Computing the Partial Correlation of ICA Models for Non-Gaussian Graph Signal Processing
Abstract
1. Introduction
1.1. Background
1.2. New Contributions and Paper Organization
2. The Partial Correlation of ICA Models
2.1. Statement of the Problem
2.2. A General Formula for the Residual Covariance
3. Estimating the ICA Partial Correlation Coefficients
| Algorithm 1: Computing ICA-PCC. |
| 1: Input: Learning data set 2: Compute from the learning data set (any ICA algorithm is a candidate) 3: for n = 1, 2 … N 4: for m = n … N 5: Compute (Equations (23), (29) and (30)) 6: Compute 7: Compute (Equation (13)) 8: Compute 9: end for 10: end for 11: Output |
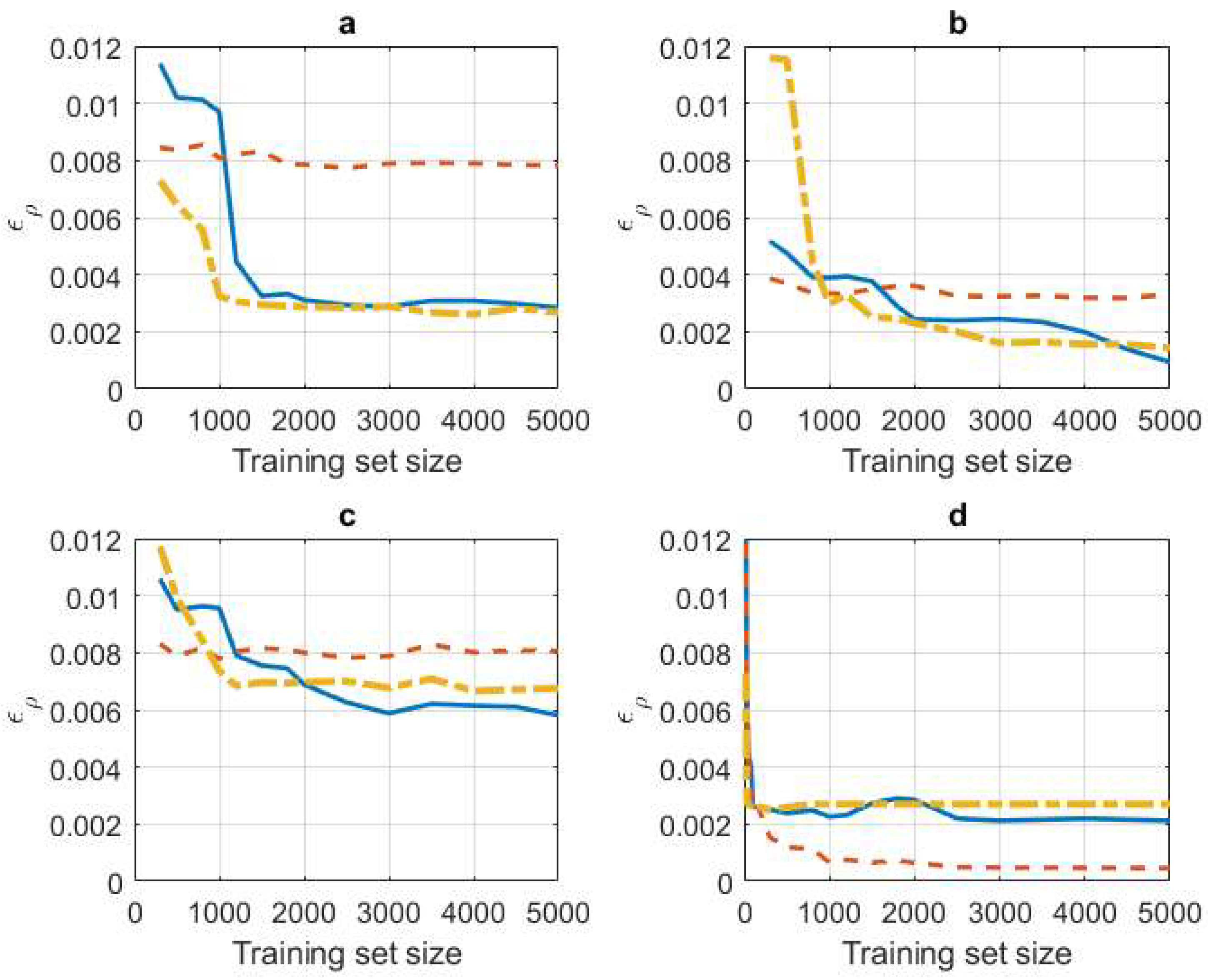
4. Experiments
4.1. Synthetic Data Experiments
4.2. A Real Data Application
5. Conclusions and Extensions
Author Contributions
Funding
Conflicts of Interest
Appendix A. Derivation of the General Formula
- -
- Term 1
- -
- Term 2
- -
- Term 3Let us express in terms of as defined in (A7)where we have taken into account that the conditional mean is an unbiased estimator. Then, we know that and if we define as a diagonal matrix having in its main diagonal the values , we can write
- -
- Term 4
References
- Baba, K.; Shibata, R.; Sibuya, M. Partial correlation and conditional correlation as measures of conditional independence. Aust. N. Z. J. Stat. 2004, 46, 657–664. [Google Scholar] [CrossRef]
- Shuman, D.I.; Narang, S.K.; Frossard, P.; Ortega, A.; Vandergheynst, P. The emerging field of signal processing on graphs: Extending high-dimensional data analysis to networks and other irregular domains. IEEE Signal Process. Mag. 2013, 30, 83–98. [Google Scholar] [CrossRef]
- Sandryhaila, A.; Moura, J.M.F. Discrete signal processing on graphs. IEEE Trans. Signal Process. 2013, 61, 1644–1656. [Google Scholar] [CrossRef]
