Deep Learning and Artificial Intelligence for the Determination of the Cervical Vertebra Maturation Degree from Lateral Radiography
Abstract
1. Introduction and the Organisation of The Paper
2. Importance of the Work and Its Interest for the Orthodontics Community
3. The Classical Radiographic Manual Methods
3.1. Hand-Wrist Radiograph Method HWM:
3.2. Vertebrae Maturation CVM:
- Cervical stage 1 (CS1) = 2 years before mandibular growth peak:Lower borders of C2 to C4 vertebrae are flat. C3 and C4 superior borders are tapered from posterior to anterior.
- Cervical stage 2 (CS2) = 1 year before mandibular growth peak:Lower border of C2 presents a concavity. Bodies of C3 and C4 are the same.
- Cervical stage 3 (CS3) = during the year of the mandibular growth peak:Lower borders of C2 and C3 present concavities. Vertebrae are growing so C3 and C4 may be either trapezoid or rectangular shape, as superior borders are less and less tapered.
- Cervical stage 4 (CS4) = 1 or 2 years after mandibular growth peak:Lower borders of C2, C3 and C4 present concavities. Both C3 and C4 bodies are rectangular with horizontal superior borders longer than higher.
- Cervical stage 5 (CS5) = 1 year after the end of mandibular growth peak:Still concavities of lower borders of C2, C3 and C4. At least one of C3 or C4 bodies are squared and spaces between bodies are reduced.
- Cervical stage 6 (CS6) = 2 years after the end of mandibular growth peak:The concavities of lower borders of C2 to C4 have deepened. C3 and C4 bodies are both square or rectangular vertical in shape (bodies higher than wide)
Cephalometric Appraisals:
- The concavity depth of the lower vertebral border (estimated by the distance of the middle point (Cm) from the line connecting posterior to anterior points (Clp-Cla))
- The tapering of upper border of vertebral C3 and C4 bodies (estimated by the ratio between posterior and anterior bodies heights (Cup-Clp)/(Cua-Cla))
- The lengthening of vertebral bodies (estimated by the ratio between the bases length and anterior bodies borders height (Clp-Cla)/Cua-Cla)
3.3. The Difficulties of the Labeling Task
3.4. The Need for Automatisation and the Help Which It Brings
4. Proposed Method
- Appropriate needed preprocessing of the images before starting the training, evaluation and testing steps;
- A Comparison of different appropriate CNN and DL existing methods on a reduced set of images in our data base;
- Appropriate choice of a DL CNN structure, criteria, optimization algorithm and setting of all the parameters and hyper parameters for our application;
- Showing the performances of the proposed method for a different number of training, evaluating and testing images;
- Pre-clinical evaluation of the implemented method.
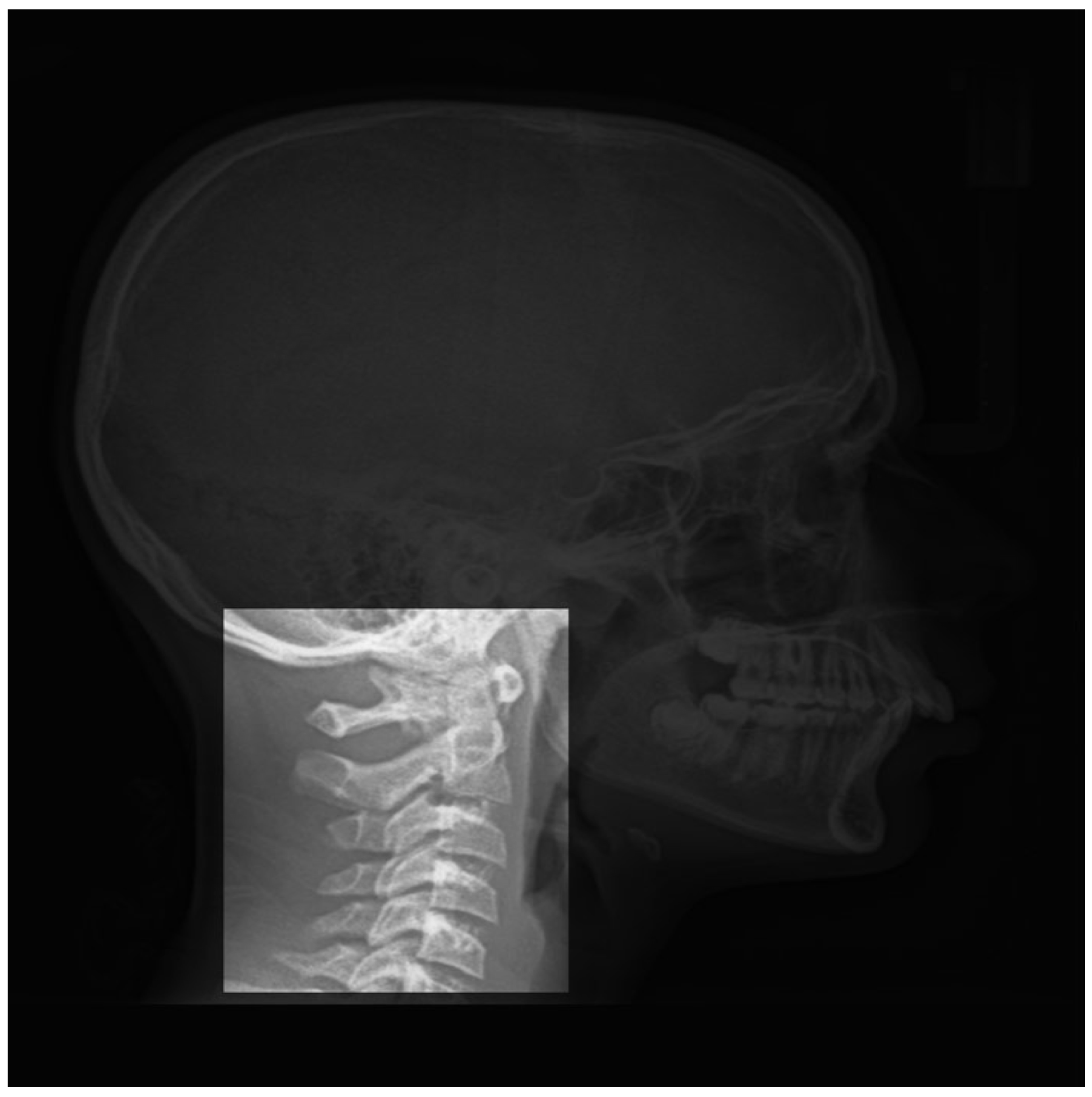
4.1. Preprocessing of the Data
4.2. Considered Deep Learning Networks
- Resnet:Resnet was introduced in the paper “Deep Residual Learning for Image Recognition ” [49]. There are several variants with different output sizes, including Resnet18, Resnet34, Resnet50, Resnet101, and Resnet152, all of which are available from torchvision models. As our dataset is small, we used Resnet18 that we adapted in our case for 6 classes.
- Alexnet:Alexnet was introduced in the paper “ImageNet Classification with Deep Convolutional Neural Networks” [51] and was the first very successful CNN on the ImageNet dataset.
- VGG:VGG was introduced in the paper “Very Deep Convolutional Networks for Large-Scale Image Recognition” [52]. Torchvision offers eight versions of VGG with various lengths and some that have batch normalizations layers.
- Squeezenet:The Squeeznet architecture is described in the paper “SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and <0.5 MB model size” [53]. It uses a different output structure than the other models mentioned here. Torchvision has two versions of Squeezenet. We used version 1.0.
- Densenet:Densenet was introduced in the paper “Densely Connected Convolutional Networks” [50]. Torchvision has four variants of Densenet. Here we used Densenet-121 and modified the output layer, which is a linear layer with 1024 input features and 6 classes, for our case.
- Inception v3:Inception v3 was first described in “Rethinking the Inception Architecture for Computer Vision” [54]. This network is unique because it has two output layers when training. The second output is known as an auxiliary output and is contained in the AuxLogits part of the network. The primary output is a linear layer at the end of the network. Note that when testing, we only consider the primary output.
