Knowledge Discovery from Medical Data and Development of an Expert System in Immunology
Abstract
:1. Introduction
1.1. The Need for Using Expert Systems in Medicine
1.2. Short Consideration of Medical Expert Systems
1.3. Aim and Scope of the Article
2. Brief Description of the Immunological Basis of Bruton’s Disease
3. Discussion Concerning CRAN Package Repositories
4. The Database Used
4.1. Description of the Exploratory Database
4.2. Database after Initial Modification
5. Exploring of the Database Held and Discussion
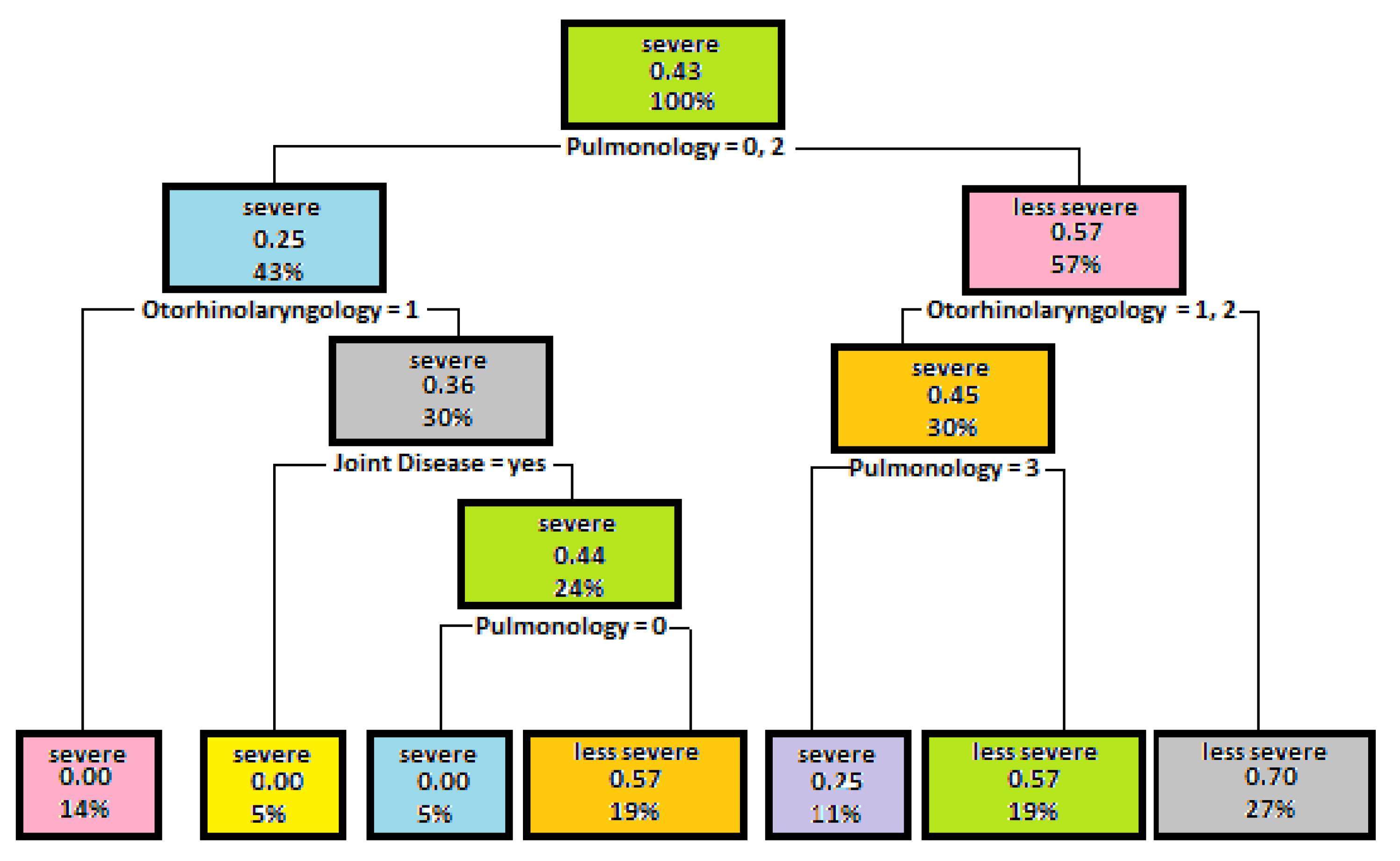
5.1. Decision Trees Generated to Determine Disease Severity
5.2. Results Decision Trees Created for Identifying the Mutation Severity
6. Establishing an Expert System Used for Diagnostic Applications
6.1. The CLIPS Language Selection
6.2. Brief Description of the Created Software
7. Verification of the Implemented Expert Systems
8. Summary
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cakir, S.; Otunctemur, A. Artificial Intelligence in Medicine. Eur. Arch. Med. Res. 2018, 34 (Suppl. S1), S1–S3. [Google Scholar] [CrossRef]
- Ishak, W.H.; Siraj, F. Artificial intelligence in medical application: An exploration. Health Inform. Eur. J. 2002, 16, 1–9. [Google Scholar]
- Mulawka, J. Expert Systems, 2nd ed.; Wydawnictwa Naukowo-Techniczne (WNT): Warsaw, Poland, 1997. (In Polish) [Google Scholar]
- Zhang, Y.; Tao, C.; Gong, Y.; Wang, K.; Zhao, Z. The International Conference on Intelligent Biology and Medicine 2018: Medical Informatics Thematic Track (MedicalInfo2018). BMC Med. Inform. Decis. Mak. 2019, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Strona, R. Prezentation: Expert Systems. Available online: https//www.r-project.org/ (accessed on 2 September 2019). (In Polish).