- Ortega, A.; Frossard, P.; Kovacevic, J.; Moura, J.M.F.; Vandergheynst, P. Graph Signal Processing: Overview, challenges and applications. Proc. IEEE 2018, 106, 808–828. [Google Scholar] [CrossRef]
- Zhang, C.; Florencio, D.; Chou, P.A. Graph Signal Processing—A Probabilistic Framework; Tech. Rep. MSR-TR-2015-31; Microsoft Research Lab: Redmond, WA, USA, 2015. [Google Scholar]
- Pávez, E.; Ortega, A. Generalized precision matrix estimation for graph signal processing. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing, Shanghai, China, 20–25 March 2016; pp. 6350–6354. [Google Scholar]
- Mazumder, R.; Hastie, T. The graphical lasso: New insights and alternatives. Electron. J. Stat. 2012, 6, 2125–2149. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.J.; Sustik, M.A.; Dhillon, I.S.; Ravikumar, P. Sparse inverse covariance matrix estimation using quadratic approximation. Adv. Neural Inf. Process. Syst. 2011, 24, 2330–2338. [Google Scholar]
- Chen, X.; Xu, M.; Wu, W.B. Covariance and precision matrix estimation for high-dimensional time series. Ann. Stat. 2013, 41, 2994–3021. [Google Scholar] [CrossRef]
- Öllerer, V.; Croux, C. Robust high-dimensional precision matrix estimation. In Modern Multivariate and Robust Methods; Nordhausen, K., Taskinen, S., Eds.; Springer: New York, NY, USA, 2015; pp. 329–354. [Google Scholar]
- Friedman, J.; Hastie, T.; Tibshirani, R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 2008, 9, 432–441. [Google Scholar] [CrossRef]
- Peng, J.; Wang, P.; Zhou, N.; Zhu, J. Partial correlation estimation by joint sparse regression model. J. Am. Stat. Assoc. 2009, 104, 735–746. [Google Scholar] [CrossRef]
- Belda, J.; Vergara, L.; Salazar, A.; Safont, G. Estimating the Laplacian matrix of Gaussian mixtures for signal processing on graphs. Signal Process. 2018, 148, 241–249. [Google Scholar] [CrossRef]
- Salazar, A.; Vergara, L. Independent Component Analysis (ICA): Algorithms, Applications and Ambiguities; Nova Science Publishers: New York, NY, USA, 2018. [Google Scholar]
