Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network
Abstract
1. Introduction
2. Literature Review
2.1. Classification Paradigms
2.2. Existing Methods
3. Methods
3.1. Dataset
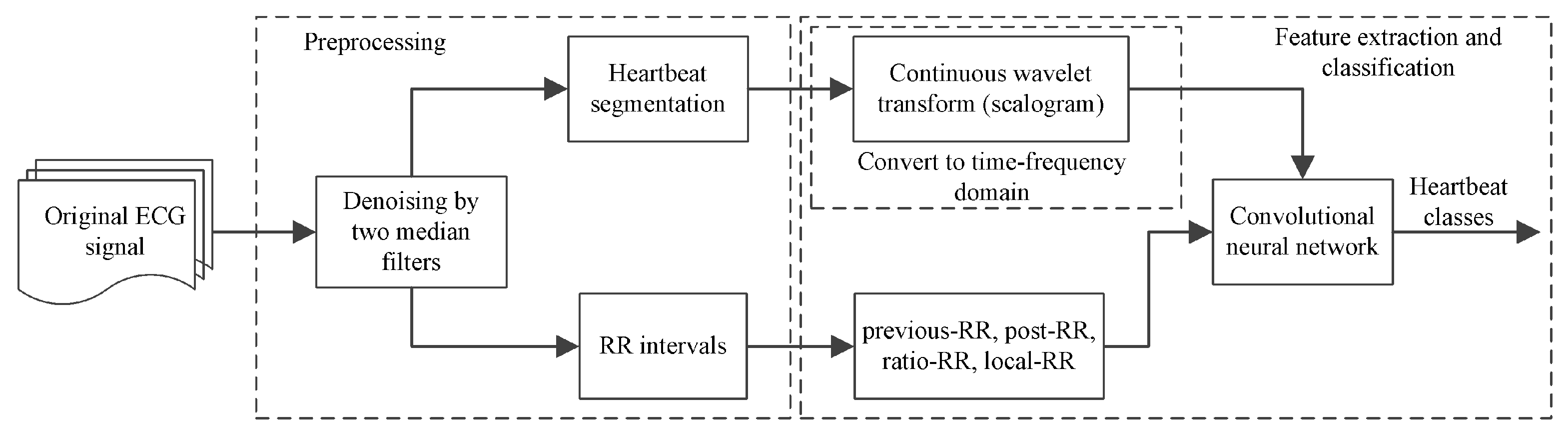
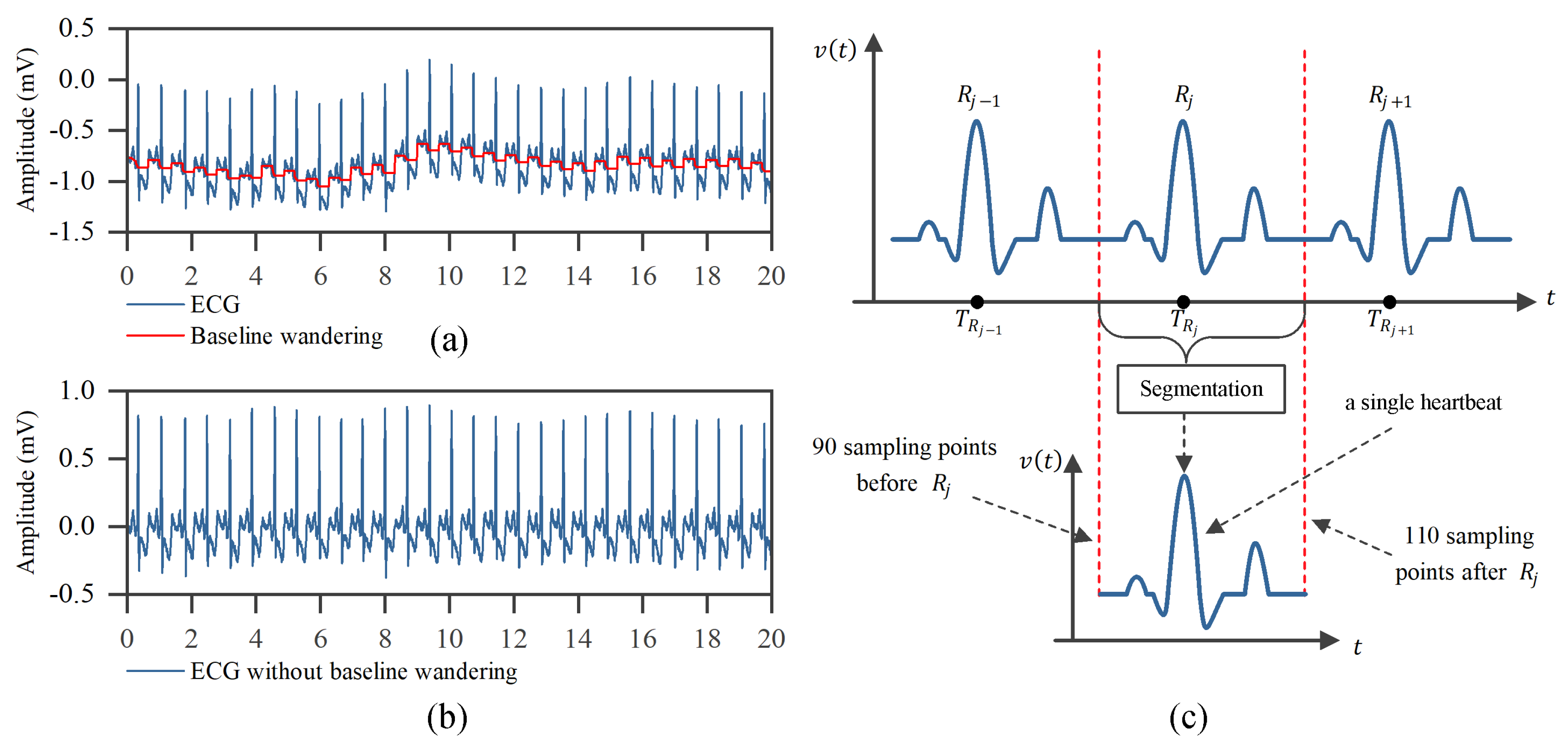
3.2. ECG Preprocessing and Heartbeat Segmentation
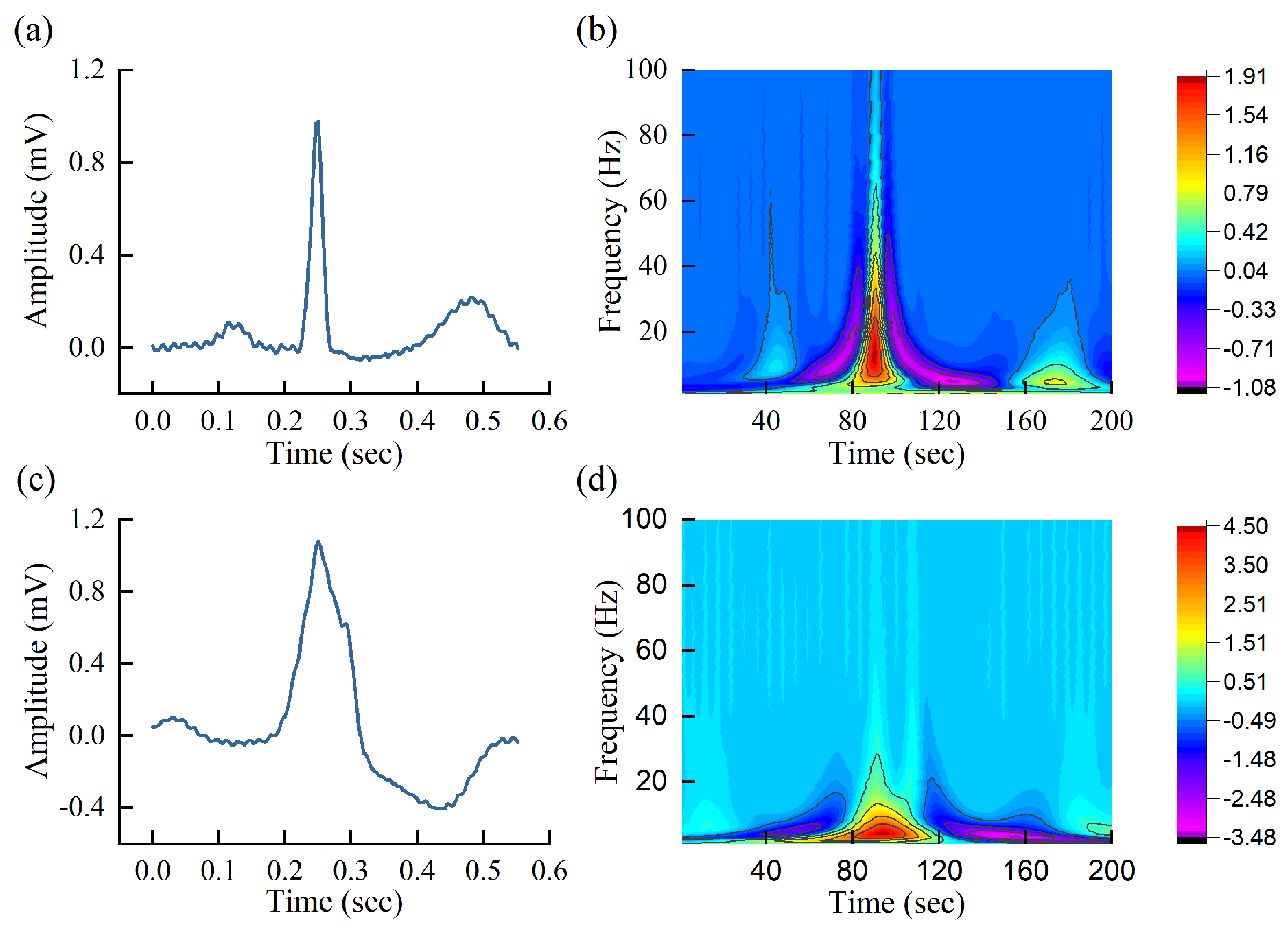
3.3. Time-Frequency Scalogram via CWT
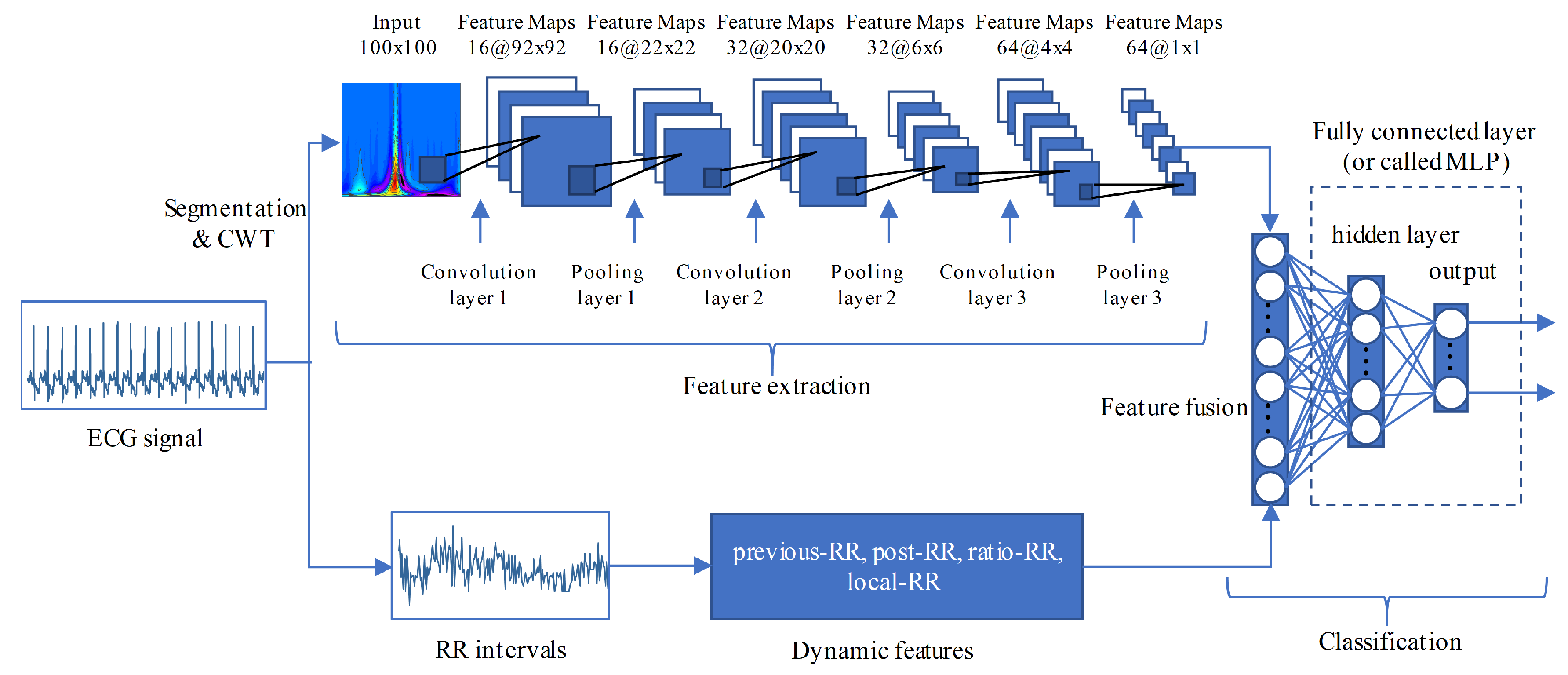
3.4. ECG Classification Based on CNN
3.5. Training Setting
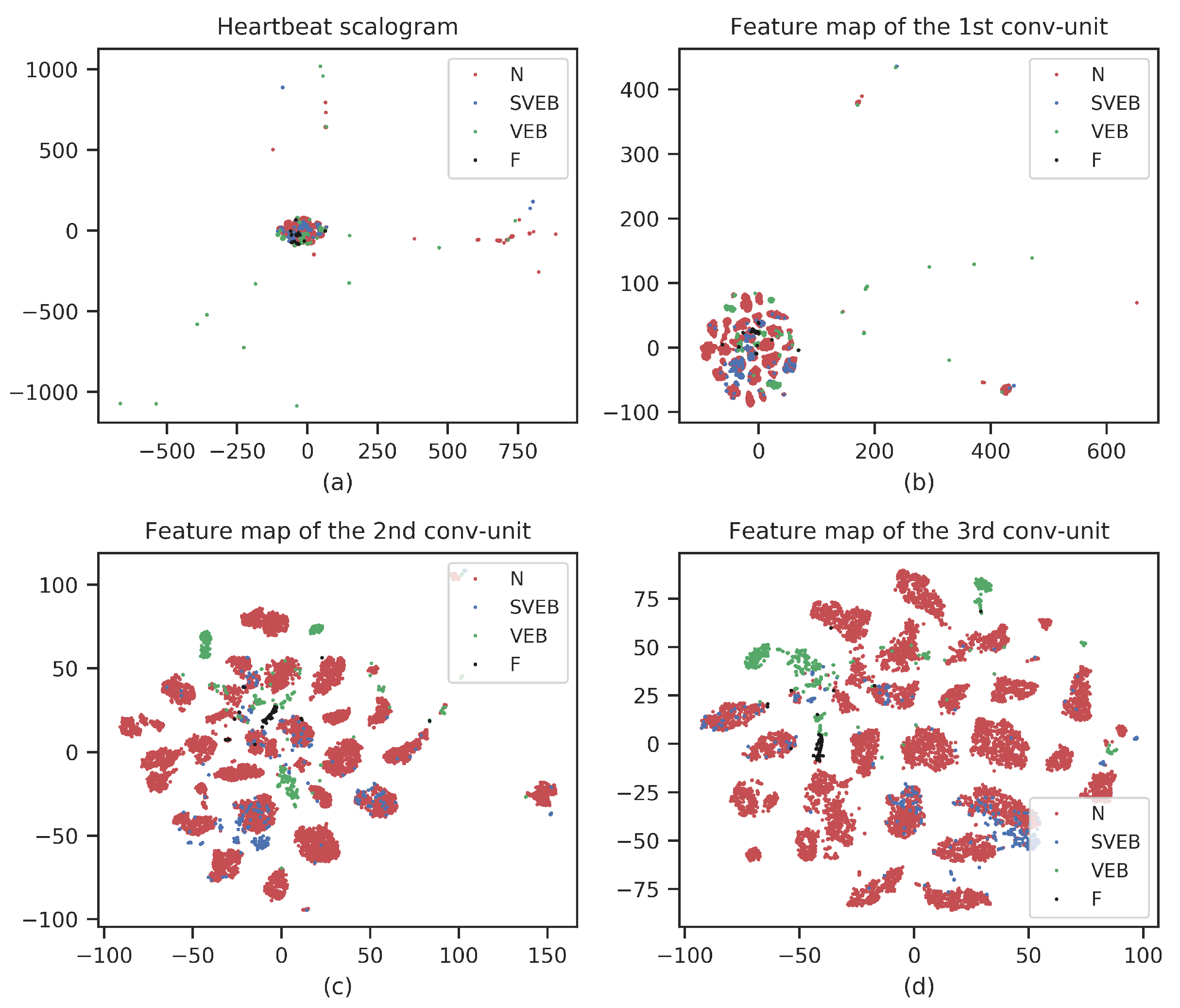
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sannino, G.; De Pietro, G. A deep learning approach for ECG-based heartbeat classification for arrhythmia detection. Future Gener. Comput. Syst. 2018, 86, 446–455. [Google Scholar] [CrossRef]
- Luo, K.; Li, J.; Wang, Z.; Cuschieri, A. Patient-specific deep architectural model for ECG classification. J. Healthc. Eng. 2017, 2017, 4108720. [Google Scholar] [CrossRef]
- Yang, S.; Shen, H. Heartbeat classification using discrete wavelet transform and kernel principal component analysis. In Proceedings of the IEEE 2013 Tencon-Spring, Sydney, Australia, 17–19 April 2013; pp. 34–38. [Google Scholar]
- De Chazal, P.; O’Dwyer, M.; Reilly, R.B. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. 2012, 59, 2930–2941. [Google Scholar] [PubMed]
