Heart Rate Dynamics in Patients with Obstructive Sleep Apnea: Heart Rate Variability and Entropy
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.1.1. Participants
2.1.2. PSG and Sleep Outcomes
2.2. Signal Preprocessing
2.3. Conventional TIME and Frequency Domain HRV Measures
2.4. Nonlinear Measures
2.4.1. Sample Entropy (SampEn)
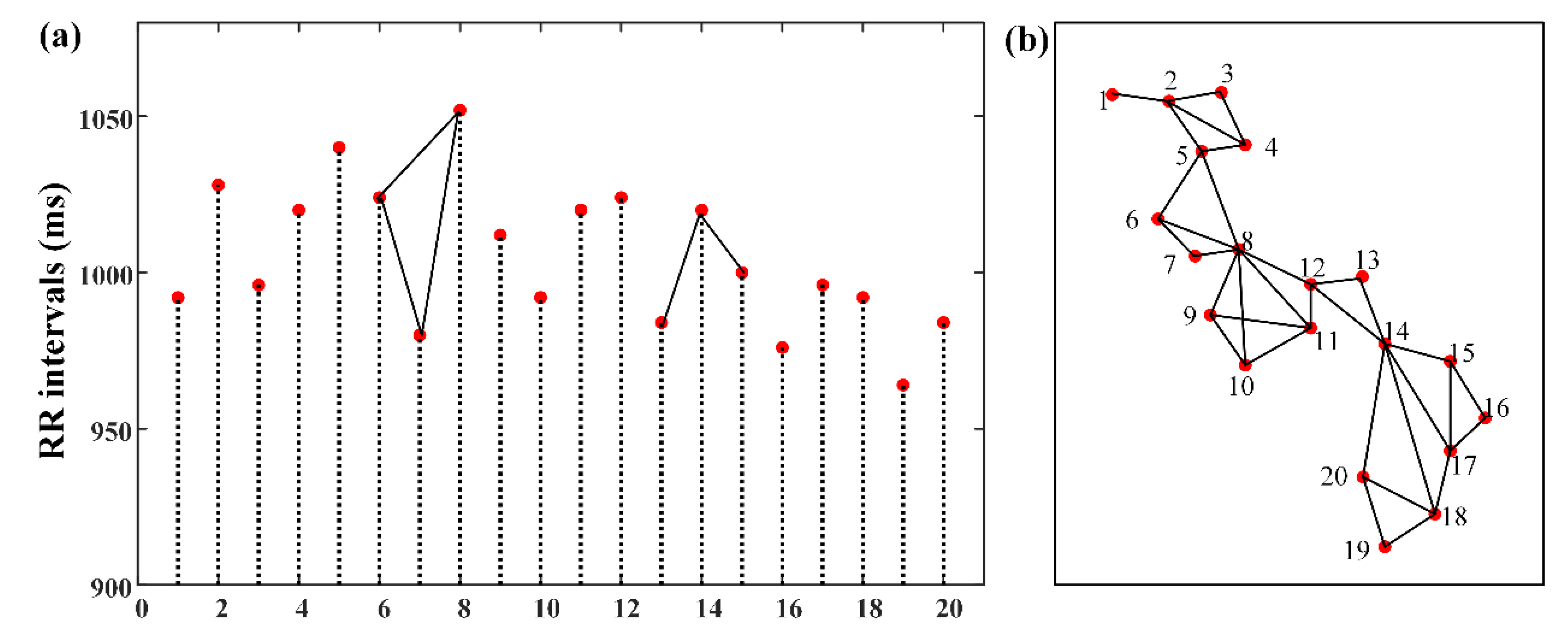
2.4.2. Shannon Entropy of the Degree Distribution (EDD)
2.5. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Corben, L.A.; Michael, H.; Janet, C.; Geneieve, T.; Delatycki, M.B. Increased prevalence of sleep-disordered breathing in Friedreich ataxia. Neurology 2013, 81, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.G.; Johnson, D.C. Frequency of sleep apnea in stroke and TIA patients: A meta-analysis. J. Clin. Sleep Med. 2010, 6, 131–137. [Google Scholar] [PubMed]
- Lettieri, C.J.; Eliasson, A.H.; Andrada, T.; Khramtsov, A.; Raphaelson, M.; Kristo, D.A. Obstructive sleep apnea syndrome: Are we missing an at-risk population? J. Clin. Sleep Med. 2005, 128, 381–385. [Google Scholar] [CrossRef]
- Sateia, M.J. International Classification of Sleep Disorders-Third Edition. Chest 2014, 146, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Acharya, U.R.; Ng, E.Y.K.; Fujita, H. A review of ecg-based diagnosis support systems for obstructive sleep apnea. J. Mech. Med. Biol. 2016, 16, 1640004. [Google Scholar] [CrossRef]
- Pumprla, J.; Howorka, K.; Groves, D.; Chester, M.; Nolan, J. Functional assessment of heart rate variability: Physiological basis and practical applications. Int. J. Cardiol. 2002, 84, 1–14. [Google Scholar] [CrossRef]
- Yan, M.; Tseng, P.H.; Ahn, A.; Wu, M.S.; Ho, Y.L.; Chen, M.F.; Peng, C.K. Cardiac Autonomic Alteration and Metabolic Syndrome: An Ambulatory ECG-based Study in A General Population. Sci. Rep. 2017, 7, 44363. [Google Scholar]
- Crespo, A.; Campo, F.D.; Gómez, J.; Álvarez, D.; Marcos, J.; Hornero, R. Nonlinear analysis of heart rate variability in patients with sleep apnea hypopnea syndrome (SAHS). A severity study. Sleep Med. 2013, 14, e262–e263. [Google Scholar] [CrossRef]
- Cysarz, D.; Bettermann, H.; Van, L.P. Entropies of short binary sequences in heart period dynamics. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2163–H2172. [Google Scholar] [CrossRef]