- Common, P.; Jutten, C. Handbook of Blind Source Separation: Independent Component Analysis and Applications; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Hyvarinen, A. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef]
- Lee, T.W. Independent Component Analysis: Theory and Applications; Kluwer: Norwell, MA, USA, 1998. [Google Scholar]
- Chai, R.; Naik, G.R.; Nguyen, T.N.; Ling, S.H.; Tran, Y.; Craig, A.; Nguyen, H.T. Driver fatigue classification with independent component by entropy rate bound minimization analysis in an EEG-based system. IEEE J. Biomed. Health Inform. 2017, 21, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, S.; Huang, T.; Zhang, Z.; Hu, Y.; Zhang, T. Infrared spectrum blind deconvolution algorithm via learned dictionaries and sparse representation. Appl. Opt. 2016, 55, 2813–2818. [Google Scholar] [CrossRef]
- Naik, G.R.; Selvan, S.E.; Nguyen, H.T. Single-Channel EMG Classification with Ensemble-Empirical-Mode-Decomposition-Based ICA for Diagnosing Neuromuscular Disorders. IEEE Trans. Neural Syst. Rehab. Eng. 2016, 24, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, S.; Li, Y.; Naik, G.R. Edge effect elimination in single-mixture blind source separation. Circuits Syst. Signal Process. 2013, 32, 2317–2334. [Google Scholar] [CrossRef]
- Yuejie, Ch. Guaranteed blind sparse spikes deconvolution via lifting and convex optimization. IEEE J. Select. Top. Signal Process. 2016, 10, 782–794. [Google Scholar]
- Pendharkara, G.; Naik, G.R.; Nguyen, H.T. Using blind source separation on accelerometry data to analyze and distinguish the toe walking gait from normal gait in ITW children. Biomed. Signal Process. Control 2014, 13, 41–49. [Google Scholar] [CrossRef]
- Guo, Y.; Naik, G.R.; Nguyen, H.T. Single channel blind source separation based local mean decomposition for biomedical applications. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Osaka, Japan, 3–7 July 2013; pp. 6812–6815. [Google Scholar]