- Xu, S.S.; Mak, M.W.; Cheung, C.C. Towards end-to-end ECG classification with raw signal extraction and deep neural networks. IEEE J. Biomed. Health Inform. 2018, 23, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Sellami, A.; Hwang, H. A robust deep convolutional neural network with batch-weighted loss for heartbeat classification. Expert Syst. Appl. 2019, 122, 75–84. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef]
- Daubechies, I. The wavelet transform, time-frequency localization and signal analysis. IEEE Trans. Inf. Theory 1990, 36, 961–1005. [Google Scholar] [CrossRef]
- Portnoff, M. Time-frequency representation of digital signals and systems based on short-time Fourier analysis. IEEE Trans. Acoust. Speech Signal Process. 1980, 28, 55–69. [Google Scholar] [CrossRef]
- Guo, M.F.; Zeng, X.D.; Chen, D.Y.; Yang, N.C. Deep-learning-based earth fault detection using continuous wavelet transform and convolutional neural network in resonant grounding distribution systems. IEEE Sens. J. 2017, 18, 1291–1300. [Google Scholar] [CrossRef]
- Wei, Y.; Xia, W.; Lin, M.; Huang, J.; Ni, B.; Dong, J.; Zhao, Y.; Yan, S. HCP: A flexible CNN framework for multi-label image classification. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 1901–1907. [Google Scholar] [CrossRef] [PubMed]
- Yue-Hei Ng, J.; Hausknecht, M.; Vijayanarasimhan, S.; Vinyals, O.; Monga, R.; Toderici, G. Beyond short snippets: Deep networks for video classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 4694–4702. [Google Scholar]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Jian, L.; Shuang, S.; Guozhong, S.; Yu, F. Classification