- Russell, S.J.; Norvig, P. Artificial Intelligence. A Modern Approach; Prentice Hall: Englewood Cliffs, NJ, USA, 2002. [Google Scholar]
- Giarratano, J.C.; Riley, G.D. Expert Systems. Principles and Programming; Thomson: Boston, MA, USA, 2005. [Google Scholar]
- Harmon, P.; Maus, R.; Morrissey, W. Expert Systems, Tools and Applications; John Wiley: New York, NY, USA, 1988. [Google Scholar]
- Jackson, P. Introduction to Expert Systems; Addison-Wesley: New York, NY, USA, 1999. [Google Scholar]
- Blachnik, M. Expert Systems. Available online: http://mblachnik.pl/lib/exe/fetch.php/dydaktyka/zajęcia/se/lab12_clips.pdf (accessed on 11 July 2019).
- TMYCIN Expert System Tool. Available online: https://www.cs.utexas.edu/users/novak/tmycin.html (accessed on 28 May 2021).
- Banks, G. Artificial intelligence in medical diagnosis: The INTERNIST/CADUCEUS approach. Crit. Rev. Med. Inform. 1986, 1, 23–54. [Google Scholar] [PubMed]
- Proven Radiology AI. Available online: https://www.aidoc.com (accessed on 28 May 2021).
- Coiera, E. Guide to Health Informatics, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2003; pp. 332–343. [Google Scholar]
- One-Stop Resource for Data on Infectious Diseases. Available online: https://www.gideononline.com/ (accessed on 28 May 2021).
- A Model-Based Consultation System for the Long-Term Management of Glaucoma. Available online: https://www.ijcai.org/Proceedings/77-2/Papers/060.pdf (accessed on 28 May 2021).
