SPAER: Sparse Deep Convolutional Autoencoder Model to Extract Low Dimensional Imaging Biomarkers for Early Detection of Breast Cancer Using Dynamic Thermography
Abstract
1. Introduction
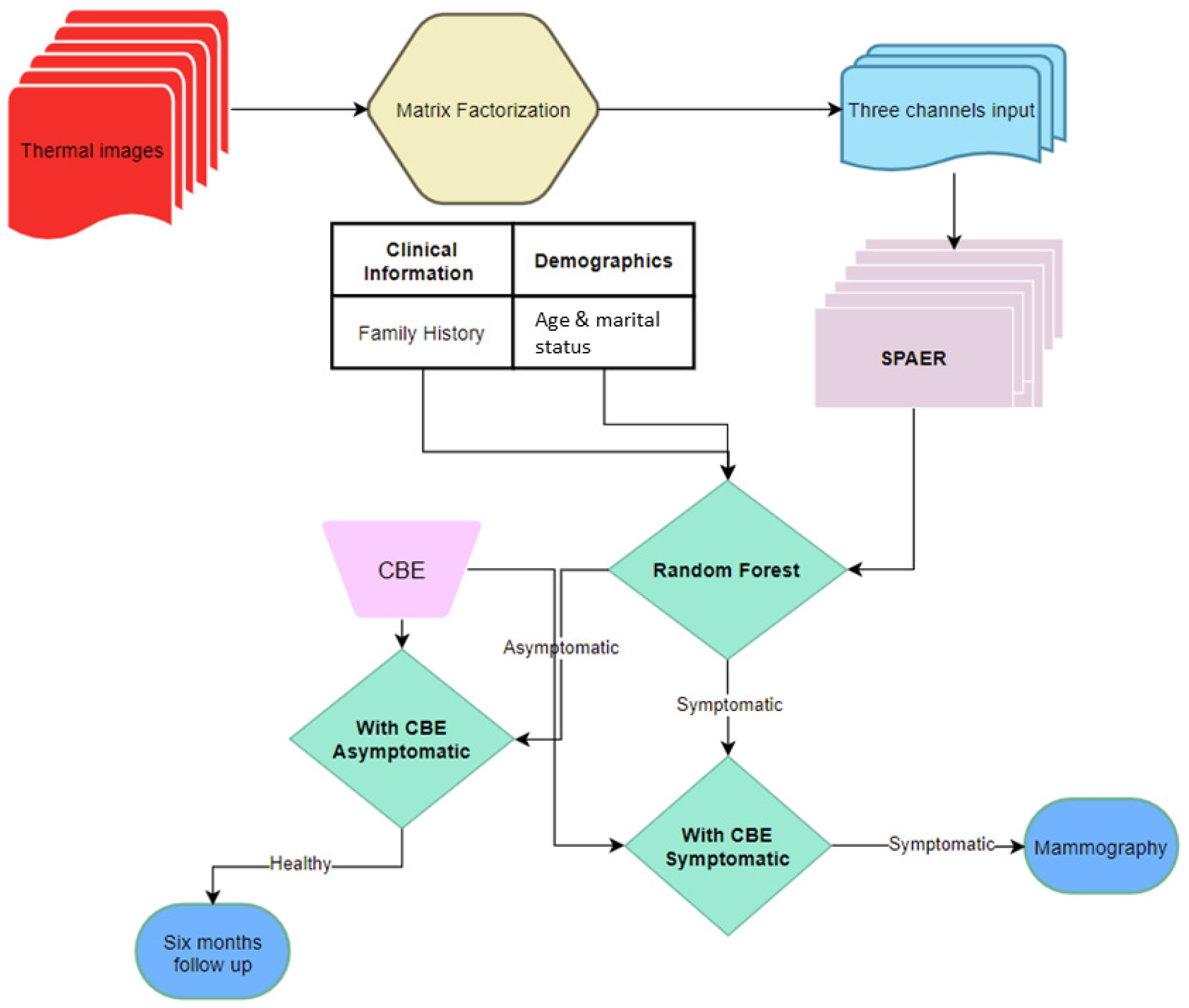
2. Materials and Methods