of ECG Arrhythmia Using CNN, SVM and LDA. In Proceedings of the Artificial Intelligence and Security, 5th International Conference, ICAIS 2019; Proceedings: Lecture Notes in Computer Science (LNCS 11633), New York, NY, USA, 26–28 July 2019; pp. 191–201. [Google Scholar] [CrossRef]
- Chen, S.; Hua, W.; Li, Z.; Li, J.; Gao, X. Heartbeat classification using projected and dynamic features of ECG signal. Biomed. Signal Process. Control 2017, 31, 165–173. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Luo, X.; Choi, K.S.; Wu, X. Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 2014, 46, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G.; Moreira, G.; Menotti, D.; Luz, E. Inter-Patient ECG Heartbeat Classification with Temporal VCG Optimized by PSO. Sci. Rep. 2017, 7, 10543. [Google Scholar] [CrossRef]
- De Chazal, P.; Reilly, R.B. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2535–2543. [Google Scholar] [CrossRef]
- Hu, Y.H.; Palreddy, S.; Tompkins, W.J. A patient-adaptable ECG beat classifier using a mixture of experts approach. IEEE Trans. Biomed. Eng. 1997, 44, 891–900. [Google Scholar]
- Ince, T.; Kiranyaz, S.; Gabbouj, M. A generic and robust system for automated patient-specific classification of ECG signals. IEEE Trans. Biomed. Eng. 2009, 56, 1415–1426. [Google Scholar] [CrossRef]