- Sullivan, K.; Stiehm, E.R. Stiehm’s Immune Deficiencies, 2nd ed.; Elsevier: London, UK, 2020. [Google Scholar]
- Pac, M.; Bernatowska, E. Comprehensive activities to increase recognition of primary immunodeficiency and access to immunoglobulin replacement therapy in Poland. Eur. J. Pediatr. 2016, 175, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Reust, C.E. Evaluation of Primary Immunodeficiency Disease in Children. Am. Fam. Physician 2013, 87, 773–778. [Google Scholar] [PubMed]
- Tóth, B.; Volokha, A.; Mihas, A.; Pac, M.; Bernatowska, E.; Kondratenko, I.; Polyakov, A.; Erdős, M.; Pasic, S.; Bataneant, M.; et al. Genetic and demographic features of X-linked agammaglobulinemia in Eastern and Central Europe: A cohort study. Mol. Immunol. 2009, 46, 2140–2146. [Google Scholar] [CrossRef]
- Pac, M.; Bernatowska, E.; Jackowska, T.; Mikołuć, B. Clinical analysis of X-linked agammaglobulinaemia and common variable immunodeficiency in children—What should paediatricians know? Pediatr. Pol. 2019, 94, 76–80. [Google Scholar] [CrossRef]
- Bruton, O. Agammaglobulinemia. Pediatrics 1952, 9, 722–728. [Google Scholar] [PubMed]
