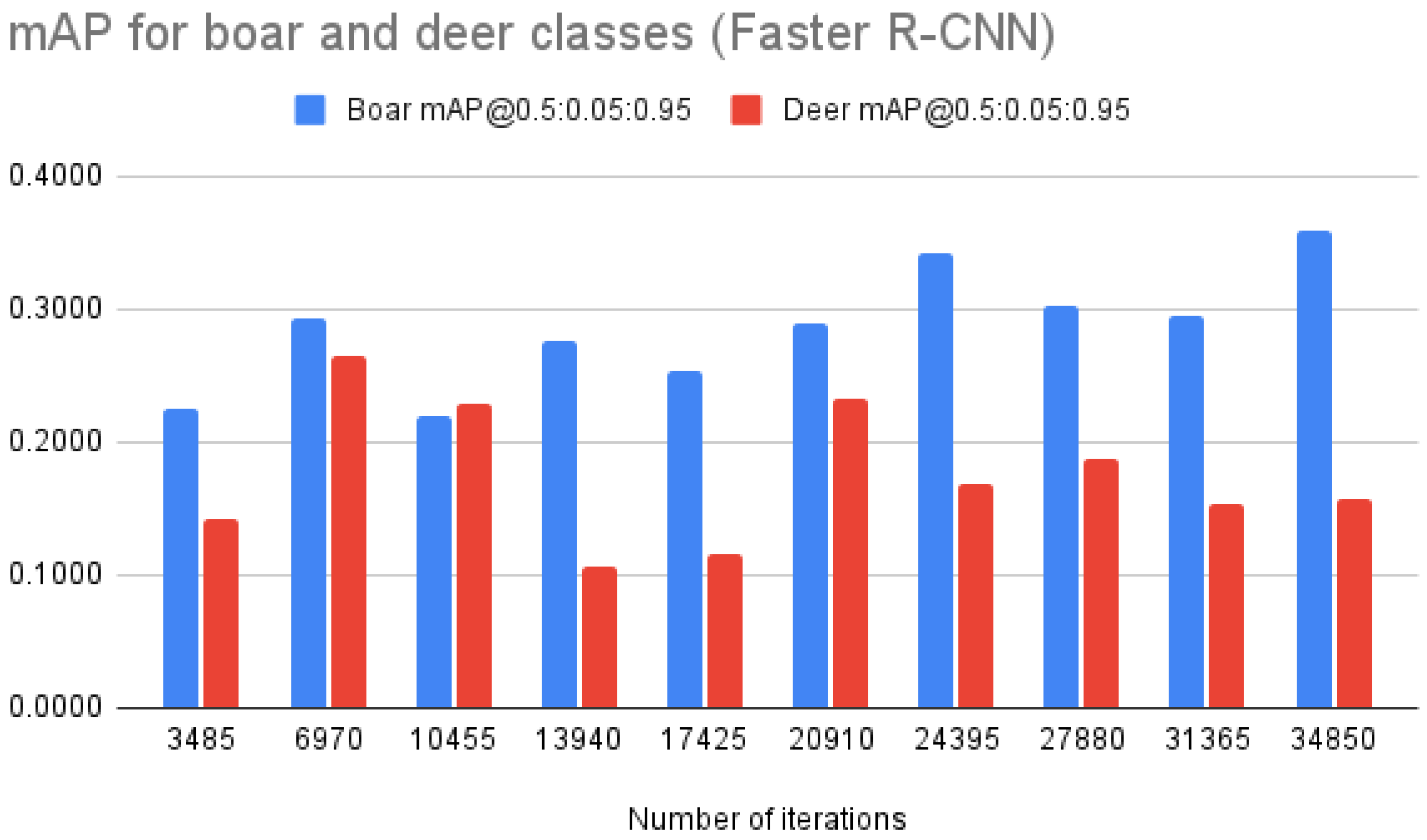
| mAP “deer” | 0.1420 | 0.2584 | 0.2288 | 0.1062 | 0.1164 | 0.2336 | 0.1684 | 0.1877 | 0.1539 | 0.1576 | |
| mAP “boar” | 0.2244 | 0.2810 | 0.2187 | 0.2764 | 0.2532 | 0.2900 | 0.3417 | 0.3021 | 0.2942 | 0.3589 | |
| mAP@0.5 | 0.3229 | 0.4561 | 0.3934 | 0.3305 | 0.3148 | 0.4562 | 0.4073 | 0.4065 | 0.3776 | 0.4204 | |
| mAP “deer” | 0.2800 | 0.4737 | 0.4337 | 0.2154 | 0.2098 | 0.4332 | 0.3065 | 0.3414 | 0.2956 | 0.2996 | |
| mAP “boar” | 0.3657 | 0.4385 | 0.3531 | 0.4456 | 0.4197 | 0.4791 | 0.5080 | 0.4715 | 0.4596 | 0.5411 | |
| mAP @0.75 | 0.1932 | 0.2860 | 0.2229 | 0.1926 | 0.1959 | 0.2855 | 0.2758 | 0.2571 | 0.2488 | 0.2756 | |
| mAP “deer” | 0.1367 | 0.2671 | 0.2222 | 0.0970 | 0.1175 | 0.2218 | 0.1659 | 0.1881 | 0.1454 | 0.1536 | |
| mAP “boar” | 0.2496 | 0.3048 | 0.2235 | 0.2881 | 0.2743 | 0.3492 | 0.3857 | 0.3260 | 0.3521 | 0.3976 | |
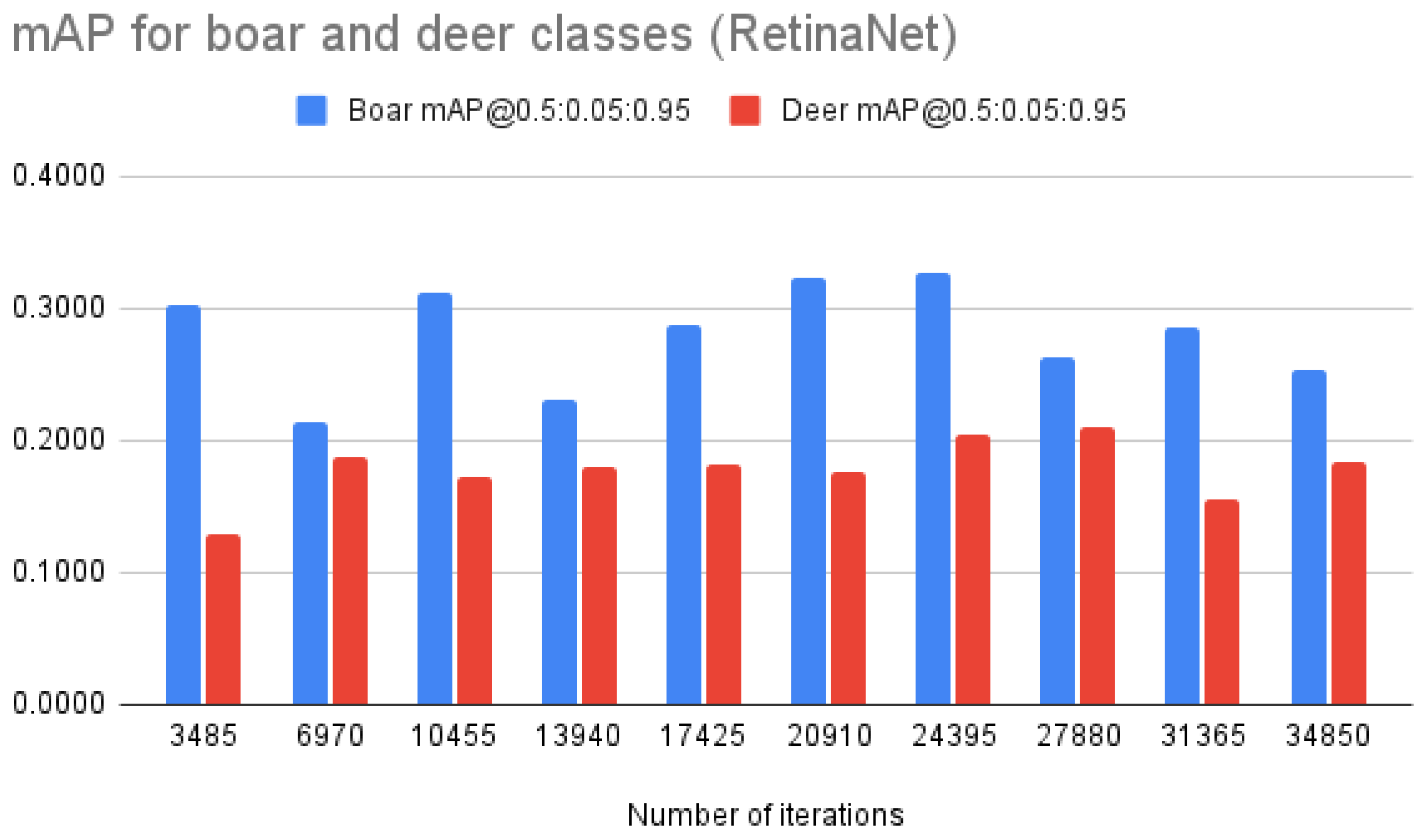
| RetinaNet | mAP @0.5:0.05:0.95 | 0.2158 | 0.2016 | 0.2413 | 0.2046 | 0.2346 | 0.2494 | 0.2659 | 0.2364 | 0.2202 | 0.2192 |
| mAP “deer” | 0.1287 | 0.1884 | 0.1715 | 0.1791 | 0.1827 | 0.1757 | 0.2053 | 0.2098 | 0.1551 | 0.1844 | |
| mAP “boar” | 0.3029 | 0.2148 | 0.3111 | 0.2301 | 0.2865 | 0.3231 | 0.3266 | 0.2630 | 0.2853 | 0.2540 | |