- Ye, C.; Kumar, B.V.; Coimbra, M.T. An automatic subject-adaptable heartbeat classifier based on multiview learning. IEEE J. Biomed. Health Inform. 2015, 20, 1485–1492. [Google Scholar] [CrossRef]
- Mar, T.; Zaunseder, S.; Martínez, J.P.; Llamedo, M.; Poll, R. Optimization of ECG classification by means of feature selection. IEEE Trans. Biomed. Eng. 2011, 58, 2168–2177. [Google Scholar] [CrossRef]
- Mondéjar-Guerra, V.; Novo, J.; Rouco, J.; Penedo, M.G.; Ortega, M. Heartbeat classification fusing temporal and morphological information of ECGs via ensemble of classifiers. Biomed. Signal Process. Control 2019, 47, 41–48. [Google Scholar] [CrossRef]
- He, J.; Rong, J.; Sun, L.; Wang, H.; Zhang, Y.; Ma, J. A framework for cardiac arrhythmia detection from IoT-based ECGs. World Wide Web 2020, 23, 2835–2850. [Google Scholar] [CrossRef]
- Wu, Z.; Lan, T.; Yang, C.; Nie, Z. A Novel Method to Detect Multiple Arrhythmias Based on Time-Frequency Analysis and Convolutional Neural Networks. IEEE Access 2019, 7, 170820–170830. [Google Scholar] [CrossRef]
- Hijazi, S.; Kumar, R.; Rowen, C. Using Convolutional Neural Networks for Image Recognition; Cadence Design Systems Inc.: San Jose, CA, USA, 2015; pp. 1–12. [Google Scholar]
- Xiong, Z.; Shen, Q.; Wang, Y.; Zhu, C. Paragraph vector representation based on word to vector and CNN learning. Comput. Mater. Contin. 2018, 55, 213–227. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- Ketkar, N. Introduction to pytorch. In Deep Learning with Python; Springer: Berlin/Heidelberg, Germany, 2017; pp. 195–208. [Google Scholar]