2.1. Low-Rank Matrix Approximation
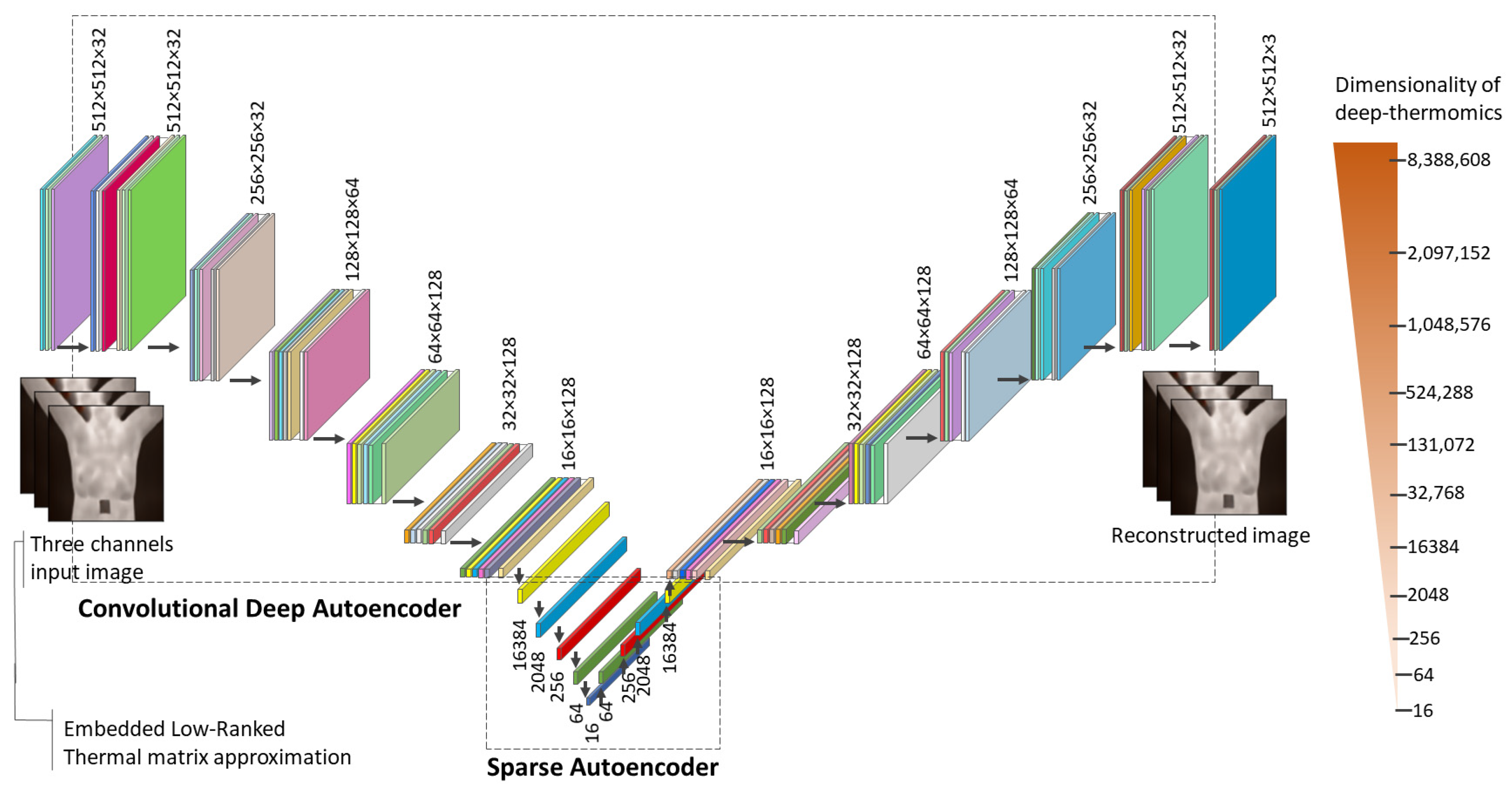
2.2. Sparse Latent Space Deep Thermomics
3. Results
3.1. Results of Low-Rank Thermal Matrix Approximation
3.2. Sparse Autoencoder and Deep Thermomics
3.3. Classification Outcome
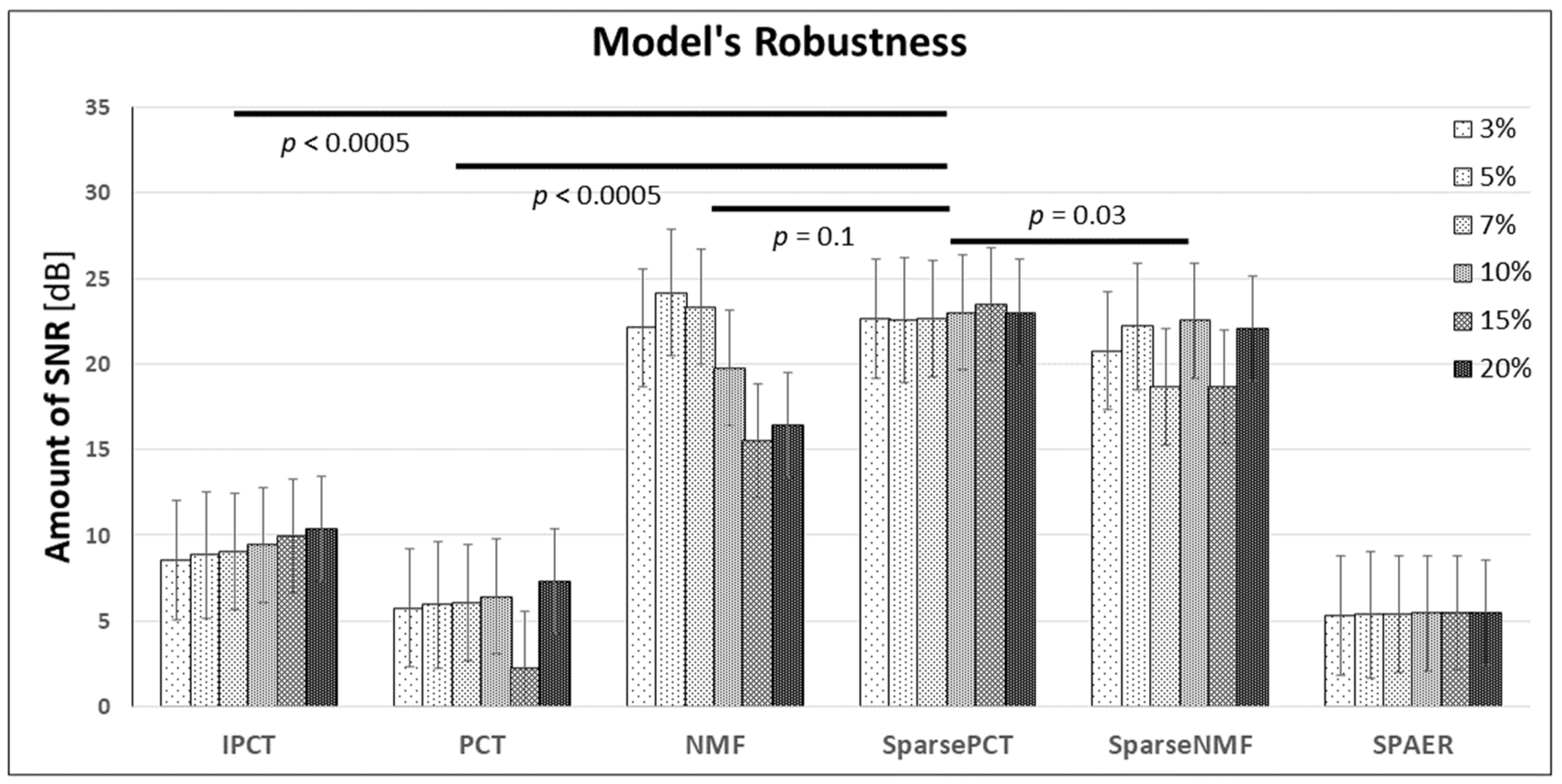
3.4. System’s Robustness
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. A Cancer J Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Report on Cancer. September 2018. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 3 March 2021).