- Al-Angari, H.M.; Sahakian, A.V. Use of sample entropy approach to study heart rate variability in obstructive sleep apnea syndrome. IEEE Trans. Biomed. Eng. 2007, 54, 1900–1904. [Google Scholar] [CrossRef]
- Ravelo-García, A.; Navarro-Mesa, J.; Casanova-Blancas, U.; Martin-Gonzalez, S.; Quintana-Morales, P.; Guerra-Moreno, I.; Canino-Rodríguez, J.; Hernández-Pérez, E. Application of the Permutation Entropy over the Heart Rate Variability for the Improvement of Electrocardiogram-based Sleep Breathing Pause Detection. Entropy 2015, 17, 914–927. [Google Scholar] [CrossRef]
- Pan, W.Y.; Su, M.C.; Wu, H.T.; Su, T.J.; Lin, M.C.; Sun, C.K. Multiscale entropic assessment of autonomic dysfunction in patients with obstructive sleep apnea and therapeutic impact of continuous positive airway pressure treatment. Sleep Med. 2016, 20, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Lacasa, L.; Luque, B.; Ballesteros, F.; Luque, J.; Nuño, J.C. From time series to complex networks: The visibility graph. Proc. Natl. Acad. Sci. USA 2008, 105, 4972–4975. [Google Scholar] [CrossRef] [PubMed]
- Hou, F.Z.; Li, F.W.; Wang, J.; Yan, F.R. Visibility graph analysis of very short-term heart rate variability during sleep. Phys. A Stat. Mech. Appl. 2016, 458, 140–145. [Google Scholar] [CrossRef]
- Hou, F.Z.; Wang, J.; Wu, X.C.; Yan, F.R. A dynamic marker of very short-term heartbeat under pathological states via network analysis. EPL 2014, 107, 58001. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Cui, L.; Mueller, R.; Tao, S.; Redline, S. The National Sleep Research Resource: Towards a sleep data commons. J. Am. Med Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Redline, S.; Sanders, M.H.; Lind, B.K.; Quan, S.F.; Iber, C.; Gottlieb, D.J.; Bonekat, W.H.; Rapoport, D.M.; Smith, P.L.; Kiley, J.P. Methods for obtaining and analyzing unattended polysomnography data for a multicenter study. Sleep Heart Health Research Group. Sleep 1998, 21, 759–767. [Google Scholar]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.T.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The Sleep Heart Health Study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar]