- Chen, Y. Learning Classifiers from Imbalanced, only Positive and Unlabeled Data Sets; Department of Computer Science, Iowa State University: Ames, IA, USA, 2009. [Google Scholar]
- Zhong, Z.; Li, J.; Ma, L.; Jiang, H.; Zhao, H. Deep residual networks for hyperspectral image classification. In Proceedings of the 2017 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Fort Worth, TX, USA, 23–28 July 2017; pp. 1824–1827. [Google Scholar]

| AAMI Classes | Normal (N) | Supraventricular Ectopic Beat (SVEB) | Ventricular Ectopic Beat (VEB) | Fusion Beat (F) | Unknown Beat (Q) |
|---|---|---|---|---|---|
| MIT-BIH arrhythmia types | Normal beat (NOR)—N | Atrial premature beat (AP)—A | Ventricular escape beat (VE)—E | Fusion of ventricular and normal beat (fVN)—F | Unclassifiable beat (U)—Q |
| Right bundle branch block beat (RBBB)—R | Premature or ectopic supraventricular beat (SP)—S | Premature ventricular contraction (PVC)—V | Fusion of paced and normal beat (fPN)—f | ||
| Left bundle branch block beat (LBBB)—L | Nodal (junctional) premature beat (NP)—J | Paced beat (P)—/ | |||
| Atrial escape beat (AE)—e | Aberrated arial premature beat (aAP)—a | ||||
| Nodal (junctional) escape beat (NE)—j |
| No. | Layer Name | Kernel Size | Filter | Padding | Stride | Output Shape | Parameters |
|---|---|---|---|---|---|---|---|
| 1 | Input1 * | - | - | - | - | 100 × 100 × 1 | - |