- Plebani, A.; Soresina, A.; Rondelli, R.; Amato, G.M.; Azzari, C.; Cardinale, F.; Cazzola, G.; Consolini, R.; De Mattia, D.; Dell’Erba, G.; et al. Clinical, Immunological, and Molecular Analysis in a Large Cohort of Patients with X-linked Agammaglobulinaemia: An Italian Multicenter Study. Clin. Immunol. 2002, 104, 221–230. [Google Scholar] [CrossRef]
- Lopez-Granados, E.; de Diego, R.P.; Cerdán, A.F.; Casariego, G.F.; Rodríguez, M.C.G. A genotype-phenotype correlation study in a group of 54 patients with X-linked agammaglobulinemia. J. Allergy Clin. Immunol. 2005, 116, 690–697. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.W.; Chen, T.X.; Jiang, L.P.; Chan, K.W.; Yang, W.Y.; Lee, B.W.; Chiang, W.C.; Chen, X.Y.; Fok, S.S.; Lee, T.L.; et al. Clinical Characteristics and genotype-phenotype correlation in 62 patients with X-linked agammaglobulinemia. J. Clin. Immunol. 2010, 30, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Aadam, Z.; Kechout, N.; Barakat, A.; Chan, K.W.; Ben-Ali, M.; Ben-Mustapha, I.; Zidi, F.; Ailal, F.; Attal, N.; Doudou, F.; et al. X-linked agammaglobulinemia in a large series of North African patients: Fequency, clinical features and novel BTK mutations. J. Clin. Immunol. 