- Chernick, M.R. Wavelet Methods for Time Series Analysis; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Percival, D.B.; Mofjeld, H.O. Analysis of subtidal coastal sea level fluctuations using wavelets. Publ. Am. Stat. Assoc. 1997, 92, 868–880. [Google Scholar] [CrossRef]
- Fornito, A.; Zalesky, A.; Breakspear, M. Graph analysis of the human connectome: Promise, progress, and pitfalls. Neuroimage 2013, 80, 426–444. [Google Scholar] [CrossRef]
- Marzbanrad, F.; Jelinek, H.; Ng, E.; Tamayo, M.; Hambly, B.; Mclachlan, C.; Matthews, S.; Palaniswami, M.; Khandoker, A. The effect of automated preprocessing of RR interval tachogram on discrimination capability of Heart Rate Variability parameters. In Proceedings of the 2013 Computing in Cardiology Conference, Zaragoza, Spain, 22–25 September 2013. [Google Scholar]
- Fleisher, L.A.; Fleckenstein, J.F.; Frank, S.M.; Thuluvath, P.J. Heart Rate Variability as a Predictor of Autonomic Dysfunction in Patients Awaiting Liver Transplantation. Dig. Dis. Sci. 2000, 45, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Galland, B.C.; Hayman, R.M.; Taylor, B.J.; Bolton, D.P.; Sayers, R.M.; Williams, S.M. Factors affecting heart rate variability and heart rate responses to tilting in infants aged 1 and 3 months. Pediatric Res. 2000, 48, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Owen, H.; Reynolds, K.J. Heart rate variability indices for very short-term (30 beat) analysis. Part 1: Survey and toolbox. J. Clin. Monit. Comput. 2013, 27, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.; Bigger, J.T.; Camm, A.J.; Kleiger, R.E.; Malliani, A.; Moss, A.J.; Schwartz, P.J. Heart rate variabilityStandards of measurement, physiological interpretation, and clinical use. Ann. Noninvasive Electrocardiol. 1996, 93, 1043–1065. [Google Scholar]
- Tsuji, H.; Venditti, F.J.; Manders, E.S.; Evans, J.C.; Larson, M.G.; Feldman, C.L.; Levy, D. Reduced heart rate variability and mortality risk in an elderly cohort. The Framingham Heart Study. Circulation 1994, 90, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed]
- Pincus, S.M.; Goldberger, A.L. Physiological Time-Series Analysis: What Does Regularity Quantify? Am. J. Physiol. 1994, 266, 1643–1656. [Google Scholar] [CrossRef]
- Montesinos, L.; Castaldo, R.; Pecchia, L. On the use of approximate entropy and sample entropy with centre of pressure time-series. J. Neuroeng. Rehabil. 2018, 15, 116. [Google Scholar] [CrossRef]