| 2 | Conv2D | 7 × 7 | 16 | 0 | 1 | 94 × 94 × 16 | 784 |
| 3 | Batch Normalization | - | - | - | - | 94 × 94 × 16 | 64 |
| 4 | ReLu | - | - | - | - | 94 × 94 × 16 | - |
| 5 | Max pooling | 5 × 5 | - | 0 | 5 | 18 × 18 × 16 | - |
| 6 | Conv2D | 3 × 3 | 32 | 0 | 1 | 16 × 16 × 32 | 4608 |
| 7 | Batch Normalization | - | - | - | - | 16 × 16 × 32 | 128 |
| 8 | ReLu | - | - | - | - | 16 × 16 × 32 | - |
| 9 | Max pooling | 3 × 3 | - | 0 | 3 | 5 × 5 × 32 | - |
| 10 | Conv2D | 3 × 3 | 64 | 0 | 1 | 3 × 3 × 64 | 18,432 |
| 11 | Batch Normalization | - | - | - | - | 3 × 3 × 64 | 256 |
| 12 | ReLu | - | - | - | - | 3 × 3 × 64 | - |
| 13 | Global Max pooling | 3 × 3 | - | - | - | 1 × 1 × 64 | - |
| 14 | Flatten | - | - | - | - | 64 | - |
| 15 | Input2 ** | - | - | - | - | 4 | - |
| 16 | Concatenate | - | - | - | - | 68 | - |
| 17 | Dense | - | - | - | - | 32 | 2208 |
| 18 | Dense | - | - | - | - | 4 | 132 |
| Methods | SVEB | VEB | ||||||
|---|---|---|---|---|---|---|---|---|
| Positive Predictive Value | Sensitivity | F1-score | Accuracy | Positive Predictive Value | Sensitivity | F1-score | Accuracy | |
| Liu et al. [15] | 39.87% | 33.12% | 36.18% | 95.49% | 76.51% | 90.2% | 82.79% | 97.45% |
| Chen et al. [16] | 38.40% | 29.50% | 33.36% | 95.34% | 85.25% | 70.85% | 77.38% | 97.32% |
| Zhang et al. [17] | 35.98% | 79.06% | 49.46% | 93.33% | 92.75% | 85.48% | 88.96% | 98.63% |
| Ye et al. [5] | 52.34% | 61.02% | 56.34% | 96.27% | 61.45% | 81.82% | 70.19% | 95.52% |