2016, 36, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Morzy, T. Eksploration of Data: Methods and Algorithms; Panstwowe Wydawnictwo Naukowe (PWN): Warsaw, Poland, 2013. (In Polish) [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference and Prediction, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- CLIPS Rule Based Programming Language. Available online: https://sourceforge.net/projects/clipsrules/ (accessed on 28 May 2021).
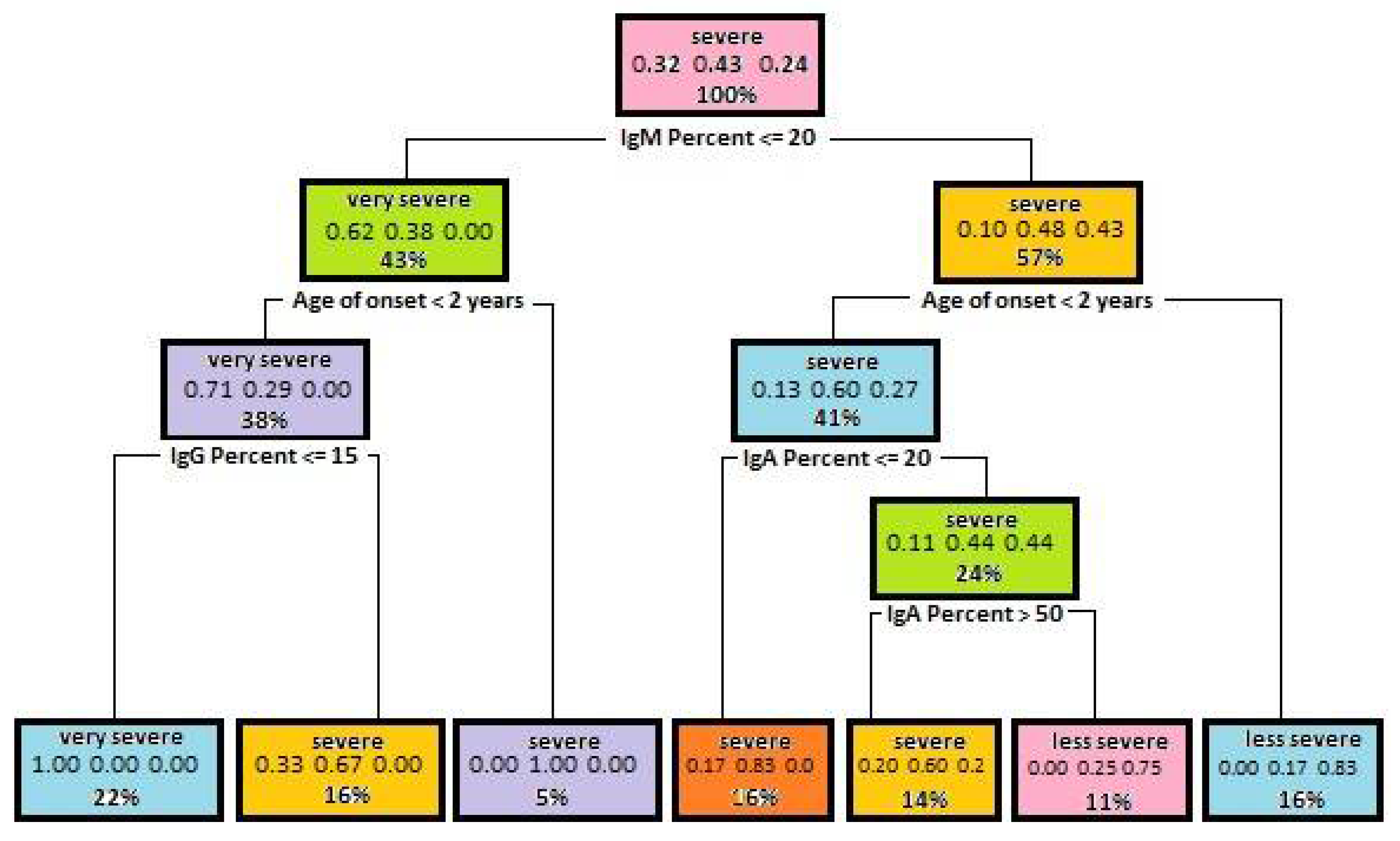
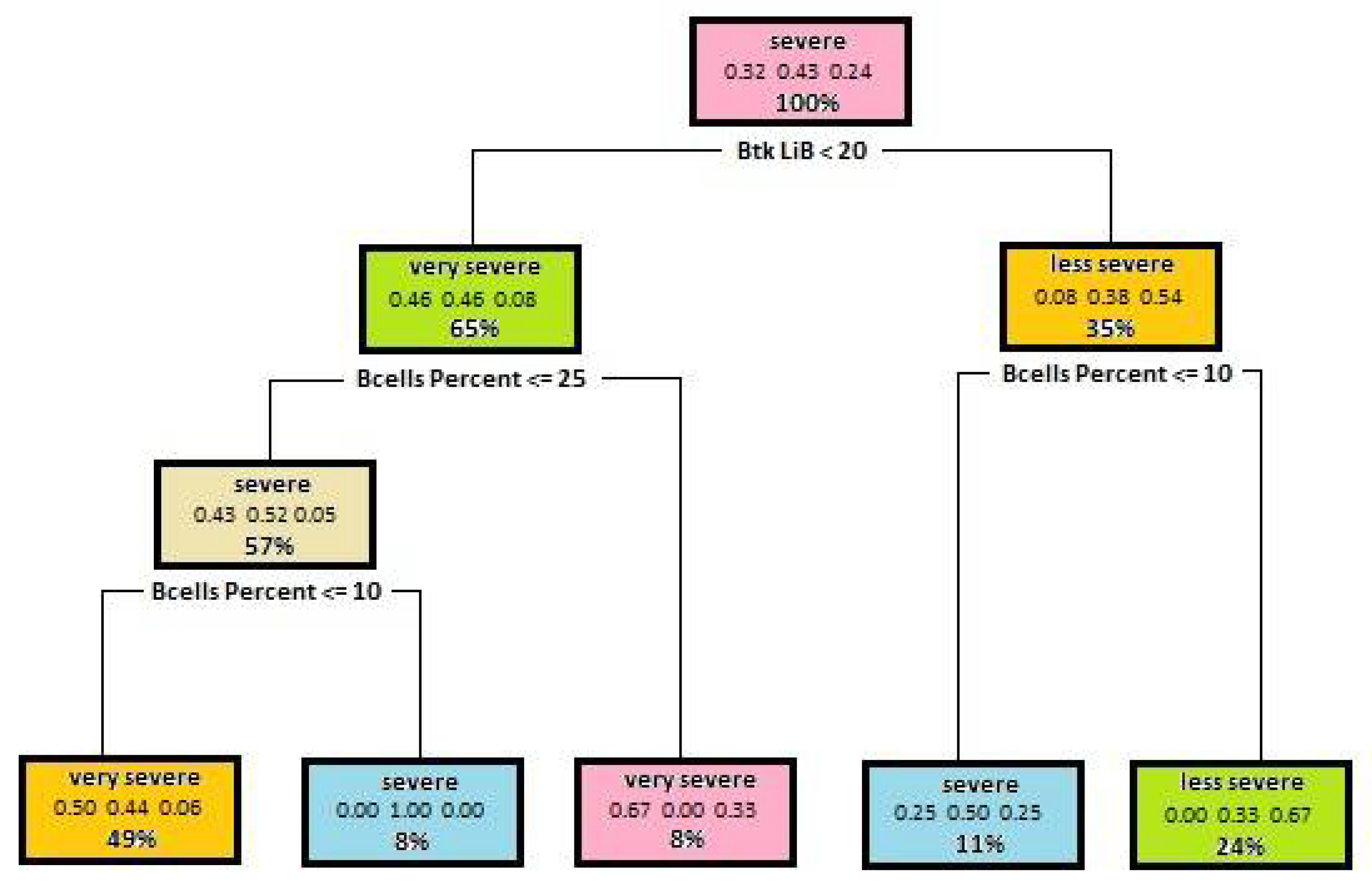
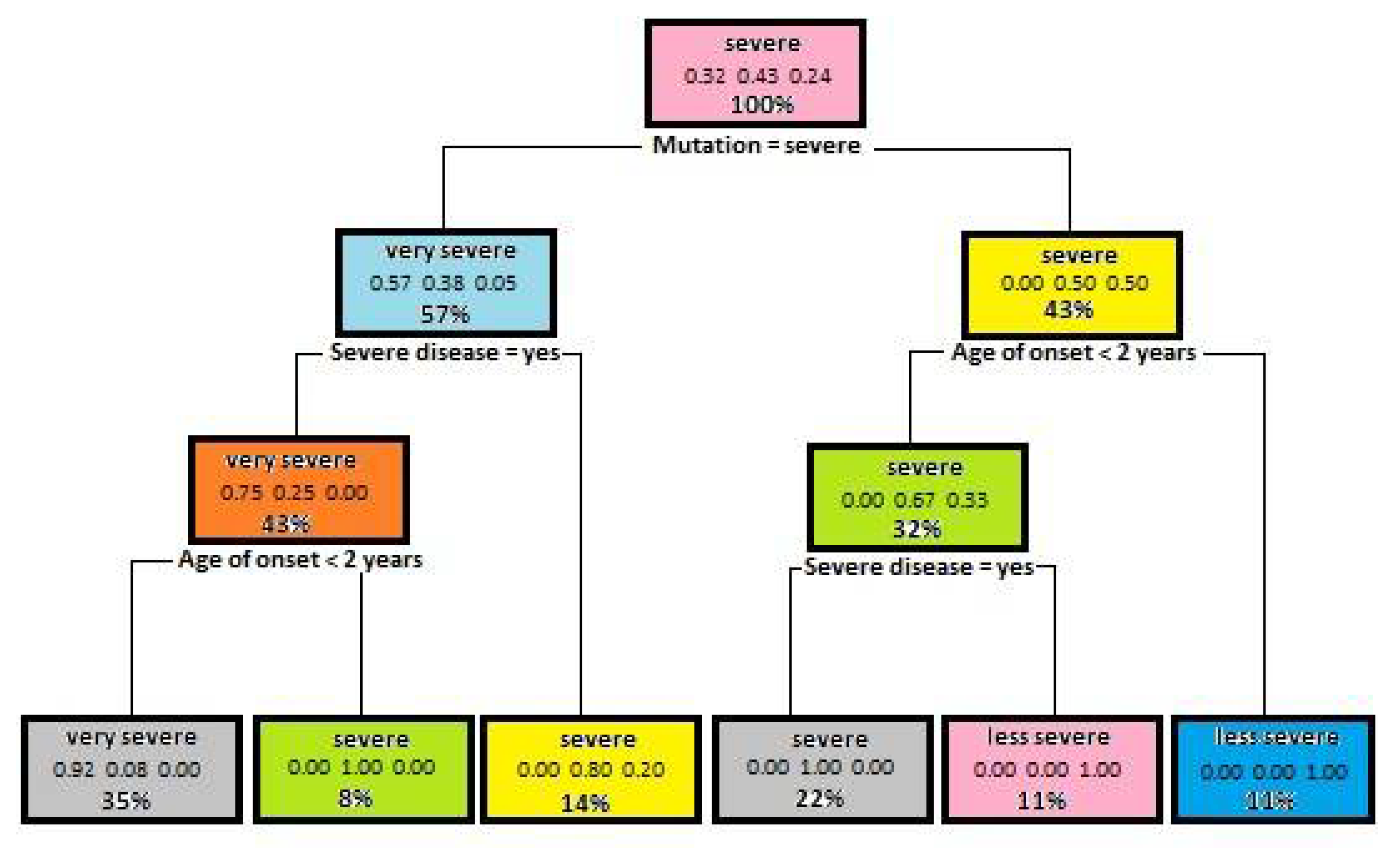
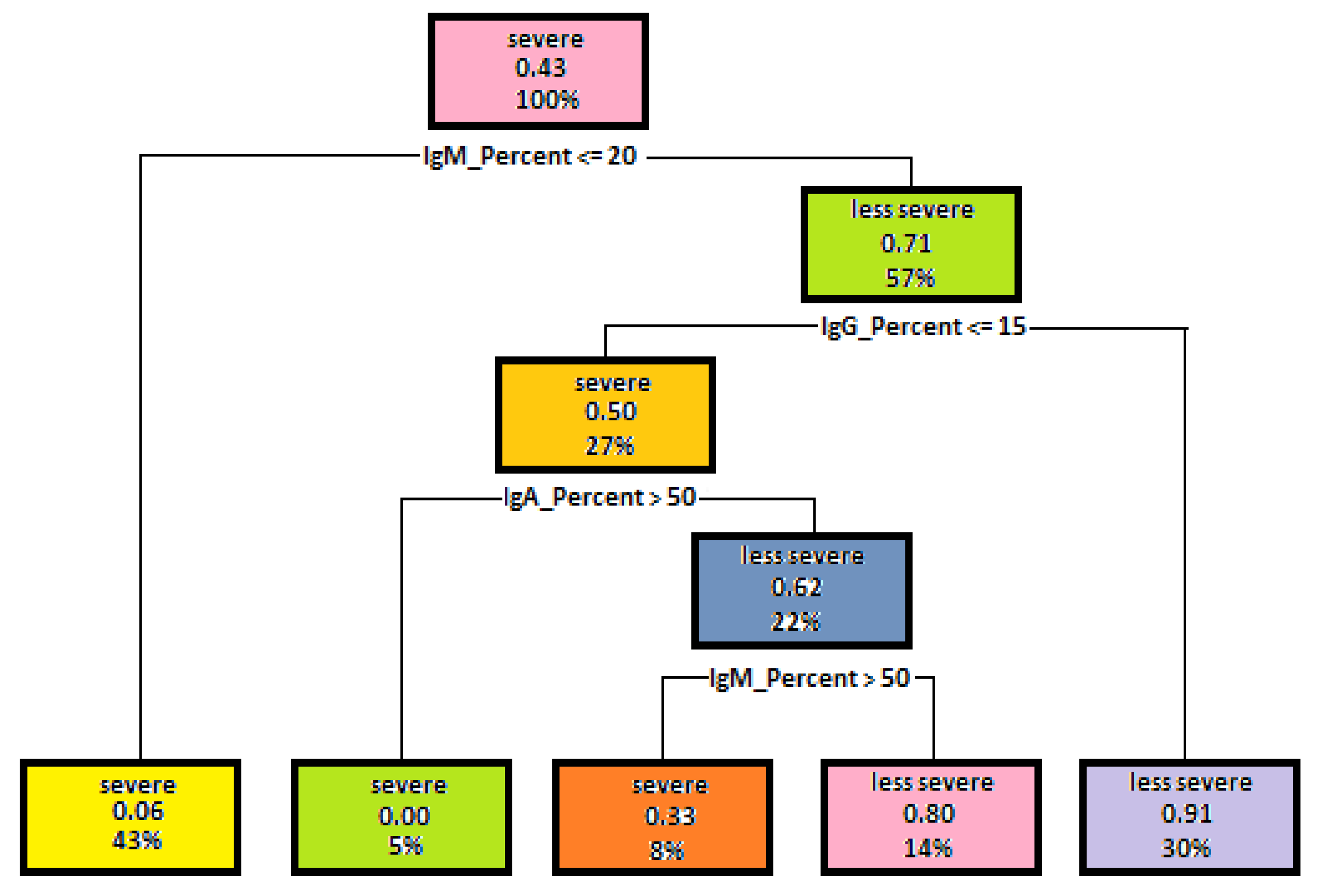

| Attribute | Gini Index for Disease Severity | Gini Index for Mutation Severity |
|---|---|---|
| Age of onset | 2.3768844 | 1.3391285 |
| Percentage of IgG | 3.1353790 | 3.5060282 |
| Percentage of IgM | 6.2144411 | 4.2951580 |
| Percentage of IgA | 3.1072242 | 2.0747686 |
| Percentage of B cells | 2.6674329 | 1.9705842 |
| Btk in B cells | 4.9642351 | 4.5973863 |