| mAP@0.5 | 0.3740 | 0.3725 | 0.4133 | 0.3574 | 0.4134 | 0.4198 | 0.4364 | 0.4173 | 0.3738 | 0.3922 | |
| mAP “deer” | 0.2727 | 0.3776 | 0.3361 | 0.3473 | 0.3530 | 0.3437 | 0.3814 | 0.4021 | 0.3017 | 0.3789 | |
| mAP “boar” | 0.4752 | 0.3673 | 0.4904 | 0.3675 | 0.4737 | 0.4959 | 0.4913 | 0.4325 | 0.4458 | 0.4054 | |
| mAP @0.75 | 0.1996 | 0.1909 | 0.2483 | 0.2179 | 0.2666 | 0.2678 | 0.2890 | 0.2421 | 0.2341 | 0.2236 | |
| mAP “deer” | 0.1028 | 0.1473 | 0.1499 | 0.1642 | 0.1844 | 0.1631 | 0.2152 | 0.1929 | 0.1442 | 0.1556 | |
| mAP “boar” | 0.2963 | 0.2345 | 0.3467 | 0.2716 | 0.3487 | 0.3724 | 0.3628 | 0.2913 | 0.3240 | 0.2915 | |
| Metrics | Model | ||
|---|---|---|---|
| RetinaNet Pre-Trained | RetinaNet Not Pre-Trained | RetinaNet Pre-Trained (Non-Oversampled Dataset) | |
| mAP @0.5:0.05:0.95 | 0.2158 | 0.1695 | 0.2290 |
| mAP “deer” | 0.1287 | 0.1492 | 0.1953 |
| mAP “boar” | 0.3029 | 0.1897 | 0.2626 |
| mAP@0.5 | 0.3740 | 0.2989 | 0.4029 |
| mAP “deer” | 0.2727 | 0.2900 | 0.3758 |
| mAP “boar” | 0.4752 | 0.3078 | 0.4299 |
| mAP @0.75 | 0.1996 | 0.1688 | 0.2265 |
| mAP “deer” | 0.1028 | 0.1441 | 0.1714 |