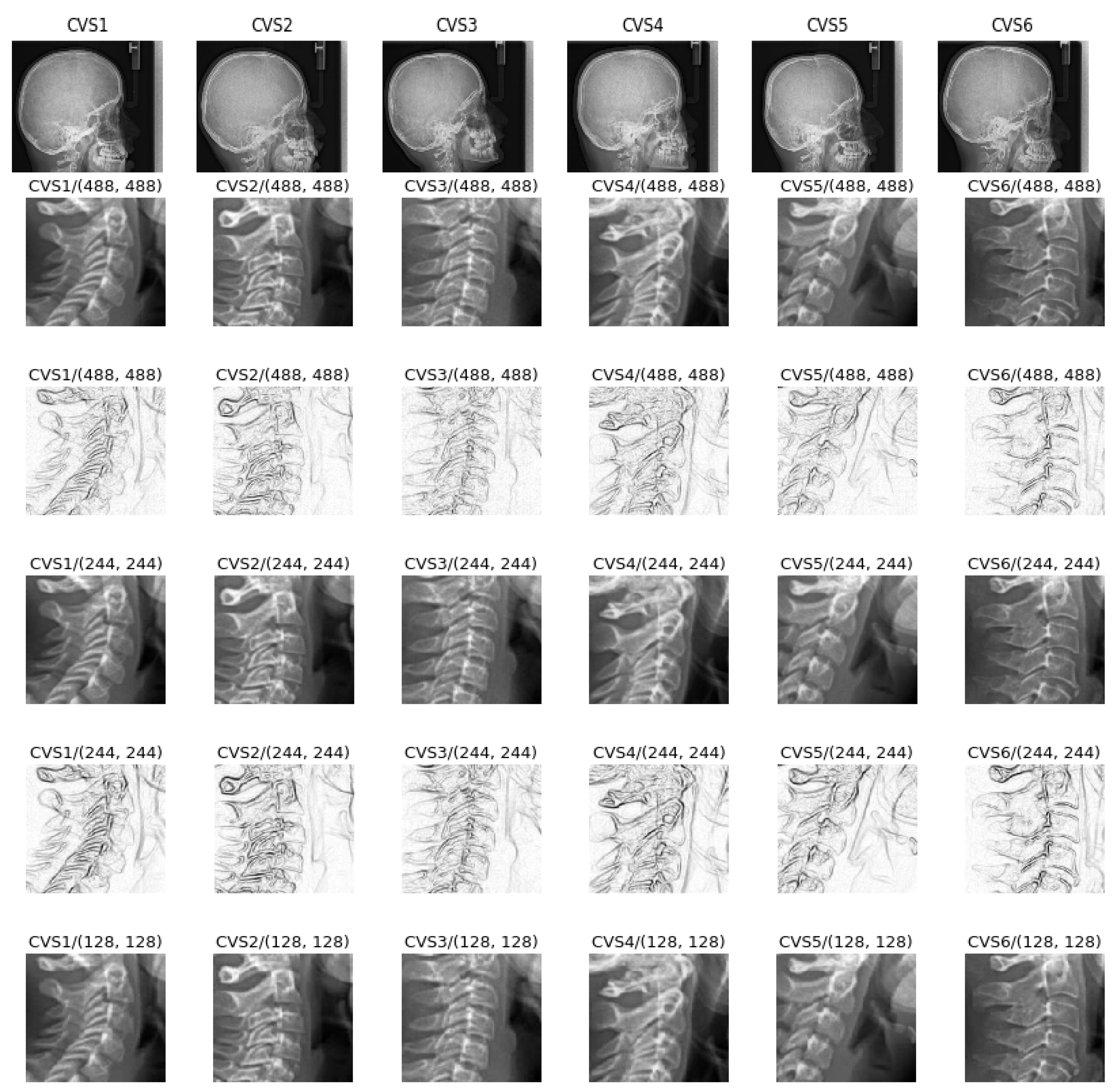
4.3. Comparison of a Simple Network on Different Preprocessed Images
4.4. Tools and Implementation
- SciKit-Image https://scikit-image.org/:All simple imag processing such as reading and writing images, Cropping, resizing and Filtering.
- SciKit-Learn https://scikit-learn.org/stable/: Data shuffling, Kmeans and Gaussian Mixture clustering, Principal Component Analysis and performance metrics.
- Keras wth tensorflow backend https://keras.io/:VGG16, VGG19 and ResNet50 convolution network models with ImageNet weights.
- PyTorch https://pytorch.org/:GPU and CPU vision algorithms, optimization, scheduler, feature transfer, etc.
- For a given Training data set, Create Model (keras.layers);
- Configure Model (model.compile);
- Train model (model.fit);
- For given Evaluating and Testing data, evaluate the trained model (loss = model.evaluate);
- Get prediction (pred = model.predict).
4.5. Prediction Results with Different Networks
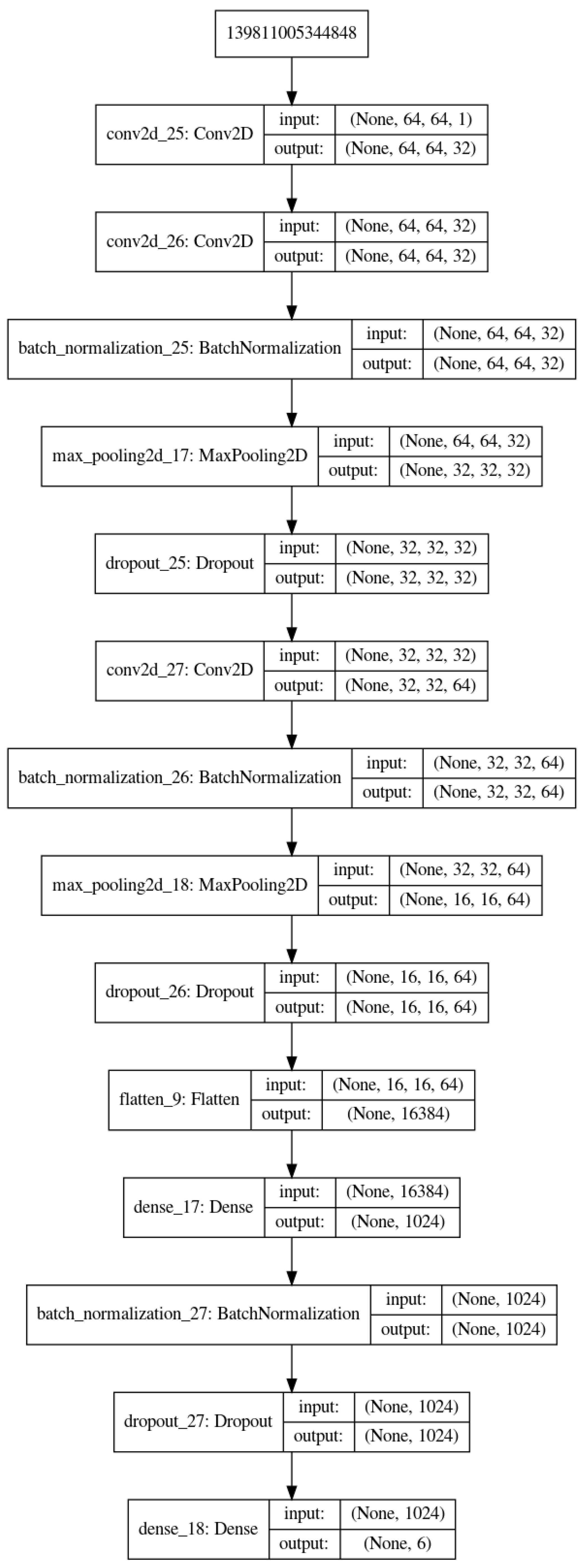
5. Proposed Method, Structure and Metrics
- Almost all the pre-trained networks with feature transfer did not really work better. This is due to the fact that these networks are trained with great databases with photographic images which are more diverse than the radiographic images we have.
- A cropping of images of the size 512 × 512 to the specific informative part of cervical vertebra and possibly resizing them to 256 × 256 for reducing the computational costs are enough. However, to improve more, we tried different pre-processing of the images: mean, median and entropic filter to the images.
- The proposed method is the same as in Figure 3 with slightly different parameters which are optimized for our application.