- Crystal, P.; Strano, S.D.; Shcharynski, S.; Koretz, M.J. Using sonography to screen women with mammographically dense breasts. Am. J. Roentgenol. 2003, 181, 177–182. [Google Scholar] [CrossRef]
- Kerlikowske, K.; Grady, D.; Barclay, J.; Sickles, E.A.; Ernster, V. Effect of age, breast density, and family history on the sensitivity of first screening mammography. JAMA 1996, 276, 33–38. [Google Scholar] [CrossRef]
- Hong, S.; Song, S.Y.; Park, B.; Suh, M.; Choi, K.S.; Jung, S.E.; Jun, J.K. Effect of digital mammography for breast cancer screening: A comparative study of more than 8 million Korean women. Radiology 2020, 294, 247–255. [Google Scholar] [CrossRef]
- Kennedy, D.A.; Lee, T.; Seely, D. A comparative review of thermography as a breast cancer screening technique. Integr. Cancer Ther. 2009, 8, 9–16. [Google Scholar] [CrossRef]
- De Gonzalez, A.B.; Reeves, G. Mammographic screening before age 50 years in the UK: Comparison of the radiation risks with the mortality benefits. Br. J. Cancer 2005, 93, 590–596. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fletcher, S.W.; Elmore, J.G. Mammographic screening for breast cancer. N. Engl. J. Med. 2003, 348, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Osako, T.; Iwase, T.; Takahashi, K.; Iijima, K.; Miyagi, Y.; Nishimura, S.; Kasumi, F. Diagnostic mammography and ultrasonography for palpable and nonpalpable breast cancer in women aged 30 to 39 years. Breast Cancer 2007, 14, 255–259. [Google Scholar] [CrossRef]
- Duffy, S.W.; Vulkan, D.; Cuckle, H.; Parmar, D.; Sheikh, S.; Smith, R.A.; Myles, J. Effect of mammographic screening from age 40 years on breast cancer mortality (UK Age trial): Final results of a randomised, controlled trial. Lancet Oncol. 2020, 21, P1165–P1172. [Google Scholar] [CrossRef]
- Nguyen, Q.D.; Nguyen, N.T.; Dixon, I.I.I.L.; Monetto, F.E.P.; Robinson, A.S. Spontaneously Disappearing Calcifications in the Breast: A Rare Instance Where a Decrease in Size on Mammogram Is Not Good. Cureus 2020, 12, e8753. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.A. Screening for breast cancer with MRI. In Seminars in Ultrasound, CT and MRI; WB Saunders: Berlin/Heidelberg, Germany, 2003; Volume 24, pp. 45–54. [Google Scholar]
- Ahern, C.H.; Shih, Y.T.; Dong, W.; Parmigiani, G.; Shen, Y. Cost-effectiveness of alternative strategies for integrating MRI into breast cancer screening for women at high risk. Br. J. Cancer 2014, 111, 1542–1551. [Google Scholar] [CrossRef]
- Barton, M.B.; Harris, R.; Fletcher, S.W. Does this patient have breast cancer? The screening clinical breast examination: Should it be done? How? JAMA 1999, 282, 1270–1280. [Google Scholar] [CrossRef]
- Oestreicher, N.; Lehman, C.D.; Seger, D.J.; Buist, D.S.; White, E. The incremental contribution of clinical breast examination to invasive cancer detection in a mammography screening program. Am. J. Roentgenol. 2005, 184, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Feig, S.A.; Shaber, G.S.; Schwartz, G.F.; Patchefsky, A.; Libshitz, H.I.; Edeiken, J.; Wallace, J.D. Thermography, mammography, and clinical examination in breast cancer screening: Review of 16,000 studies. Radiology 1977, 122, 123–127. [Google Scholar] [CrossRef]