| Garcia et al. [18] | 53.00% | 62.00% | 57.15% | - | 59.40% | 87.30% | 70.70% | - |
| Our method | 89.54% | 74.56% | 81.37% | 98.74% | 93.25% | 95.65% | 94.43% | 99.27% |
| Classes | Metrics | Methods | |||||
|---|---|---|---|---|---|---|---|
| Liu et al. | Chen et al. | Zhang et al. | Ye et al. | Garcia et al. | Our Method | ||
| N | Positive predictive value | 96.66% | 95.42% | 98.98% | 97.55% | 98.00% | 98.17% |
| Sensitivity | 94.06% | 98.42% | 88.94% | 88.61% | 94.00% | 99.42% | |
| F1-score | 95.34% | 96.90% | 93.69% | 92.87% | 95.96% | 98.79% | |
| SVEB | Positive predictive value | 39.87% | 38.40% | 35.98% | 52.34% | 53.00% | 89.54% |
| Sensitivity | 33.12% | 29.50% | 79.06% | 61.02% | 62.00% | 74.56% | |
| F1-score | 36.18% | 33.36% | 49.46% | 56.34% | 57.15% | 81.37% | |
| VEB | Positive predictive value | 76.51% | 85.25% | 92.75% | 61.45% | 59.40% | 93.25% |
| Sensitivity | 90.20% | 70.85% | 85.48% | 81.82% | 87.30% | 95.65% | |
| F1-score | 82.79% | 77.38% | 88.96% | 70.19% | 70.70% | 94.43% | |
| F | Positive predictive value | 12.99% | 0.00% | 13.73% | 2.50% | - | 2.04% |
| Sensitivity | 40.72% | 0.00% | 93.81% | 19.69% | - | 0.26% | |
| F1-score | 19.70% | 0.00% | 23.96% | 4.43% | - | 0.46% | |
| Average | Positive predictive value | 56.51% | 54.77% | 60.36% | 53.46% | 52.60% | 70.75% |
| Sensitivity | 63.53% | 49.69% | 86.82% | 62.79% | 60.83% | 67.47% | |
| F1-score | 58.50% | 51.91% | 64.02% | 55.96% | 55.95% | 68.76% | |