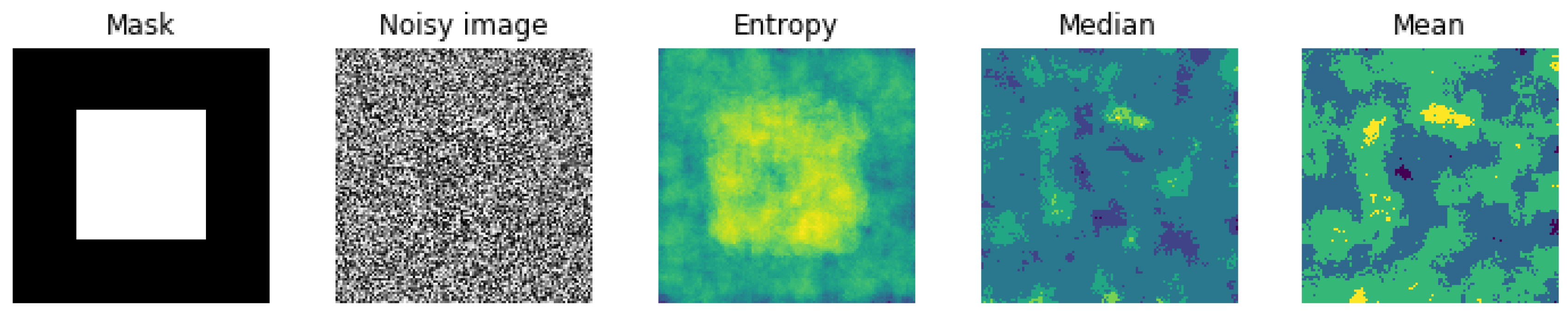
5.1. Entropic Filtering
5.2. Optimisation Criteria
5.3. Optimisation Algorithms
5.4. Metrics Used to Measure the Quality of the Classification
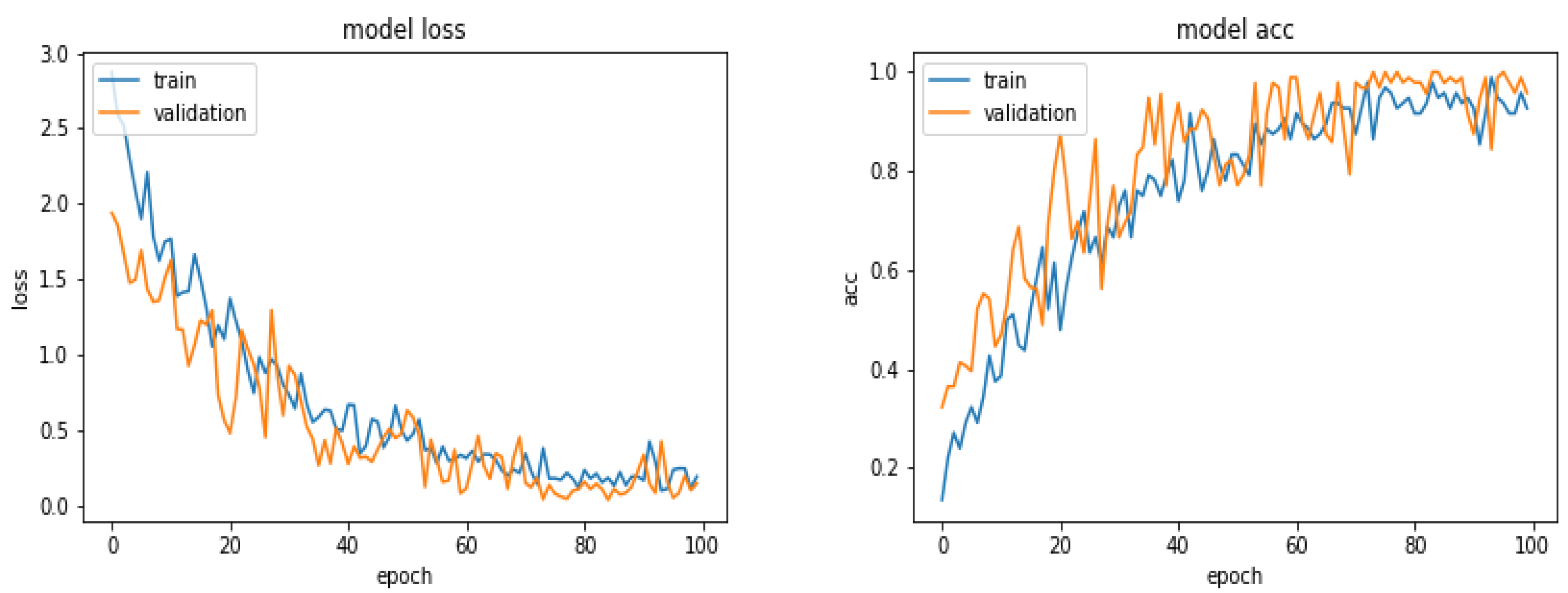
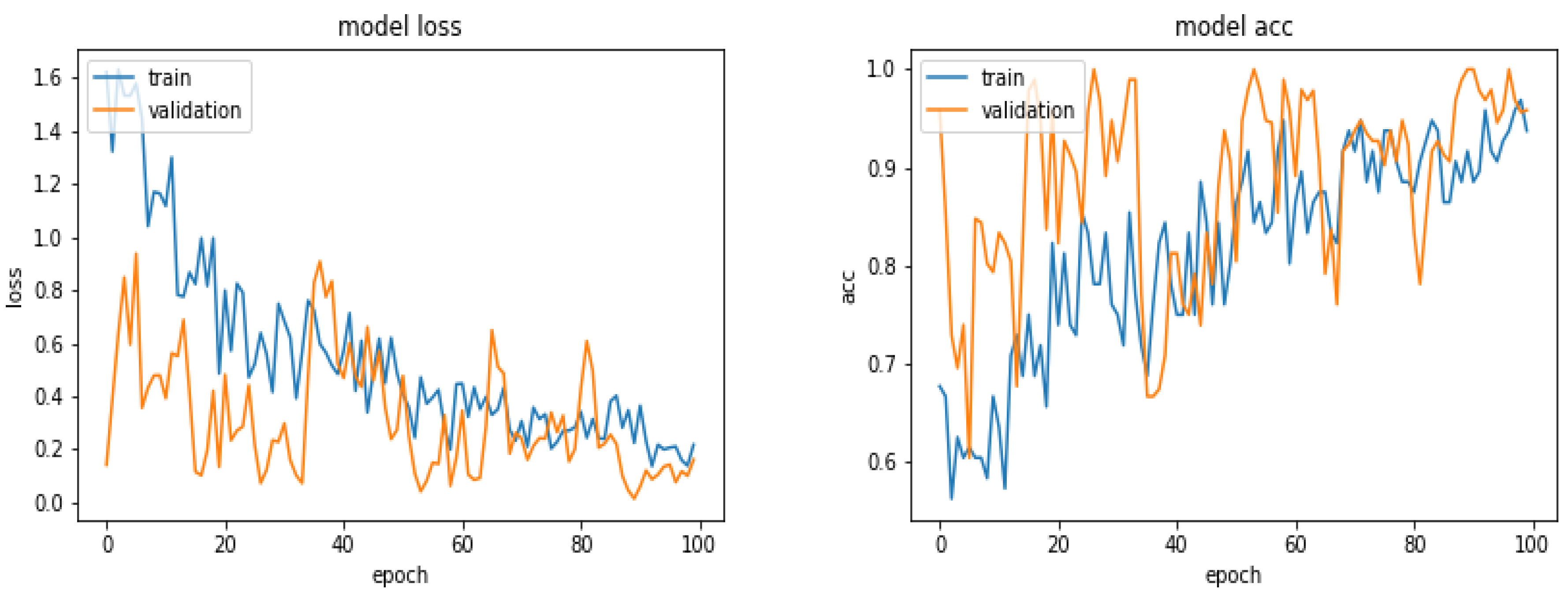
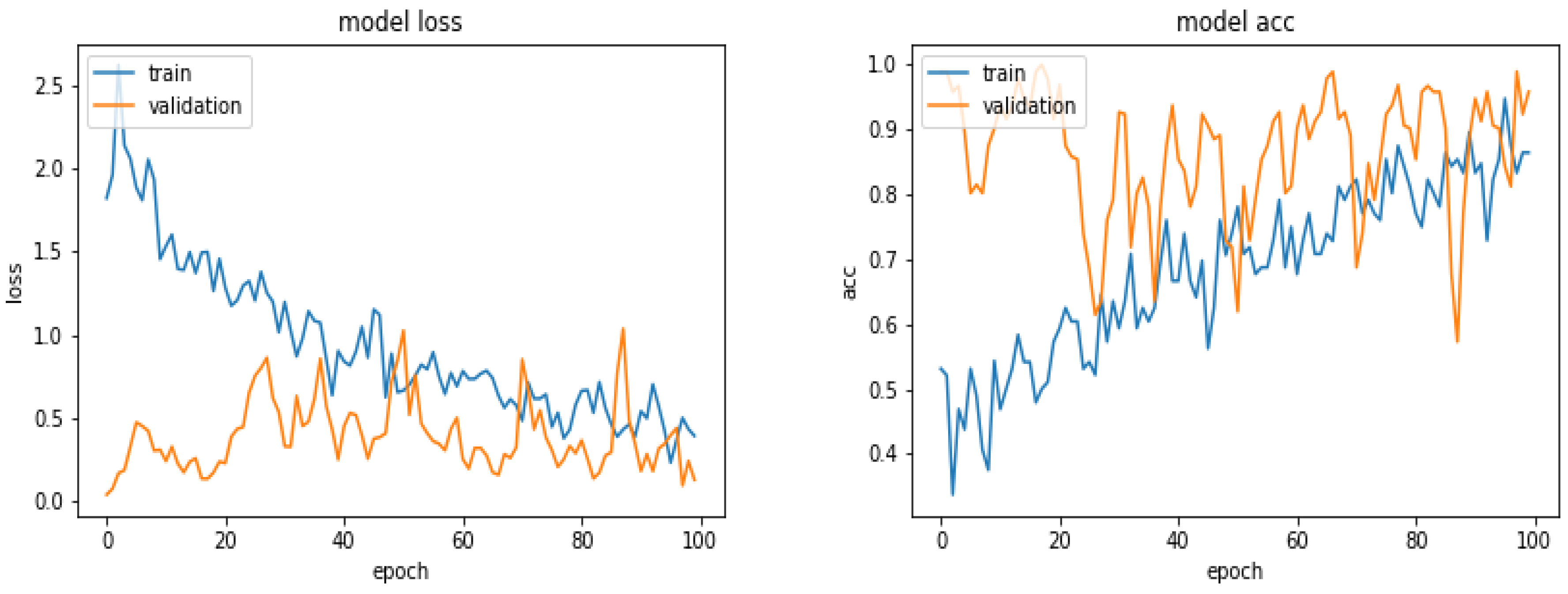
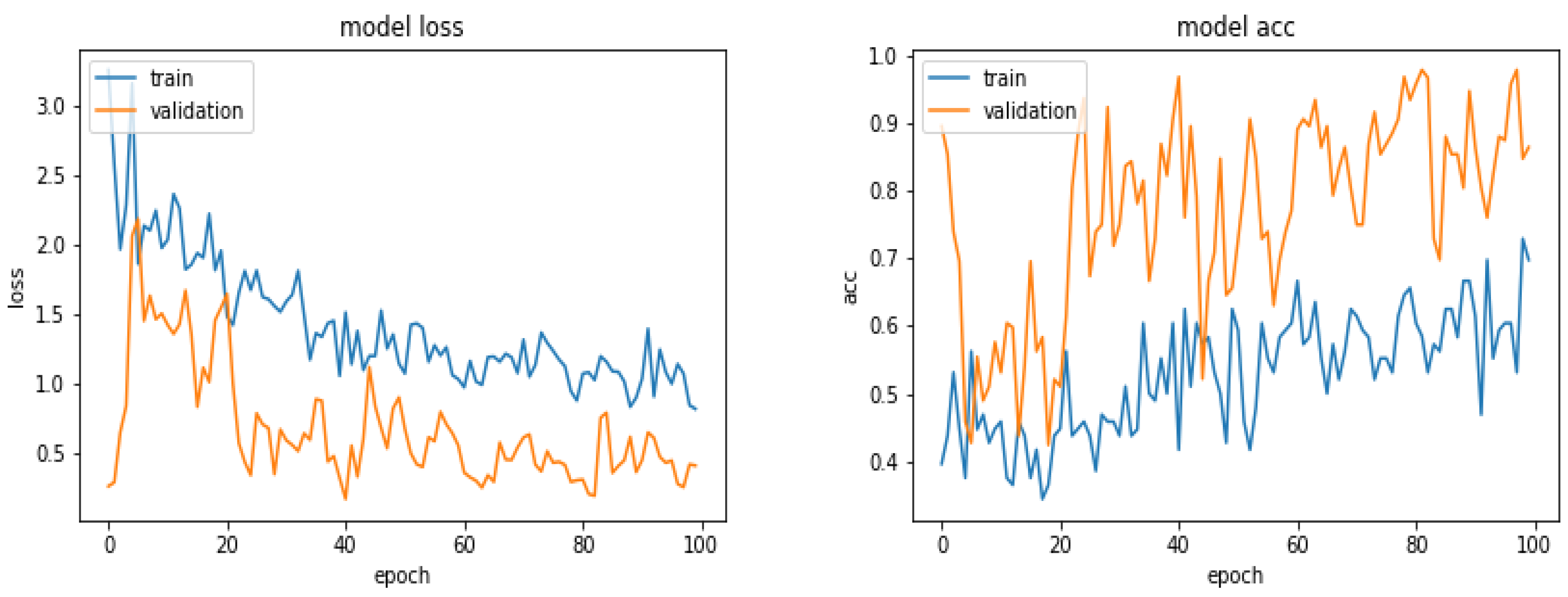
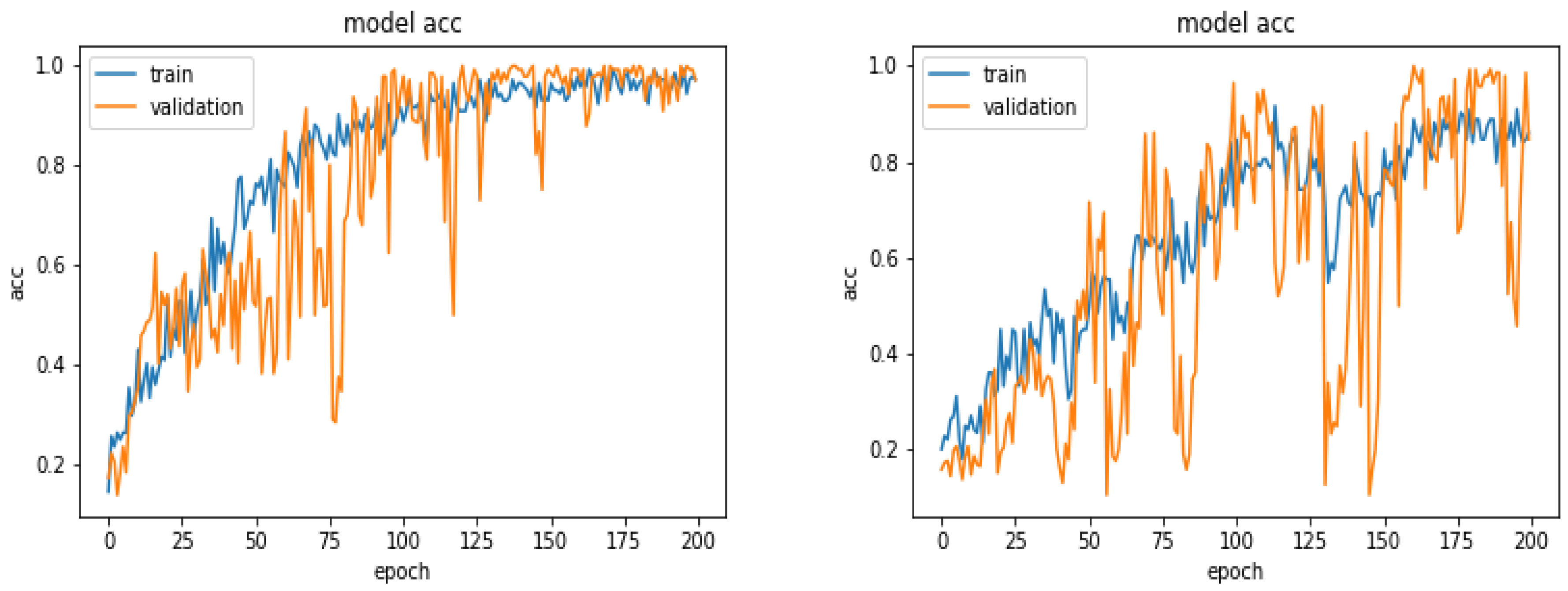
5.5. Results
- set a lower and an upper bounds for LR. For example: (1 × 10:1 × 10);
- start the training of the network;
- increase the LR in an exponential way after each batch update;
- for each LR, record the loss at each batch update;
- train for a few epoches and plot the loss as a function of the LR;
- examine the plot and identify the optimal LR;
- update the LR and train the network for the full set of training data.
- apply the trained network to the evaluation and test data and look for a possibly over fitting problem;
- If all are right, then between those images in the evaluation and test set data, move those which are classified correctly with a very high probability to the training set and update the evaluation and test sets with new images until the end of the data base.