- Keyserlingk, J.R.; Ahlgren, P.D.; Yu, E.; Belliveau, N.; Yassa, M. Functional infrared imaging of the breast. IEEE Eng. Med. Biol. Mag. 2000, 19, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Anbar, M. Clinical thermal imaging today. IEEE Eng. Med. Biol. Mag. 1998, 17, 25–33. [Google Scholar] [CrossRef]
- Gautherie, M. Thermopathology of breast cancer: Measurement and analysis of in vivo temperature and blood flow. Ann. New York Acad. Sci. 1980, 335, 383–415. [Google Scholar] [CrossRef] [PubMed]
- Recinella, A.N.; Gonzalez-Hernandez, J.L.; Kandlikar, S.G.; Dabydeen, D.; Medeiros, L.; Phatak, P. Clinical Infrared Imaging in the Prone Position for Breast Cancer Screening—Initial Screening and Digital Model Validation. J. Eng. Sci. Med Diagn. Ther. 2020, 3, 011005. [Google Scholar] [CrossRef]
- Folkman, J.; Merler, E.; Abernathy, C.; Williams, G. Isolation of a tumor factor responsible for angiogenesis. J. Exp. Med. 1971, 133, 275. [Google Scholar] [CrossRef] [PubMed]
- Algire, G.H.; Chalkley, H.W.; Legallais, F.Y.; Park, H.D. Vascular reactions to normal and malignant tissue in vivo. I vascular reactions of mice to wound and to normal and neoplastic transplants. J. Natl. Cancer Inst. 1945, 6, 7385. [Google Scholar] [CrossRef]
- Longatto Filho, A.; Lopes, J.M.; Schmitt, F.C. Angiogenesis and breast cancer. J. Oncol. 2010, 2010. [Google Scholar] [CrossRef]
- Kerbel, R.S. Tumor angiogenesis. N. Engl. J. Med. 2008, 358, 2039–2049. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Bonapace, L.; Coissieux, M.M.; Wyckoff, J.; Mertz, K.D.; Varga, Z.; Junt, T.; Bentires-Alj, M. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature 2014, 515, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Relf, M.; LeJeune, S.; Scott, P.A.; Fox, S.; Smith, K.; Leek, R.; Harris, A.L. Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor β-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer Res. 1997, 57, 963–969. [Google Scholar]
- Kopeć, M.; Abramczyk, H. Angiogenesis-a crucial step in breast cancer growth, progression and dissemination by Raman imaging. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 198, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Abramczyk, H.; Surmacki, J.; Kopeć, M.; Olejnik, A.K.; Kaufman-Szymczyk, A.; Fabianowska-Majewska, K. Epigenetic changes in cancer by Raman imaging, fluorescence imaging, AFM and scanning near-field optical microscopy (SNOM). Acetylation in normal and human cancer breast cells MCF10A, MCF7 and MDA-MB-231. Analyst 2016, 141, 5646–5658. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.F.; Saade, D.C.M.; Sequeiros, G.O.; Silva, A.C.; Paiva, A.C.; Bravo, R.S.; Conci, A. A New Database for Breast Research with Infrared Image. J. Med Imaging Health Inform. 2014, 4, 92–100. [Google Scholar] [CrossRef]
- Figueiredo, A.A.A.; Fernandes, H.C.; Guimaraes, G. Experimental approach for breast cancer center estimation using infrared thermography. Infrared Phys. Technol. 2018, 95, 100–112. [Google Scholar] [CrossRef]
- Hildebrandt, C.; Raschner, C.; Ammer, K. An overview of recent application of medical infrared thermography in sports medicine in Austria. Sensors 2010, 10, 4700–4715. [Google Scholar] [CrossRef]
- Usamentiaga, R.; Mokhtari, Y.; Ibarra-Castanedo, C.; Klein, M.; Genest, M.; Maldague, X. Automated dynamic inspection using active infrared thermography. IEEE Trans. Ind. Inform. 2018, 14, 5648–5657. [Google Scholar] [CrossRef]