- Lacasa, L.; Just, W. Visibility graphs and symbolic dynamics. Phys. D Nonlinear Phenom. 2017, 374, 35–44. [Google Scholar] [CrossRef]
- La Rovere, M.T.; Pinna, G.D.; Hohnloser, S.H.; Marcus, F.I.; Mortara, A.; Nohara, R.; Bigger, J.T.; Camm, A.J.; Schwartz, P.J. Baroreflex sensitivity and heart rate variability in the identification of patients at risk for life-threatening arrhythmias: Implications for clinical trials. Circulation 2001, 103, 2072–2077. [Google Scholar] [CrossRef]
- Sztajzel, J. Heart rate variability: A noninvasive electrocardiographic method to measure the autonomic nervous system. Swiss Med. Wkly. 2004, 134, 514–522. [Google Scholar] [PubMed]
- Lahiri, M.K.; Kannankeril, P.J.; Goldberger, J.J. Assessment of Autonomic Function in Cardiovascular Disease: Physiological Basis and Prognostic Implications. J. Am. Coll. Cardiol. 2008, 51, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Malliani, A.; Lombardi, F.; Pagani, M. Power spectrum analysis of heart rate variability: A tool to explore neural regulatory mechanisms. Br. Heart J. 1994, 71, 1. [Google Scholar] [CrossRef] [PubMed]
- Yoo Suk, K.; Sung Yul, K.; Do Yang, P.; Hee Won, W.; Gyo-Seung, H.; Hyun Jun, K. Clinical Implication of Heart Rate Variability in Obstructive Sleep Apnea Syndrome Patients. J. Craniofac. Surg. 2015, 26, 1592–1595. [Google Scholar]
- Goldberger, A.L. Is the normal heartbeat chaotic or homeostatic? News Physiol. Sci. 1991, 6, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Lipsitz, L.A.; Goldberger, A.L. Loss of ‘complexity’ and aging. Potential applications of fractals and chaos theory to senescence. JAMA 1992, 267, 1806–1809. [Google Scholar] [CrossRef] [PubMed]
- Manor, B.; Lipsitz, L.A. Physiologic complexity and aging: Implications for physical function and rehabilitation. Prog. Neuropsychopharmacol. Biol. Psychiatry 2013, 45, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.B.; Chen, W.T.; He, W.X.; Liu, H. Complexity analysis of the biomedical signal using fuzzy entropy measurement. Appl. Soft Comput. 2011, 11, 2871–2879. [Google Scholar] [CrossRef]
- Alcaraz, R.; Rieta, J.J. A review on sample entropy applications for the non-invasive analysis of atrial fibrillation electrocardiograms. Biomed. Signal Process. Control 2010, 5, 1–14. [Google Scholar] [CrossRef]