- Liming, W.; Chi, Y. Blind Deconvolution from Multiple Sparse Inputs. IEEE Signal Process. Lett. 2016, 23, 1384–1388. [Google Scholar]
- Salazar, A.; Vergara, L.; Serrano, A.; Igual, J. A General Procedure for Learning Mixtures of Independent Component Analyzers. Pattern Recognit. 2010, 43, 69–85. [Google Scholar] [CrossRef]
- Safont, G.; Salazar, A.; Vergara, L.; Gomez, E.; Villanueva, V. Probabilistic distance for mixtures of independent component analyzers. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1161–1173. [Google Scholar] [CrossRef]
- Salazar, A.; Igual, J.; Safont, G.; Vergara, L.; Vidal, A. Image applications of agglomerative clustering using mixtures of non-Gaussian distributions. In Proceedings of the 2015 International Conference on Computational Science and Computational Intelligence, Las Vegas, NV, USA, 7–9 December 2015; pp. 459–463. [Google Scholar]
- Safont, G.; Salazar, A.; Rodriguez, A.; Vergara, L. On recovering missing ground penetrating radar traces by statistical interpolation methods. Remote Sens. 2014, 6, 7546–7565. [Google Scholar] [CrossRef]
- Salazar, A.; Safont, G.; Soriano, A.; Vergara, L. Automatic credit card fraud detection based on non-linear signal processing. In Proceedings of the IEEE International Carnahan Conference on Security Technology, Boston, MA, USA, 15–18 October 2012; pp. 207–212. [Google Scholar]
- Salazar, A.; Igual, J.; Vergara, L.; Serrano, A. Learning hierarchies from ICA mixtures. In Proceedings of the IEEE International Joint Conference on Artificial Neural Networks, Orlando, FL, USA, 12–17 August 2007; pp. 2271–2276. [Google Scholar]
- Vergara, L.; Bernabeu, P. Simple approach to nonlinear prediction. Electron. Lett. 2001, 37, 928–936. [Google Scholar] [CrossRef]