- Rajic, N. Principal component thermography for flaw contrast enhancement and flaw depth characterisation in composite structures. Compos. Struct. 2002, 58, 521–528. [Google Scholar] [CrossRef]
- Marinetti, S.; Finesso, L.; Marsilio, E. Matrix factorization methods: Application to thermal ndt/e. NDT E Int. 2006, 39, 611–616. [Google Scholar] [CrossRef]
- Cramer, K.E.; Winfree, W.P. Fixed eigenvector analysis of thermographic NDE data. In Thermosense: Thermal Infrared Applications XXXIII, M; Safai, J., Brown, R., Eds.; International Society for Optics and Photonics: Bellingham, WA, USA, 2011; Volume 8013, pp. 225–235. [Google Scholar] [CrossRef]
- Yousefi, B.; Sharifipour, H.M.; Eskandari, M.; IbarraCastanedo, C.; Laurendeau, D.; Watts, R.; Klein, M.; Maldague, X.P. Incremental low rank noise reduction for robust infrared tracking of body temperature during medical imaging. Electronics 2019, 8, 1301. [Google Scholar] [CrossRef]
- Ahmed, J.; Gao, B.; Woo, W.L. Wavelet-integrated alternating sparse dictionary matrix decomposition in thermal imaging cfrp defect detection. IEEE Trans. Ind. Inform. 2018, 15, 4033–4043. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Yousefi, B.; Sfarra, S.; Castanedo, C.I.; Maldague, X.P. Comparative analysis on thermal non-destructive testing imagery applying Candid Covariance-Free Incremental Principal Component Thermography (CCIPCT). Infrared Phys. Technol. 2017, 85, 163–169. [Google Scholar] [CrossRef]
- Yousefi, B.; Sfarra, S.; Sarasini, F.; Castanedo, C.I.; Maldague, X.P. Low-rank sparse principal component thermography (sparse-pct): Comparative assessment on detection of subsurface defects. Infrared Phys. Technol. 2019, 98, 278–284. [Google Scholar] [CrossRef]
- Wu, J.-Y.; Sfarra, S.; Yao, Y. Sparse principal component thermography for subsurface defect detection in composite products. IEEE Trans. Ind. Inform. 2018, 14, 5594–5600. [Google Scholar] [CrossRef]
- Ding, C.H.; Li, T.; Jordan, M.I. Convex and semi-nonnegative matrix factorizations. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 32, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, B.; Sfarra, S.; Ibarra-Castanedo, C.; Avdelidis, N.P.; Maldague, X.P. Thermography data fusion and nonnegative matrix factorization for the evaluation of cultural heritage objects and buildings. J. Therm. Anal. Calorim. 2019, 136, 943–955. [Google Scholar] [CrossRef]
- Yousefi, B.; Ibarra-Castanedo, C.; Maldague, X.P. Infrared nondestructive testing via semi-nonnegative matrix factorization. In Proceedings of the 15th International Workshop on Advanced Infrared Technology and Applications (AITA 2019), Florence, Italy, 17–19 September 2019; p. 13. [Google Scholar]
- Yousefi, B.; Castanedo, I.C.; Maldague, P.X. Measuring heterogeneous thermal patterns in infrared-based diagnostic systems using sparse low-rank matrix approximation: Comparative study. IEEE Trans. Instrum. Meas. 2020, 70, 1–9. [Google Scholar] [CrossRef]
- Yousefi, B.; Castanedo, I.C.; Maldague, P.X. Low-rank Convex/Sparse Thermal Matrix Approximation for Infrared-based Diagnostic System. arXiv 2020, arXiv:2010.06784. [Google Scholar]