| mAP “boar” | 0.2963 | 0.1935 | 0.2815 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vecvanags, A.; Aktas, K.; Pavlovs, I.; Avots, E.; Filipovs, J.; Brauns, A.; Done, G.; Jakovels, D.; Anbarjafari, G. Ungulate Detection and Species Classification from Camera Trap Images Using RetinaNet and Faster R-CNN. Entropy 2022, 24, 353. https://doi.org/10.3390/e24030353
Vecvanags A, Aktas K, Pavlovs I, Avots E, Filipovs J, Brauns A, Done G, Jakovels D, Anbarjafari G. Ungulate Detection and Species Classification from Camera Trap Images Using RetinaNet and Faster R-CNN. Entropy. 2022; 24(3):353. https://doi.org/10.3390/e24030353
Chicago/Turabian StyleVecvanags, Alekss, Kadir Aktas, Ilja Pavlovs, Egils Avots, Jevgenijs Filipovs, Agris Brauns, Gundega Done, Dainis Jakovels, and Gholamreza Anbarjafari. 2022. "Ungulate Detection and Species Classification from Camera Trap Images Using RetinaNet and Faster R-CNN" Entropy 24, no. 3: 353. https://doi.org/10.3390/e24030353
APA StyleVecvanags, A., Aktas, K., Pavlovs, I., Avots, E., Filipovs, J., Brauns, A., Done, G., Jakovels, D., & Anbarjafari, G. (2022). Ungulate Detection and Species Classification from Camera Trap Images Using RetinaNet and Faster R-CNN. Entropy, 24(3), 353. https://doi.org/10.3390/e24030353