5.6. Results with 360 Images
5.7. Results with 600 Images
5.8. Results with 900 Images
5.9. Results with 1870 Images
5.10. Comparison of Accuracy for Different Number of Training Images
5.11. Comparison of the Results for Different Optimization Algorithms
5.12. Comparison of the Results for Different Preprocessing
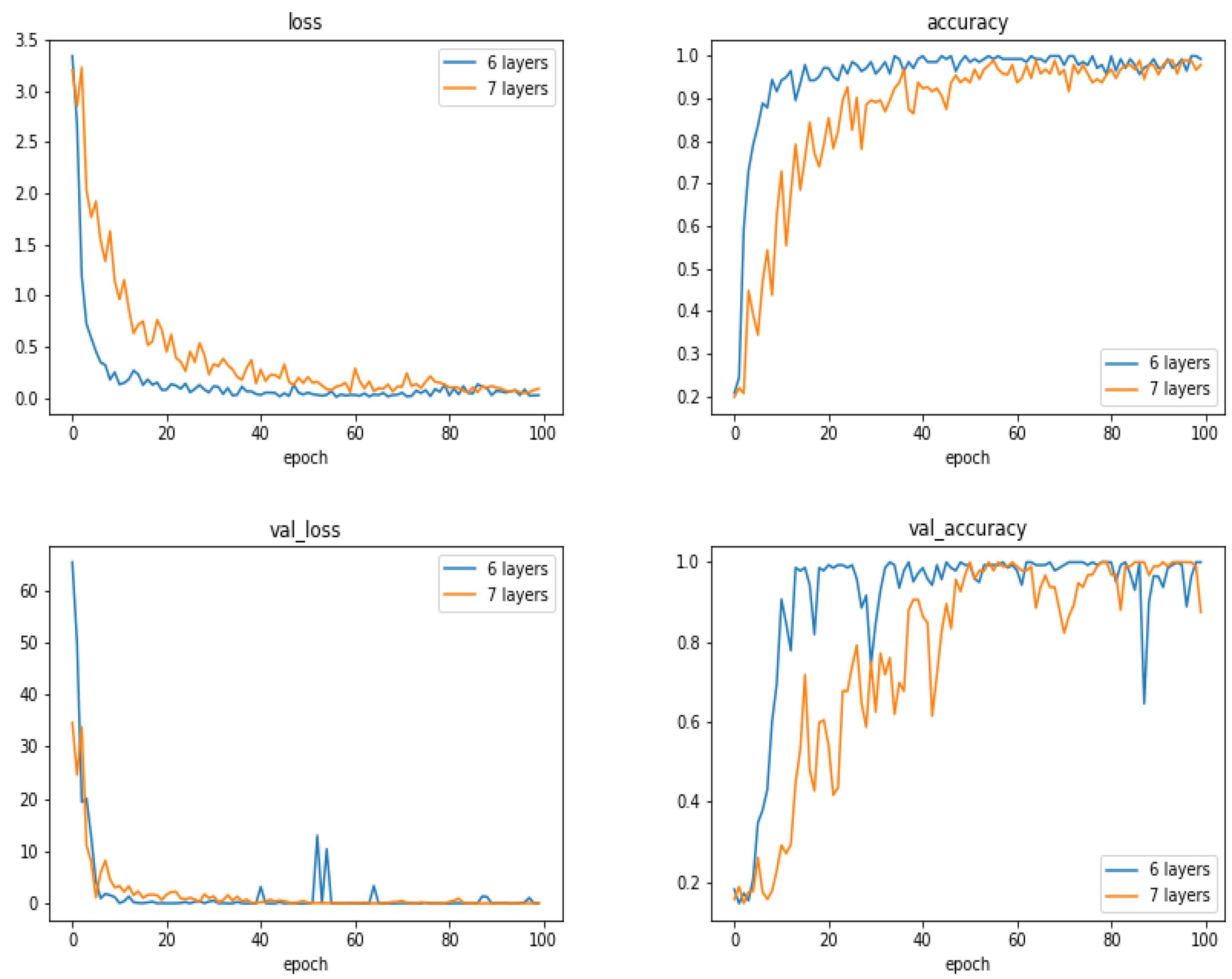
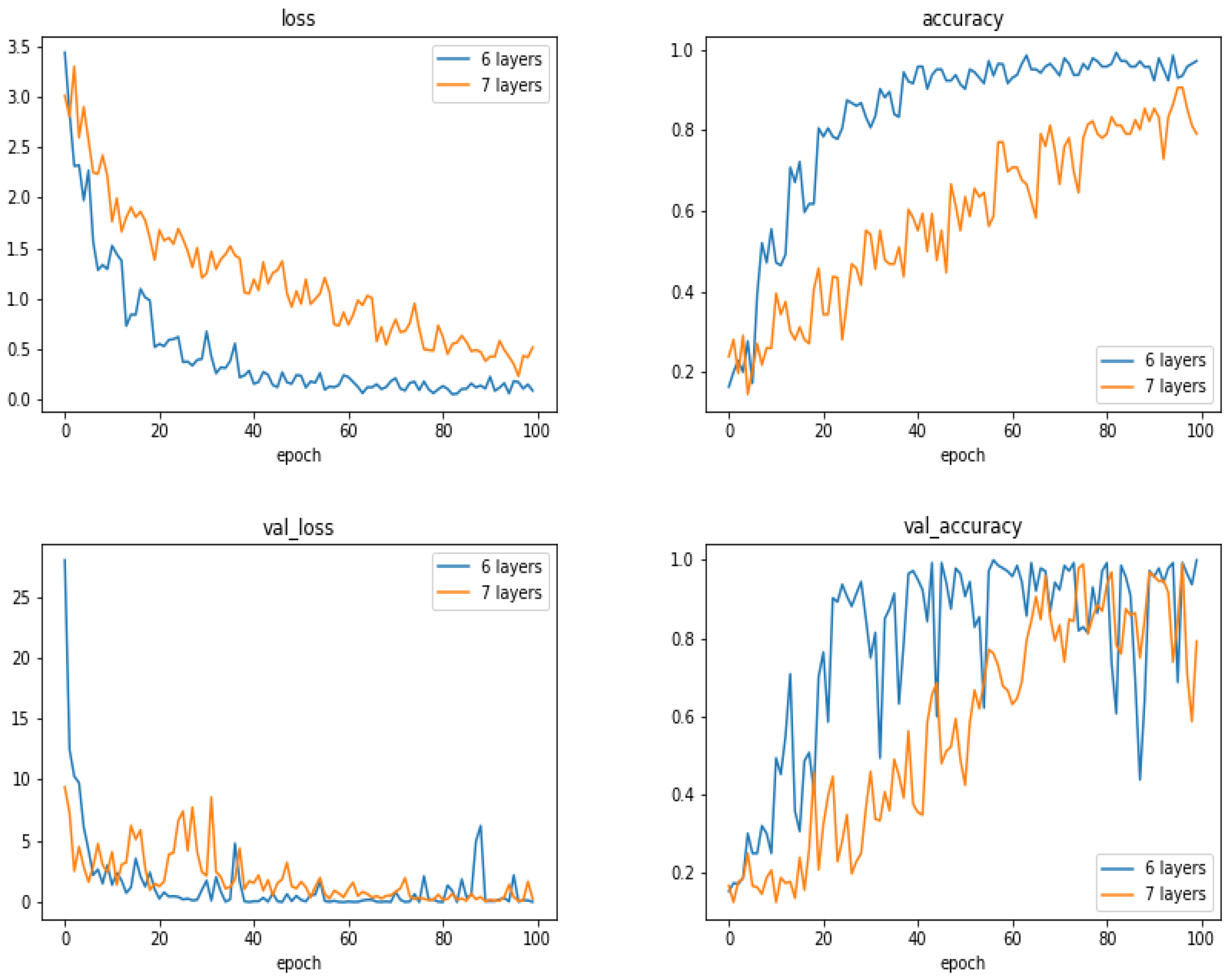
5.13. Comparison of the Results as a Function of the Number of Layes
5.14. Discussions
- Many existing CNN and DL structures have been developed particularly for photography images. They could not be used as they are for our X-ray radiography images. First, in general, they are constructed for colored images while the X-ray images are gray level. Using the pre-trained models cannot be used directly and even trying to fine tune them was not effective. Trying to train them directly was not successful, as in general, they need a very large number of images.
- We proposed a simple six-layer structure with five combination of convolution, normalization, max-pooling and dropout layers and one combination of dense, normalization and dropout layers. This structure was the most successful for our case.
- The choices of pre-processing, optimization criteria, optimization algorithms and optimization of hyper-parameters are very difficult. These need experience and deep knowledge of mathematics and techniques.
- We used categorical entropy as the optimization criteria, SGD or ADAM for optimization algorithm, and manually fixing the hyper-parameters using cross validation.