- Yousefi, B.; Akbari, H.; Maldague, X.P. Detecting Vasodilation as Potential Diagnostic Biomarker in Breast Cancer Using Deep Learning-Driven Thermomics. Biosensors 2020, 10, 164. [Google Scholar] [CrossRef]
- Chaves, E.; Gonçalves, C.B.; Albertini, M.K.; Lee, S.; Jeon, G.; Fernandes, H.C. Evaluation of transfer learning of pre-trained CNNs applied to breast cancer detection on infrared images. Appl. Opt. 2020, 59, E23–E28. [Google Scholar] [CrossRef]
- Gonçalves, C.B.; Leles, A.C.; Oliveira, L.E.; Guimaraes, G.; Cunha, J.R.; Fernandes, H. Machine learning and infrared thermography for breast cancer detection. In Proceedings of the 15th International Workshop on Advanced Infrared Technology and Applications (AITA 2019), Florence, Italy, 17–19 September 2019; p. 45. [Google Scholar]
- Ruiz, L.; Torres, M.; Gómez, A.; Díaz, S.; González, J.M.; Cavas, F. Detection and Classification of Aircraft Fixation Elements during Manufacturing Processes Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 6856. [Google Scholar] [CrossRef]
- Donoho, D.L. For most large underdetermined systems of linear equations the minimal 1-norm solution is also the sparsest solution. Communications on Pure and Applied Mathematics. Courant Inst. Math. Sci. 2006, 59, 797–829. [Google Scholar]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Gillies, R.J. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Olshausen, B.A.; Field, D.J. Sparse coding with an overcomplete basis set: A strategy employed by V1? Vis. Res. 1997, 37, 3311–3325. [Google Scholar] [CrossRef]
- Arpit, D.; Zhou, Y.; Ngo, H.; Govindaraju, V. Why regularized auto-encoders learn sparse representation? In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 19–24 June 2016; pp. 136–144. [Google Scholar]
- Google. Python 3 Google Compute engine backend, T4, and P100 GPU and 27.4 Gb RAM; Google: Mountain View, CA, USA, 2020. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Zheng, X. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th Symposium on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Usamentiaga, R.; Ibarra-Castanedo, C.; Maldague, X. More than fifty shades of grey: Quantitative characterization of defects and interpretation using snr and cnr. J. Nondestruct. Eval. 2018, 37, 25. [Google Scholar] [CrossRef]
- Chiang, H.T.; Hsieh, Y.Y.; Fu, S.W.; Hung, K.H.; Tsao, Y.; Chien, S.Y. Noise reduction in ECG signals using fully convolutional denoising autoencoders. IEEE Access 2019, 7, 60806–60813. [Google Scholar] [CrossRef]
- Sakurada, M.; Yairi, T. Anomaly detection using autoencoders with nonlinear dimensionality reduction. In Proceedings of the MLSDA 2014 2nd Workshop on Machine Learning for Sensory Data Analysis, Gold Coast, QLD, Australia, 2 December 2014; pp. 4–11. [Google Scholar]
- Bhowick, D.; Gupta, D.K.; Maiti, S.; Shankar, U. Stacked autoencoders based machine learning for noise reduction and signal reconstruction in geophysical data. arXiv 2019, arXiv:1907.03278. [Google Scholar]
- FDA. Safety Communication, FDA Warns Thermography Should Not Be Used in Place of Mammography to Detect, Diagnose, or Screen for Breast Cancer. Available online: www.fda.gov (accessed on 25 February 2019).
- FDA. Consumer Update, Breast Cancer Screening: Thermogram No Substitute for Mammogram, U.S. Food and Drug Administration. Available online: www.fda.gov (accessed on 13 January 2021).