- Myung-Kul, Y.; Ki-Young, J.; Hoon-Chul, K.; Heung Dong, K.; Young-Min, S.; Joong-Ku, K.; Il Keun, L.; Ki-Jong, P.; Oh-Young, K. Effect of a ketogenic diet on EEG: Analysis of sample entropy. Seizure J. Br. Epilepsy Assoc. 2008, 17, 561–566. [Google Scholar]
- Singh, J.P.; Larson, M.G.; Tsuji, H.; Evans, J.C.; O’Donnell, C.J.; Levy, D. Reduced heart rate variability and new-onset hypertension: Insights into pathogenesis of hypertension: The Framingham Heart Study. Hypertension 1998, 32, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.C.; Zhi, C.; Hu, K.; Stanley, H.E. Multiscale aspects of cardiac control. Phys. A Stat. Mech. Its Appl. 2004, 344, 685–704. [Google Scholar] [CrossRef][Green Version]
- Babloyantz, A.; Destexhe, A. Is the normal heart a periodic oscillator? Biol. Cybern. 1988, 58, 203–211. [Google Scholar] [CrossRef] [PubMed]
| HC | OSA | between-Group (p) | ||||||
|---|---|---|---|---|---|---|---|---|
| SHHS-1 | SHHS-2 | p | SHHS-1 | SHHS-2 | p | SHHS-1 | SHHS-2 | |
| (n = 48) | (n = 18) | (n = 60) | (n = 60) | |||||
| Demographics | ||||||||
| Age (years) | 76[74,78] | 82[78,83] | <0.001 * | 75[74,77] | 80[79,83] | <0.001 * | 0.594 | 0.914 |
| Gender(male/female) | 11/37 | 5/13 | 14/46 | 14/46 | 1 | 0.758 | ||
| BMI (kg/m2) | 26.0 ± 4.1 | 26.8 ± 4.8 | 0.589 | 26.6 ± 4.3 | 26.2 ± 4.4 | 0.083 | 0.488 | 0.608 |
| Sleep Measures | ||||||||
| TST (min) | 361 ± 69 | 359 ± 77 | 0.265 | 359 ± 45 | 355 ± 58 | 0.834 | 0.498 | 0.825 |
| SE (%) | 82.4[74.3,88.8] | 77.6[67.7,87.4] | 0.064 | 85.8[79.7,90.1] | 79[71,84.4] | <0.001 * | 0.119 | 0.688 |
| Stage 1 sleep (%) | 4.84[3.31,6.83] | 6[5,7] | 0.051 | 3.55[2.3,5.41] | 5[3,7] | 0.012 * | 0.030 * | 0.305 |
| Stage 2 sleep (%) | 54 ± 10.9 | 58.7 ± 8 | 0.136 | 55.7 ± 11.6 | 57.2 ± 10.5 | 0.294 | 0.438 | 0.572 |
| Stage 3+4 sleep (%) | 20 ± 11.9 | 13.4 ± 9.4 | 0.019 * | 21.1 ± 12.1 | 17.7 ± 9.8 | 0.001 * | 0.633 | 0.111 |
| REM sleep (%) | 20.5 ± 5.8 | 21.4 ± 5.1 | 0.791 | 19.7 ± 5.2 | 20.6 ± 6.5 | 0.369 | 0.441 | 0.63 |
| AHI (event/hour) | 2.15[0.82,3.34] | 6.92[3.27,10.6] | <0.001 * | 13.5[8.8,25.1] | 15.5[8.25,24.9] | 0.466 | <0.001 * | <0.001 * |
| Parameter | SHHS-1 | SHHS-2 | ||||
|---|---|---|---|---|---|---|
| HC | OSA | p | HC | OSA | p | |
| REM Sleep | ||||||