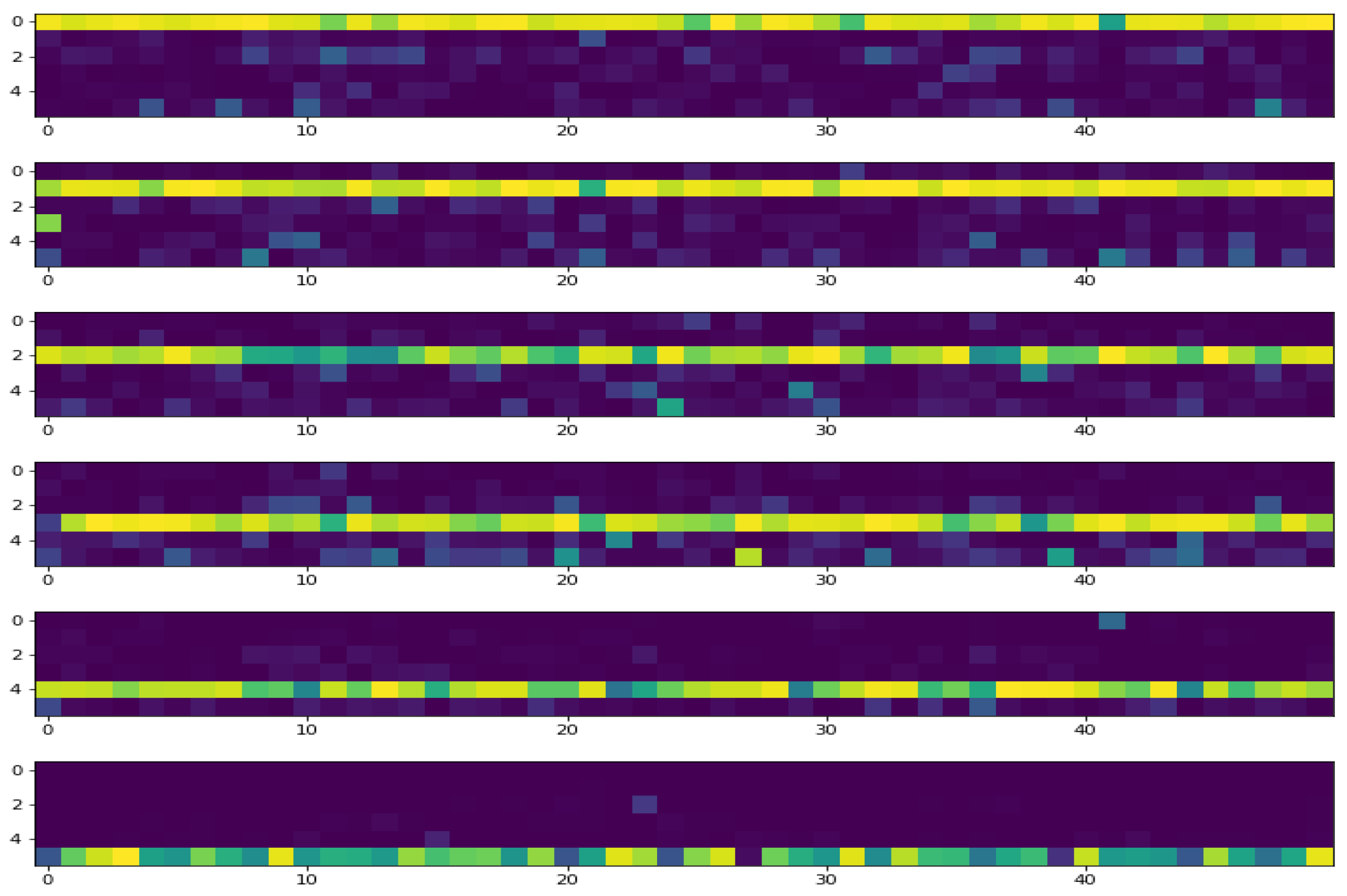
- Many performance criteria such as general accuracy, categorical accuracy, precision, recall and many other criteria, all can be derived from the confusion matrix and can be used to measure the performances of a classification method. In this paper, we mainly showed the accuracy.
- In our case, we show that when the training data are balanced, in general, the performances improve when the number of images increases. However, when the dataset is unbalanced, the situation is different.
- We did many experiments on the importance of the pre-processing of the images. Some pre-processings such as cropping on the interesting part of the image is almost always necessary, but other linear processing methods such as mean filtering or resizing did not have great impact. However, we discovered that an entropic filtering has improved the classification task.
- With small and moderate data, increasing the number layers does not improve the performances, and even, decrease them. A kind of balance between the number of parameters and the number of data has to be found. In our case, an approach of six compound layes (Convolution, Normalization, MaxPooling and DropOut) was the right one. When increased even one more layer, the performances decreased.
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baccetti, T.; Franchi, L.; McNamara, J.A. The Cervical Vertebral Maturation (CVM) Method for the Assessment of Optimal Treatment Timing in Dentofacial Orthopedics. Semin. Orthod. 2005, 11, 119–129. [Google Scholar] [CrossRef]
- Patcas, R.; Herzog, G.; Peltomäki, T.; Markic, G. New perspectives on the relationship between mandibular and statural growth. Eur. J. Orthod. 2016, 38, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.N.; Moyer, B.A.; DuBois, L.M. Skeletal maturation and craniofaciat growth. Am. J. Orthod. Dentofac. Orthod. 1990, 98, 33–40. [Google Scholar] [CrossRef]
- Hunter, C.J. The corelation of facial growth with body height and skeletal maturation at adolescence.pdf. Angle Orthod. 1966, 36, 44–54. [Google Scholar]
- Raberin, M.; Cozor, I.; Gobert-Jacquart, S. Les vertèbres cervicales: Indicateurs du dynamisme de la croissance mandibulaire? Orthod. Fr. 2012, 83, 45–58. [Google Scholar] [CrossRef]
- Franchi, L.; Baccetti, T.; McNamara, J.A. Mandibular growth as related to cervical vertebral maturation and body height. Am. J. Orthod. Dentofac. Orthop. 2000, 118, 335–340. [Google Scholar] [CrossRef]
- De Stefani, A.; Bruno, G.; Siviero, L.; Crivellin, G.; Mazzoleni, S.; Gracco, A. Évaluation radiologique de l’âge osseux avec la maturation de la phalange médiane du doigt majeur chez un patient orthodontique péripubertaire. Int Orthod. 2018, 16, 499–513. [Google Scholar] [CrossRef]
- Krisztina, M.I.; Ogodescu, A.; Réka, G.; Zsuzsa, B. Evaluation of the Skeletal Maturation Using Lower First Premolar Mineralisation. Acta Med. Marisiensis 2013, 59, 289–292. [Google Scholar] [CrossRef]
- Pyle, S.I.; Waterhouse, A.M.; Greulich, W.W. Attributes of the radiographic standard of reference for the National Health Examination Survey. Am. J. Phys. Anthropol. 1971, 35, 331–337. [Google Scholar] [CrossRef]
- Loder, R.T. Applicability of the Greulich and Pyle Skeletal Age Standards to Black and White Children of Today. Arch. Pediatr. Adolesc. Med. 1993, 147, 1329. [Google Scholar] [CrossRef]
- Bunch, P.M.; Altes, T.A.; McIlhenny, J.; Patrie, J.; Gaskin, C.M. Skeletal development of the hand and wrist: Digital bone age companion—A suitable alternative to the Greulich and Pyle atlas for bone age assessment? Skelet. Radiol. 2017, 46, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Padmanabhan, S.; Chitharanjan, A.B. Constancy of cervical vertebral maturation indicator in adults: A cross-sectional study. Int. Orthod. 2018, 16, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.J.; Bogowicz, P.; Heo, G.; Lagravère, M.O. Interrelationship and limitations of conventional radiographic assessments of skeletal maturation. Int. Orthod. 2012, 10, 135–147. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, M.T.; Yanniello, G.J. O’REILLY Mandibular growth changes and maturation of cervical vertebrae.pdf. Angle Orthod. 1988, 58, 179–184. [Google Scholar] [PubMed]
- Hassel, B.; Farman, A.G. Skeletal maturation evaluation using cervical vertebrae. Am. J. Orthod. Dentofac. Orthop. 1995, 107, 58–66. [Google Scholar] [CrossRef]
- Elhaddaoui, R.; Benyahia, H.; Azaroual, F.; Zaoui, F. Intérêt de la méthode de maturation des vertèbres cervicales (CVM) en orthopédie dento-faciale: Mise au point. Rev. Stomatol. Chir Maxillo-Faciale Chir Orale 2014, 115, 293–300. [Google Scholar] [CrossRef]
- Jaqueira, L.M.F.; Armond, M.C.; Pereira, L.J.; de Alcântara, C.E.P.; Marques, L.S. Determining skeletal maturation stage using cervical vertebrae: Evaluation of three diagnostic methods. Braz. Oral Res. 2010, 24, 433–437. [Google Scholar] [CrossRef]
- Uysal, T.; Ramoglu, S.I.; Basciftci, F.A.; Sari, Z. Chronologic age and skeletal maturation of the cervical vertebrae and hand-wrist: Is there a relationship? Am. J. Orthod. Dentofac. Orthop. 2006, 130, 622–628. [Google Scholar] [CrossRef]
- Hosni, S.; Burnside, G.; Watkinson, S.; Harrison, J.E. Comparison of statural height growth velocity at different cervical vertebral maturation stages. Am. J. Orthod. Dentofac. Orthop. 2018, 154, 545–553. [Google Scholar] [CrossRef]
- Perinetti, G.; Contardo, L.; Castaldo, A.; McNamara, J.A.; Franchi, L. Diagnostic reliability of the cervical vertebral maturation method and standing height in the identification of the mandibular growth spurt. Angle Orthod. 2016, 86, 599–609. [Google Scholar] [CrossRef]