- Celebi, E. General formula for conditional mean using higher-order statistics. Electron. Lett. 1997, 33, 2097–2099. [Google Scholar] [CrossRef]
- Lee, T.W.; Girolami, M.; Sejnowski, T.J. Independent Component Analysis Using an Extended Infomax Algorithm for Mixed Sub-Gaussian and Super-Gaussian Sources. Neural Comput. 1999, 11, 409–433. [Google Scholar] [CrossRef]
- Cardoso, J.F. Blind beamforming for non-Gaussian signals. IEE Proc. F-Radar Signal Process. 1993, 140, 362–370. [Google Scholar] [CrossRef]
- Hyvärinen, A.; Oja, E. A fast fixed-point algorithm for Independent Component Analysis. Neural Comput. 1997, 9, 1483–1492. [Google Scholar] [CrossRef]
- Salazar, A.; Vergara, L.; Miralles, R. On including sequential dependence in ICA mixture models. Signal Process. 2010, 90, 2314–2318. [Google Scholar] [CrossRef]
- Lang, E.W.; Tomé, A.; Keck, I.R.; Górriz-Sáez, J.; Puntonet, C. Brain connectivity analysis: A short survey. Comput. Intell. Neurosci. 2012, 2012. [Google Scholar] [CrossRef]
- Fiedler, M. Algebraic connectivity of graphs. Czecoslovak Math. J. 1973, 23, 298–305. [Google Scholar]
- Merris, R. Laplacian matrices of a graph: A survey. Linear Algebra Appl. 1994, 197, 143–176. [Google Scholar] [CrossRef]
- Lake, B.; Tenenbaum, J. Discovering structure by learning sparse graph. In Proceedings of the 32nd Annual Meeting of the Cognitive Science Society CogSci 2010, Portland, OR, USA, 11–14 August 2010; pp. 778–783. [Google Scholar]
- Dong, X.; Thanou, D.; Frossard, P.; Vandergheynst, P. Learning Laplacian matrix in smooth graph signal representations. IEEE Trans. Signal Process. 2016, 64, 6160–6173. [Google Scholar] [CrossRef]
- Moragues, J.; Vergara, L.; Gosálbez, J. Generalized matched subspace filter for nonindependent noise based on ICA. IEEE Trans. Signal Process. 2011, 59, 3430–3434. [Google Scholar] [CrossRef]
- Egilmez, H.E.; Pavez, E.; Ortega, A. Graph learning from data under Laplacian and structural constraints. IEEE J. Sel. Top. Signal Process. 2017, 11, 825–841. [Google Scholar] [CrossRef]