| meanNN | 949 ± 112 | 914 ± 95 | 0.082 | 969 ± 120 | 890 ± 136 | 0.031 * |
| SDNN | 37.2[28.8,47.8] | 43.5[34,52.2] | 0.107 | 40.7 ± 15.6 | 44.2 ± 14.4 | 0.381 |
| pNN50 | 2.43[0.44,6.06] | 1.52[0.53,3.9] | 0.411 | 0.77[0.2,6.37] | 2.17[0.52,7.65] | 0.226 |
| LF | 260[148,565] | 330[238,622] | 0.096 | 217[117,572] | 331[130,513] | 0.859 |
| HF | 184[60,457] | 146[76,239] | 0.471 | 127[69,330] | 180[73,514] | 0.665 |
| LFnorm | 55.2[37.9,74.9] | 69.4[51,80.9] | 0.069 | 66.4[44.4,83.4] | 68.5[45.7,88.8] | 0.665 |
| HFnorm | 39.1[26.9,46] | 31.9[17.1,41.5] | 0.647 | 42.7[19.6,53.1] | 36.9[25.1,47.5] | 0.614 |
| TP | 1139[712,2349] | 1634[938,2273] | 0.134 | 1123[710,2088] | 1621[1118,2326] | 0.516 |
| SampEn | 1.8 ± 0.39 | 1.67 ± 0.3 | 0.156 | 1.55 ± 0.38 | 1.46 ± 0.34 | 0.329 |
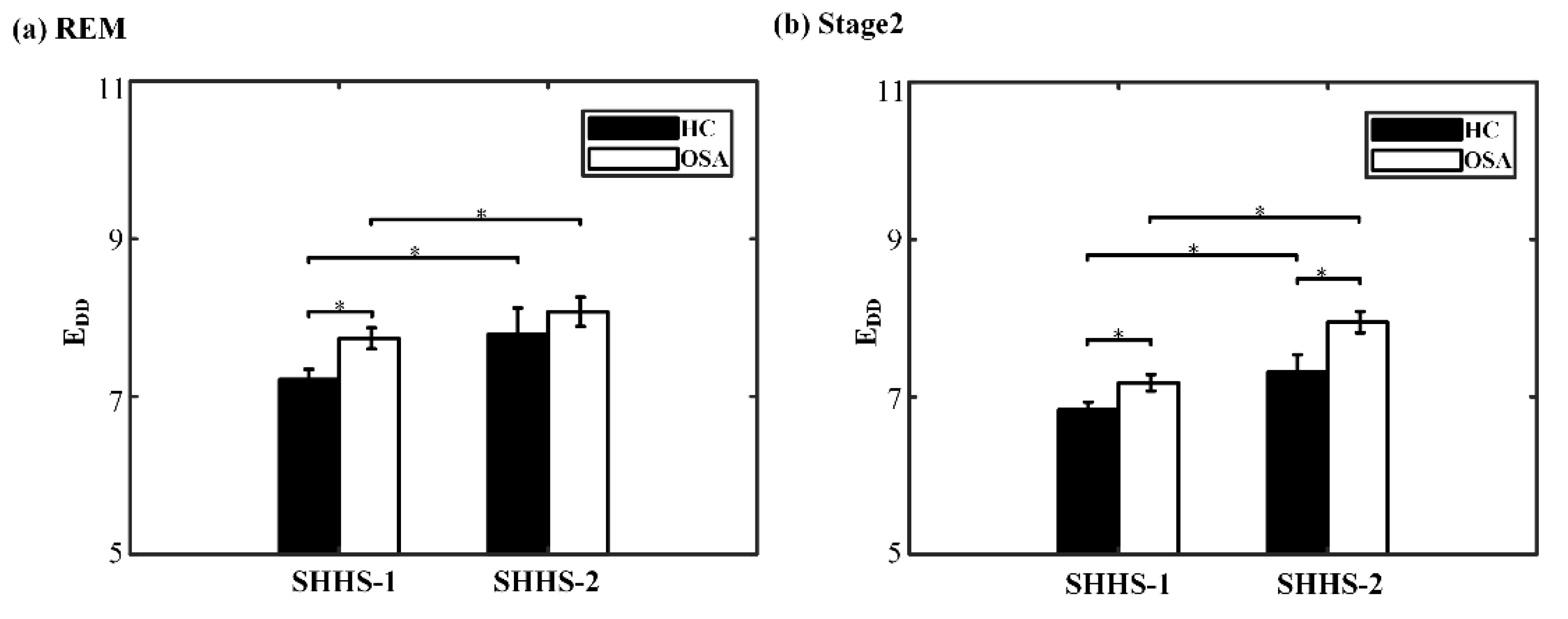
| EDD | 7.02[6.68,7.63] | 7.54[6.92,8.37] | 0.007 * | 7.79 ± 1.4 | 8.08 ± 1.44 | 0.463 |
| Stage 2 Sleep | ||||||
| meanNN | 981 ± 98 | 943 ± 111 | 0.066 | 995 ± 116 | 902 ± 97 | 0.001 * |
| SDNN | 33.3[25.7,43.7] | 37.7[29.3,48.3] | 0.309 | 33.6[27.6,42.1] | 37.1[30.7,47.4] | 0.8 |
| pNN50 | 2.15[0.77,8.29] | 2.44[0.96,5.7] | 0.497 | 2[0.5,5.03] | 2.68[1.03,6.82] | 0.383 |
| LF | 349[211,699] | 479[274,745] | 0.084 | 347[225,643] | 423[279,834] | 0.608 |
| HF | 233[116,509] | 184[126,431] | 0.522 | 171[65.1,330] | 290[145,606] | 0.128 |
| LFnorm | 52.3[40.3,67.1] | 58.8[50.9,74.5] | 0.008 * | 58.8 ± 23.1 | 60.7 ± 15 | 0.882 |
| HFnorm | 39.4 ± 12.5 | 33.3 ± 12.5 | 0.013 * | 34.9 ± 16.9 | 34.9 ± 12.2 | 0.987 |
| TP | 1078[616,1820] | 1428[842,2129] | 0.198 | 1008[608,1715] | 1496[1114,2488] | 0.1 |
| SampEn | 1.92 ± 0.29 | 1.89 ± 0.24 | 0.583 | 1.84 ± 0.26 | 1.72 ± 0.27 | 0.106 |
| EDD | 6.84 ± 0.67 | 7.18 ± 0.81 | 0.02 * | 7.32 ± 0.93 | 7.95 ± 1.04 | 0.024 * |
| Parameter | HC | OSA | ||||
|---|---|---|---|---|---|---|
| SHHS-1 | SHHS-2 | p | SHHS-1 | SHHS-2 | p | |
| REM Sleep | ||||||
| meanNN | 941 ± 106 | 969 ± 120 | 0.121 | 914 ± 95 | 890 ± 136 | 0.096 |
| SDNN | 41.5 ± 16.1 | 40.7 ± 15.6 | 0.768 | 43.7 ± 12.4 | 44.2 ± 14.4 | 0.809 |
| pNN50 | 1.35[0.28,6.41] | 0.77[0.2,6.37] | 0.811 | 1.52[0.53,3.9] | 2.17[0.52,7.65] | 0.141 |
| LF | 260[191,585] | 217[117,572] | 0.248 | 330[238,622] | 331[130,513] | 0.047 * |