- Mahajan, S. Evaluation of skeletal maturation by comparing the hand wrist radiograph and cervical vertebrae as seen in lateral cephalogram. Indian J. Dent. Res. 2011, 22, 309. [Google Scholar] [CrossRef] [PubMed]
- Sachan, K.; Tandon, P.; Sharma, V. A correlative study of dental age and skeletal maturation. Indian J. Dent. Res. 2011, 22, 882. [Google Scholar] [CrossRef] [PubMed]
- Danaei, S.M.; Karamifar, A.; Sardarian, A.; Shahidi, S.; Karamifar, H.; Alipour, A.; Ghodsi Boushehri, S. Measuring agreement between cervical vertebrae and hand-wrist maturation in determining skeletal age: Reassessing the theory in patients with short stature. Am. J. Orthod. Dentofac. Orthop. 2014, 146, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Ball, G.; Woodside, D.; Tompson, B.; Hunter, W.S.; Posluns, J. Relationship between cervical vertebral maturation and mandibular growth. Am. J. Orthod. Dentofac. Orthop. 2011, 139, e455–e461. [Google Scholar] [CrossRef]
- Ballrick, J.W.; Fields, H.W.; Beck, F.M.; Sun, Z.; Germak, J. The cervical vertebrae staging method’s reliability in detecting pre and post mandibular growth. Orthod. Waves 2013, 72, 105–111. [Google Scholar] [CrossRef]
- Perinetti, G.; Caprioglio, A.; Contardo, L. Visual assessment of the cervical vertebral maturation stages: A study of diagnostic accuracy and repeatability. Angle Orthod. 2014, 84, 951–956. [Google Scholar] [CrossRef]
- McNamara, J.A.; Franchi, L. The cervical vertebral maturation method: « A user’s guide ». Angle Orthod. 2018, 88, 133–143. [Google Scholar] [CrossRef]
- Santiago, R.C.; Cunha, A.R.; Júnior, G.C.; Fernandes, N.; Campos, M.J.S.; Costa, L.F.; Vitral, R.W.; Bolognese, A.M. New software for cervical vertebral geometry assessment and its relationship to skeletal maturation—A pilot study. Dentomaxillofac. Radiol. 2014, 43, 20130238. [Google Scholar] [CrossRef]
- Padalino, S.; Sfondrini, M.F.; Chenuil, L.; Scudeller, L.; Gandini, P. Fiabilité de l’analyse de la maturité squelettique selon la méthode CVM des vertèbres cervicales faite par un logiciel dédié. Int. Orthod. 2014, 12, 483–493. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst. 2012, 1097–1105. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.J.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3d cnn with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Kathirvel, C.T.R. Classifying Diabetic Retinopathy using Deep Learn-ing Architecture. Int. J. Eng. Res.-Tech-Nol. 2016, 5, 19–24. [Google Scholar] [CrossRef]
- Lessmann, N.; Isgum, I.; Setio, A.; de Vos, B.D.; Ciompi, F.; de Jong, P.A.; Oudkerk, M.; Willem, P.T.M.; Viergever, M.A.; van Ginneken, B. Deep convolutional neural networks for automatic coronary calcium scoring in a screening study with low-dose chest CT. In SPIE Medical Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2016; p. 978511. [Google Scholar]
- Alansary, A.; Oktay, O.; Li, Y.; Le Folgoc, L.; Hou, B.; Vaillant, G.; Kamnitsas, K.; Vlontzos, A.; Glocker, B.; Kainz, B.; et al. Evaluating reinforcement learning agents for anatomical landmarkdetection. Med. Image Anal. 2019, 53, 156–164. [Google Scholar] [CrossRef]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learningin medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. Synthetic data augmentation using GAN for improved liverlesion classification. In Proceedings of the 2018 IEEE 15th International Symposiumon Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 289–293. [Google Scholar]
- Gao, F.; Wu, T.; Li, J.; Zheng, B.; Ruan, L.; Shang, D.; Patel, B. SD-CNN: A shallow-Deep CNN for Improved Breast Cancer diagnosis. Comput. Med. Imaging Graph. 2018, 70, 53–62. [Google Scholar]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Richter, C.; Cha, K. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys. Med. Biol. 2018, 63, 095005. [Google Scholar] [CrossRef]
- Al, W.A.; Yun, I.D. Reinforcing Medical Image Classifier to Improve Generalization on Small Datasets. Available online: https://arxiv.org/abs/1909.05630v2 (accessed on 15 October 2019).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. Available online: https://arxiv.org/abs/1512.03385 (accessed on 10 October 2019).
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. Available online: https://arxiv.org/abs/1608.06993 (accessed on 10 October 2019).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Available online: https://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf (accessed on 10 October 2019).
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. Available online: https://arxiv.org/pdf/1409.1556.pdf (accessed on 10 October 2019).
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level Accuracy with 50× Fewer Parameters and <0.5 MB Model Size. Available online: https://arxiv. org/abs/1602.07360 (accessed on 10 October 2019).
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J. Rethinking the Inception Architecture for Computer Vision. Available online: https://arxiv.org/pdf/1512.00567v1.pdf (accessed on 10 October 2019).
- Ruder, S. An Overview of Gradient Descent Optimization Algorithms. Available online: http://ruder.io/optimizing-gradient-descent (accessed on 10 October 2019).
- Ruder, S. An Overview of Gradient Descent Optimization Algorithms. Available online: https://arxiv.org/abs/1609. 04747 (accessed on 10 October 2019).
- Special Issue “MaxEnt 2019—The 39th International Workshop on Bayesian Inference and Maximum Entropy Methods in Science and Engineering”, accessed on line 29 September 2019.