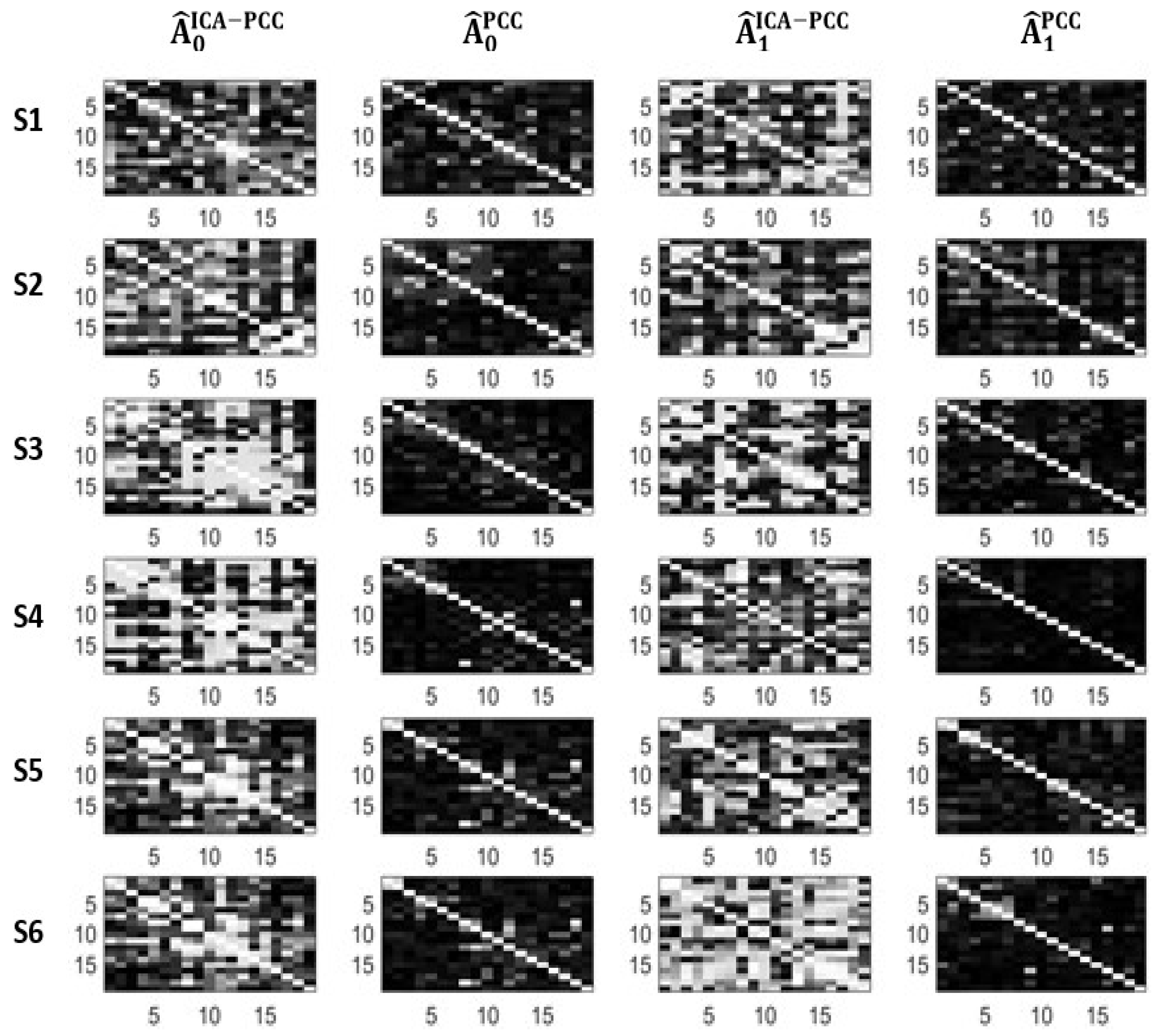
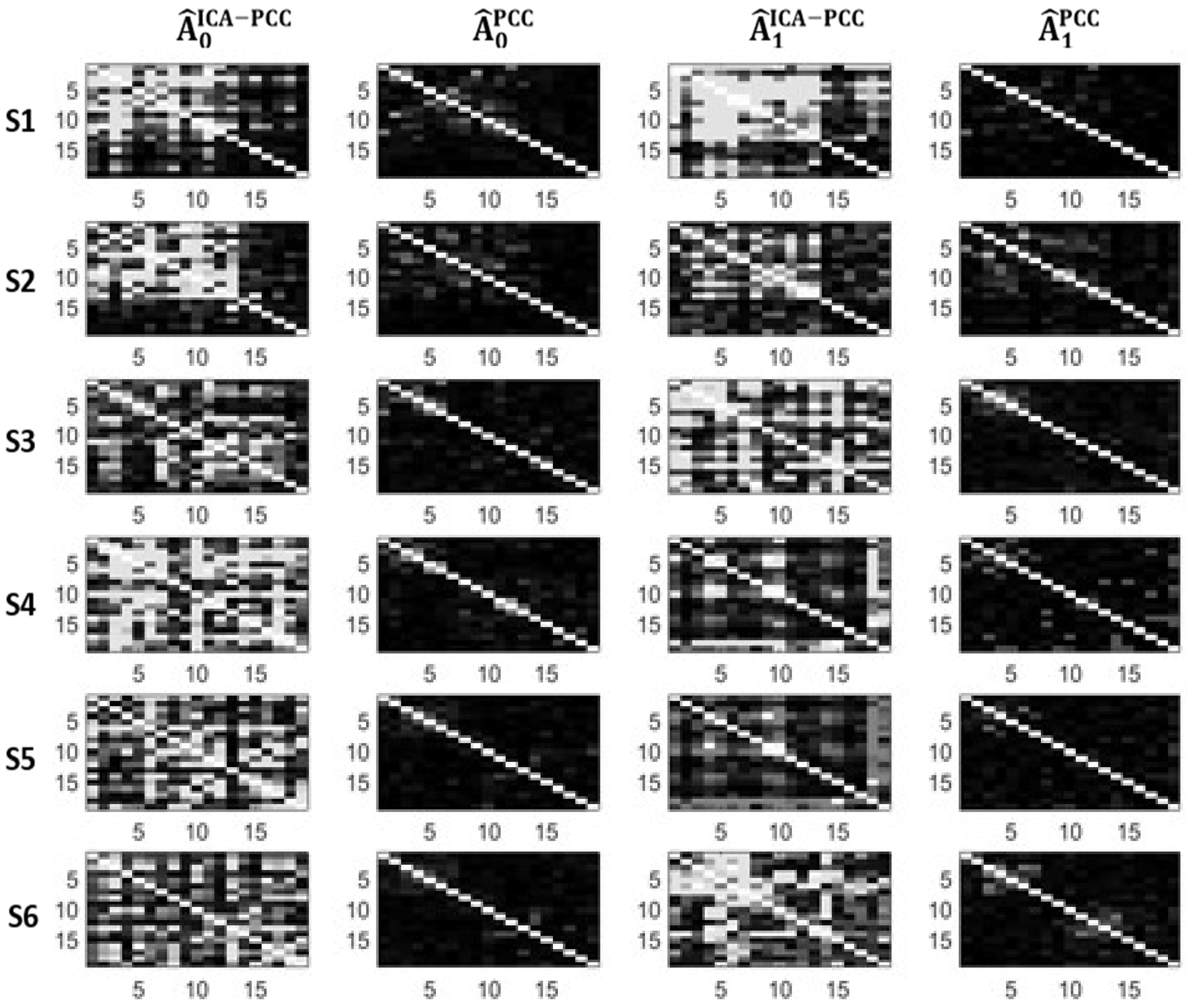
| Subj. | ||||||||
|---|---|---|---|---|---|---|---|---|
| S1 | 6.46 | 4.58 | 0.30 | 0.33 | 0.03 | 0.03 | 0.03 | 0.00 |
| S2 | 8.05 | 5.29 | 0.74 | 0.39 | 0.35 | 0.04 | 0.04 | 0.00 |
| S3 | 9.84 | 6.76 | 0.57 | 0.28 | 0.29 | 0.03 | 0.02 | 0.01 |
| S4 | 9.04 | 8.87 | 0.39 | 0.66 | 0.27 | 0.04 | 0.02 | 0.02 |
| S5 | 9.61 | 15.13 | 0.31 | 0.44 | 0.13 | 0.02 | 0.03 | 0.01 |
| S6 | 9.14 | 13.82 | 0.24 | 0.36 | 0.12 | 0.02 | 0.02 | 0.00 |
| Subj. | ||||||||
|---|---|---|---|---|---|---|---|---|
| S1 | 16.32 | 22.51 | 0.31 | 0.51 | 0.20 | 0.02 | 0.02 | 0.00 |
| S2 | 10.52 | 9.09 | 0.34 | 0.60 | 0.26 | 0.02 | 0.02 | 0.00 |
| S3 | 9.91 | 7.05 | 0.68 | 0.48 | 0.20 | 0.02 | 0.03 | 0.01 |
| S4 | 8.39 | 11.69 | 0.37 | 0.74 | 0.37 | 0.03 | 0.02 | 0.01 |
| S5 | 7.72 | 13.15 | 0.22 | 0.71 | 0.49 | 0.02 | 0.03 | 0.01 |
| S6 | 11.86 | 9.24 | 0.43 | 0.56 | 0.13 | 0.02 | 0.03 | 0.01 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belda, J.; Vergara, L.; Safont, G.; Salazar, A. Computing the Partial Correlation of ICA Models for Non-Gaussian Graph Signal Processing. Entropy 2019, 21, 22. https://doi.org/10.3390/e21010022
Belda J, Vergara L, Safont G, Salazar A. Computing the Partial Correlation of ICA Models for Non-Gaussian Graph Signal Processing. Entropy. 2019; 21(1):22. https://doi.org/10.3390/e21010022
Chicago/Turabian StyleBelda, Jordi, Luis Vergara, Gonzalo Safont, and Addisson Salazar. 2019. "Computing the Partial Correlation of ICA Models for Non-Gaussian Graph Signal Processing" Entropy 21, no. 1: 22. https://doi.org/10.3390/e21010022
APA StyleBelda, J., Vergara, L., Safont, G., & Salazar, A. (2019). Computing the Partial Correlation of ICA Models for Non-Gaussian Graph Signal Processing. Entropy, 21(1), 22. https://doi.org/10.3390/e21010022