| HF | 133[48,523] | 127[69,330] | 0.586 | 146[76,239] | 180[73,514] | 0.162 |
| LFnorm | 59.2[52.4,81.3] | 66.4[44.4,83.4] | 0.879 | 69.4[60,80.9] | 68.5[45.7,88.8] | 0.877 |
| HFnorm | 38[30.4,45.8] | 42.7[19.6,53.1] | 0.913 | 31.9[17.1,41.5] | 36.9[25.1,47.5] | 0.033 * |
| TP | 1065[872,2496] | 1123[710,2088] | 0.372 | 1634[938,2273] | 1621[1118,2326] | 0.729 |
| SampEn | 1.81 ± 0.31 | 1.55 ± 0.38 | 0.001 * | 1.67 ± 0.3 | 1.46 ± 0.34 | <0.001 * |
| EDD | 7.27 ± 1.08 | 7.79 ± 1.4 | 0.02 * | 7.54[6.92,8.37] | 8[7.11,8.94] | 0.017 * |
| Stage 2 Sleep | ||||||
| meanNN | 980 ± 90 | 995 ± 116 | 0.543 | 943 ± 111 | 902 ± 97 | 0.006 * |
| SDNN | 29.7[24.9,46] | 33.6[27.6,42.1] | 0.396 | 37.7[29.3,48.3] | 37.1[30.7,47.4] | 0.591 |
| pNN50 | 2.49[0.5,8.58] | 2[0.5,5.03] | 0.372 | 2.44[0.96,5.7] | 2.68[1.03,6.82] | 0.627 |
| LF | 305[207,608] | 347[225,643] | 0.267 | 479[274,745] | 423[279,834] | 0.489 |
| HF | 190[95,544] | 171[65,330] | 0.372 | 184[126,431] | 290[145,606] | 0.227 |
| LFnorm | 54.2 ± 17.7 | 58.8 ± 23.1 | 0.14 | 58.8[50.9,74.5] | 61.8[50.4,69.8] | 0.752 |
| HFnorm | 39.7 ± 13.8 | 34.9 ± 16.9 | 0.044 * | 33.3 ± 12.5 | 34.9 ± 12.2 | 0.387 |
| TP | 744[615,1959] | 1008[608,1715] | 0.446 | 1428[842,2129] | 1496[1114,2488] | 0.691 |
| SampEn | 1.95 ± 0.25 | 1.84 ± 0.26 | 0.177 | 1.89 ± 0.24 | 1.72 ± 0.27 | <0.001 * |
| EDD | 6.75 ± 0.7 | 7.32 ± 0.93 | 0.004 * | 7.18 ± 0.81 | 7.95 ± 1.04 | <0.001 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Fu, M.; Xu, F.; Hou, F.; Ma, Y. Heart Rate Dynamics in Patients with Obstructive Sleep Apnea: Heart Rate Variability and Entropy. Entropy 2019, 21, 927. https://doi.org/10.3390/e21100927
Zhang L, Fu M, Xu F, Hou F, Ma Y. Heart Rate Dynamics in Patients with Obstructive Sleep Apnea: Heart Rate Variability and Entropy. Entropy. 2019; 21(10):927. https://doi.org/10.3390/e21100927
Chicago/Turabian StyleZhang, Lulu, Mingyu Fu, Fengguo Xu, Fengzhen Hou, and Yan Ma. 2019. "Heart Rate Dynamics in Patients with Obstructive Sleep Apnea: Heart Rate Variability and Entropy" Entropy 21, no. 10: 927. https://doi.org/10.3390/e21100927
APA StyleZhang, L., Fu, M., Xu, F., Hou, F., & Ma, Y. (2019). Heart Rate Dynamics in Patients with Obstructive Sleep Apnea: Heart Rate Variability and Entropy. Entropy, 21(10), 927. https://doi.org/10.3390/e21100927