| Predicted Label | ||||||
|---|---|---|---|---|---|---|
| N | SVEB | VEB | F | Total | ||
| True label | N | 43,962 | 147 | 79 | 30 | 44,218 |
| SVEB | 329 | 1369 | 123 | 15 | 1836 | |
| VEB | 124 | 13 | 3079 | 3 | 3219 | |
| F | 366 | 0 | 21 | 1 | 388 | |
| Total | 44,781 | 1529 | 3302 | 49 | 49,661 | |
| Mother Wavelet | Positive Predictive Value | Sensitivity | F1-Score | Accuracy |
|---|---|---|---|---|
| Mexican hat wavelet (mexh) | 70.75% | 67.47% | 68.76% | 98.74% |
| Morlet wavelet (morl) | 61.68% | 67.13% | 63.54% | 97.65% |
| Gaussian wavelet (gaus8) | 67.23% | 66.97% | 65.63% | 98.14% |
| Gaussian wavelet (gaus4) | 65.33% | 68.18% | 66.56% | 98.30% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Lu, C.; Sun, Y.; Yang, M.; Liu, C.; Ou, C. Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network. Entropy 2021, 23, 119. https://doi.org/10.3390/e23010119
Wang T, Lu C, Sun Y, Yang M, Liu C, Ou C. Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network. Entropy. 2021; 23(1):119. https://doi.org/10.3390/e23010119
Chicago/Turabian StyleWang, Tao, Changhua Lu, Yining Sun, Mei Yang, Chun Liu, and Chunsheng Ou. 2021. "Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network" Entropy 23, no. 1: 119. https://doi.org/10.3390/e23010119
APA StyleWang, T., Lu, C., Sun, Y., Yang, M., Liu, C., & Ou, C. (2021). Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network. Entropy, 23(1), 119. https://doi.org/10.3390/e23010119