| DMR: Database for Mastology Research. | ||
|---|---|---|
| Age | Median (±IQR) | 60 (25,120) |
| Race | Caucasian | 77 (37%) |
| African | 57 (27.4%) | |
| Pardo | 72 (34.6%) | |
| Mulatto | 1 (0.5%) | |
| Indigenous | 1 (0.5%) | |
| Martial status | Married | 104 (50%) |
| Single | 66 (31.7%) | |
| Widow | 26 (12.5%) | |
| Divorced | 12 (5.7%) | |
| Diagnosis 1 | Healthy 2 | 128 (61.5%) |
| Symptomatic | 80 (38.5%) | |
| Sick 3 | 36 (17.3%) | |
| Family history | Diabetes | 52 (25%) |
| Hypertensive | 5 (2.4%) | |
| Leukemia | 1 (0.5%) | |
| None | 150 (72.1%) | |
| Hormone therapy (HT) | Hormone replacement | 38 (18.3%) |
| None | 170 (81.7%) | |
| Accuracy of Different Multivariate Models for Breast Cancer Diagnosis | ||||||
|---|---|---|---|---|---|---|
| Methods | Classification Accuracy 2 (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | T-Test 3 t-Statistic, Two-Tailed p-Value |
| IPCT-SPAER | 74.3 (69.4–80.1) | 80 | 81.3 | 72.7 | 86.7 | 5, < 0.0005 |
| PCT-SPAER | 74.3 (70.4–79.1) | 85.0 | 85.9 | 79.1 | 90.2 | 4.5, < 0.0005 |
| NMF-SPAER | 77.7 (70.9–82.1) | 78.7 | 82.1 | 73.3 | 86.1 | 1.1, < 0.27 |
| Sparse PCT-SPAER | 74.3 (69.9–79.6) | 86.3 | 80.5 | 73.4 | 90.4 | 4.9, < 0.0005 |
| Sparse NMF-SPAER | 72.3 (68.5–76.2) | 81.3 | 91.4 | 85.5 | 88.6 | 7.8, < 0.0005 |
| Clinical 1 | 72.8 (70.4–75.3) | 73.8 | 70.3 | 60.8 | 81.1 | 7.1, < 0.0005 |
| IPCT-SPAER + Clinical | 76.2 (71.4–79.6) | 86.3 | 82.8 | 75.8 | 90.6 | 2.6, 0.009 |
| PCT-SPAER + Clinical | 77.7 (73.3–82.1) | 83.7 | 84.4 | 77.01 | 89.3 | 0.6, 0.5 |
| NMF-SPAER + Clinical | 78.2 (74.3–82.5) | 80 | 87.5 | 80 | 87.5 | - |
| Sparse PCT-SPAER + Clinical | 77.7 (72.3–81.6) | 81.3 | 88.3 | 81.3 | 88.3 | 0.2, 0.8 |
| SparseNMF-SPAER + Clinical | 74.8 (70.9–77.2) | 87.5 | 81.3 | 74.5 | 91.2 | 4.8, < 0.0005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousefi, B.; Akbari, H.; Hershman, M.; Kawakita, S.; Fernandes, H.C.; Ibarra-Castanedo, C.; Ahadian, S.; Maldague, X.P.V. SPAER: Sparse Deep Convolutional Autoencoder Model to Extract Low Dimensional Imaging Biomarkers for Early Detection of Breast Cancer Using Dynamic Thermography. Appl. Sci. 2021, 11, 3248. https://doi.org/10.3390/app11073248
Yousefi B, Akbari H, Hershman M, Kawakita S, Fernandes HC, Ibarra-Castanedo C, Ahadian S, Maldague XPV. SPAER: Sparse Deep Convolutional Autoencoder Model to Extract Low Dimensional Imaging Biomarkers for Early Detection of Breast Cancer Using Dynamic Thermography. Applied Sciences. 2021; 11(7):3248. https://doi.org/10.3390/app11073248
Chicago/Turabian StyleYousefi, Bardia, Hamed Akbari, Michelle Hershman, Satoru Kawakita, Henrique C. Fernandes, Clemente Ibarra-Castanedo, Samad Ahadian, and Xavier P. V. Maldague. 2021. "SPAER: Sparse Deep Convolutional Autoencoder Model to Extract Low Dimensional Imaging Biomarkers for Early Detection of Breast Cancer Using Dynamic Thermography" Applied Sciences 11, no. 7: 3248. https://doi.org/10.3390/app11073248
APA StyleYousefi, B., Akbari, H., Hershman, M., Kawakita, S., Fernandes, H. C., Ibarra-Castanedo, C., Ahadian, S., & Maldague, X. P. V. (2021). SPAER: Sparse Deep Convolutional Autoencoder Model to Extract Low Dimensional Imaging Biomarkers for Early Detection of Breast Cancer Using Dynamic Thermography. Applied Sciences, 11(7), 3248. https://doi.org/10.3390/app11073248