| Btk in monocytes | 2.4322656 | 2.1629096 |
| Family history | 0.2389207 | 0.2670715 |
| Severe diseases | 1.7318287 | 1.3331821 |
| Less severe diseases | 0.6482581 | 1.3391285 |
| Rheumatology | 0.2090056 | 0.2530046 |
| Pulmonology | 0.9603348 | 0.7524785 |
| Otorhinolaryngology | 0.3649750 | 0.5219833 |
| Gastroenterology | 0.4077892 | 0.4276909 |
| Immunology | 0.3829554 | 0.5736574 |
| Mutation severity | 2.8436096 | --- |
| Age of Onset | IgM | IgG | IgA | Expected Result | |
|---|---|---|---|---|---|
| Data set 1 | 19 | 12 | 19 | 17 | Very severe course of the disease |
| Data set 2 | 19 | 12 | 73 | 17 | Severe course of the disease |
| Data set 3 | 47 | 12 | 73 | 17 | Severe course of the disease |
| Data set 4 | 47 | 92 | 73 | 17 | Less severe course of the disease |
| Data set 5 | 23 | 92 | 73 | 17 | Severe course of the disease |
| Data set 6 | 23 | 92 | 73 | 46 | Less severe course of the disease |
| Data set 7 | 23 | 92 | 73 | 76 | Severe course of the disease |
| Age of Onset | IgM | IgG | IgA | Expected Result | |
|---|---|---|---|---|---|