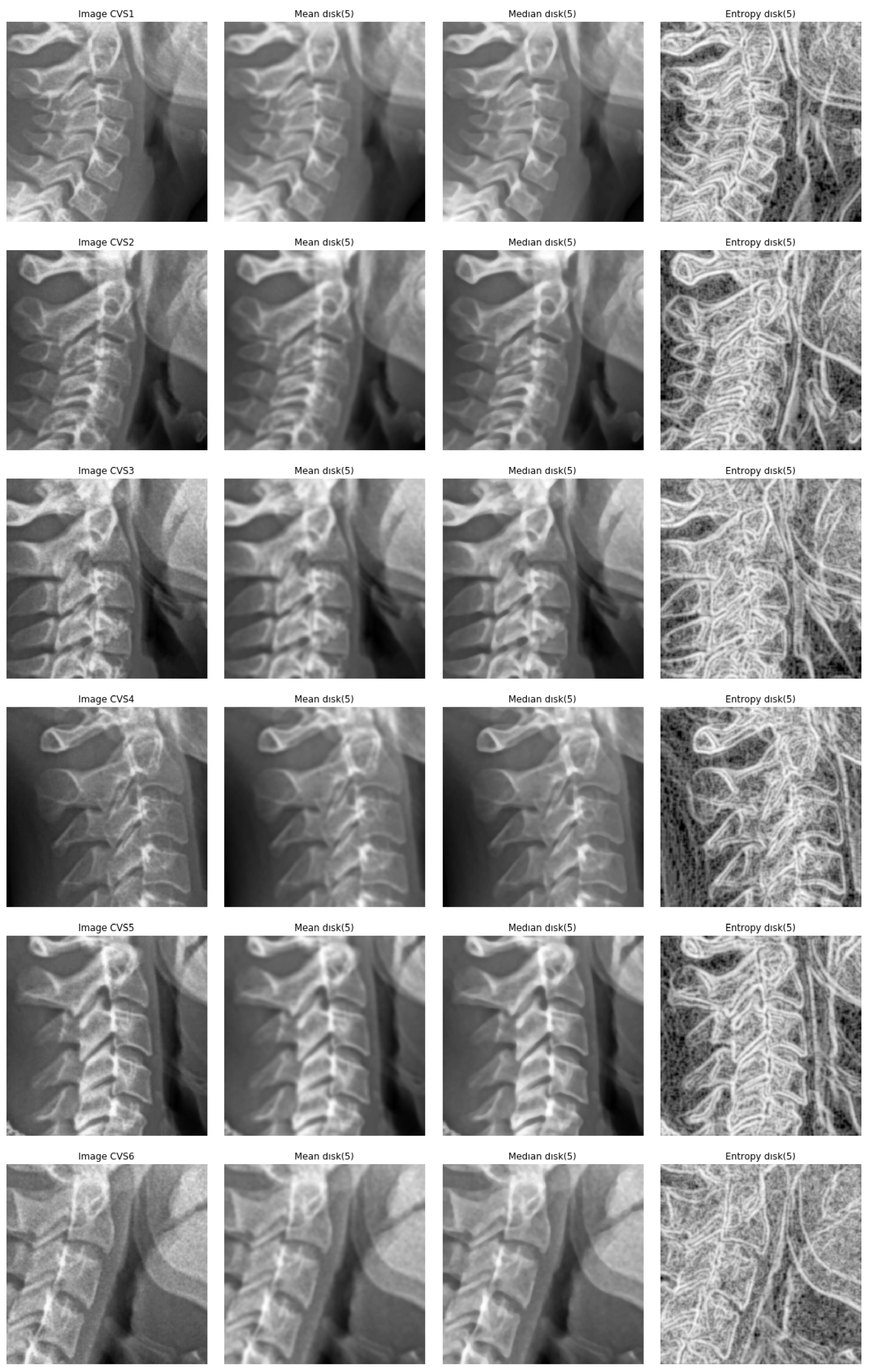

| Class | CVS1 | CVS2 | CVS3 | CVS4 | CVS5 | CVS6 |
|---|---|---|---|---|---|---|
| Test0 | 0.910 | 0.820 | 0.654 | 0.617 | 0.863 | 0.939 |
| Test1 | 0.890 | 0.734 | 0.661 | 0.580 | 0.849 | 0.924 |
| Test2 | 0.890 | 0.734 | 0.661 | 0.580 | 0.849 | 0.924 |
| Test3 | 0.932 | 0.824 | 0.644 | 0.561 | 0.839 | 0.849 |
| Test4 | 0.880 | 0.890 | 0.568 | 0.602 | 0.861 | 0.932 |
| No. of Images/Classes | CVS1 | CVS2 | CVS3 | CVS4 | CVS5 | CVS6 |
|---|---|---|---|---|---|---|
| 360 images | 0.917 | 0.977 | 0.970 | 0.980 | 0.870 | 0.850 |
| 600 images | 1.000 | 0.997 | 1.000 | 0.993 | 1.000 | 0.997 |
| 900 images | 0.980 | 0.890 | 0.993 | 0.963 | 0.933 | 0.927 |
| 1870 images | 1.000 | 0.997 | 0.987 | 0.987 | 0.987 | 0.967 |
| No. of Images / Classes | CVS1 | CVS2 | CVS3 | CVS4 | CVS5 | CVS6 |
|---|---|---|---|---|---|---|
| 360 images | 0.963 | 0.927 | 0.973 | 0.970 | 0.980 | 0.933 |
| 600 images | 1.000 | 1.000 | 1.000 | 0.997 | 1.000 | 0.997 |
| 900 images | 0.997 | 0.997 | 0.997 | 1.000 | 0.993 | 0.993 |
| 1870 images | 0.990 | 0.970 | 0.953 | 0.980 | 0.977 | 0.990 |
| No. of Images/Classes | CVS1 | CVS2 | CVS3 | CVS4 | CVS5 | CVS6 |
|---|---|---|---|---|---|---|
| 300 images: | ||||||
| No preprocessing | 1.000 | 0.983 | 0.987 | 0.997 | 0.923 | 0.990 |
| Entropy filter | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 600 images: | ||||||
| No preprocessing | 0.978 | 0.975 | 0.952 | 0.973 | 0.937 | 0.925 |
| Entropy filter | 0.995 | 0.997 | 0.990 | 0.997 | 0.995 | 0.995 |
| 900 images: | ||||||
| No preprocessing | 0.794 | 0.826 | 0.635 | 0.833 | 0.783 | 0.817 |
| Entropy filter | 0.984 | 0.984 | 0.988 | 0.996 | 0.993 | 0.996 |
| 1870 images: | ||||||
| No preprocessing | 0.879 | 0.880 | 0.651 | 0.833 | 0.885 | 0.911 |
| Entropy filter | 0.882 | 0.857 | 0.637 | 0.852 | 0.897 | 0.919 |
| No. of Layers/Classes | CVS1 | CVS2 | CVS3 | CVS4 | CVS5 | CVS6 |
|---|---|---|---|---|---|---|
| 300 images: | ||||||
| 6 layers: | 0.913 | 0.850 | 0.802 | 0.912 | 0.895 | 0.913 |
| 7 layers: | 0.892 | 0.805 | 0.890 | 0.793 | 0.885 | 0.897 |
| 900 images: | ||||||
| 6 layers: | 0.998 | 0.998 | 1.000 | 0.988 | 0.994 | 0.991 |
| 7 layers: | 0.930 | 0.939 | 0.952 | 0.924 | 0.966 | 0.969 |
| Case with 6 Layers | Case with 7 Layers | ||||||
|---|---|---|---|---|---|---|---|
| Confusion Matrix | Confusion Matrix | ||||||
| [[101 6 4 25 12 2] | [[101 6 4 25 12 2] | ||||||
| [ 0 110 3 30 5 2] | [ 0 110 3 30 5 2] | ||||||
| [ 0 0 122 22 6 0] | [ 0 0 122 22 6 0] | ||||||
| [ 0 0 0 148 2 0] | [ 0 0 0 148 2 0] | ||||||
| [ 1 1 1 8 139 0] | [ 1 1 1 8 139 0] | ||||||
| [ 0 0 0 16 2 132]] | [ 0 0 0 16 2 132]] | ||||||
| Classification Report | Classification Report | ||||||
| precision | recall | f1-score | support | precision | recall | f1-score | support |
| CVS1 0.99 | 0.67 | 0.80 | 150 | CVS1 0.99 | 0.67 | 0.80 | 150 |
| CVS2 0.94 | 0.73 | 0.82 | 150 | CVS2 0.94 | 0.73 | 0.82 | 150 |
| CVS3 0.94 | 0.81 | 0.87 | 150 | CVS3 0.94 | 0.81 | 0.87 | 150 |
| CVS4 0.59 | 0.99 | 0.74 | 150 | CVS4 0.59 | 0.99 | 0.74 | 150 |
| CVS5 0.84 | 0.93 | 0.88 | 150 | CVS5 0.84 | 0.93 | 0.88 | 150 |
| CVS6 0.97 | 0.88 | 0.92 | 150 | CVS6 0.97 | 0.88 | 0.92 | 150 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makaremi, M.; Lacaule, C.; Mohammad-Djafari, A. Deep Learning and Artificial Intelligence for the Determination of the Cervical Vertebra Maturation Degree from Lateral Radiography. Entropy 2019, 21, 1222. https://doi.org/10.3390/e21121222
Makaremi M, Lacaule C, Mohammad-Djafari A. Deep Learning and Artificial Intelligence for the Determination of the Cervical Vertebra Maturation Degree from Lateral Radiography. Entropy. 2019; 21(12):1222. https://doi.org/10.3390/e21121222
Chicago/Turabian StyleMakaremi, Masrour, Camille Lacaule, and Ali Mohammad-Djafari. 2019. "Deep Learning and Artificial Intelligence for the Determination of the Cervical Vertebra Maturation Degree from Lateral Radiography" Entropy 21, no. 12: 1222. https://doi.org/10.3390/e21121222
APA StyleMakaremi, M., Lacaule, C., & Mohammad-Djafari, A. (2019). Deep Learning and Artificial Intelligence for the Determination of the Cervical Vertebra Maturation Degree from Lateral Radiography. Entropy, 21(12), 1222. https://doi.org/10.3390/e21121222