| Data set 1 | 23 | 17 | 73 | 29 | Severe mutation |
| Data set 2 | 19 | 36 | 58 | 17 | Less severe mutation |
| Data set 3 | 3 | 81 | 14 | 62 | Severe mutation |
| Data set 4 | 120 | 81 | 7 | 35 | Severe mutation |
| Data set 5 | 23 | 41 | 7 | 35 | Less severe mutation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pac, M.; Mikutskaya, I.; Mulawka, J. Knowledge Discovery from Medical Data and Development of an Expert System in Immunology. Entropy 2021, 23, 695. https://doi.org/10.3390/e23060695
Pac M, Mikutskaya I, Mulawka J. Knowledge Discovery from Medical Data and Development of an Expert System in Immunology. Entropy. 2021; 23(6):695. https://doi.org/10.3390/e23060695
Chicago/Turabian StylePac, Małgorzata, Irina Mikutskaya, and Jan Mulawka. 2021. "Knowledge Discovery from Medical Data and Development of an Expert System in Immunology" Entropy 23, no. 6: 695. https://doi.org/10.3390/e23060695
APA StylePac, M., Mikutskaya, I., & Mulawka, J. (2021). Knowledge Discovery from Medical Data and Development of an Expert System in Immunology. Entropy, 23(6), 695. https://doi.org/10.3390/e23060695






