Synergistic Killing and Re-Sensitization of Pseudomonas aeruginosa to Antibiotics by Phage-Antibiotic Combination Treatment
Abstract
1. Introduction
2. Results & Discussion
2.1. Phage Cocktail PAM2H Re-Sensitizes MDR P. aeruginosa Strains to Antibiotics
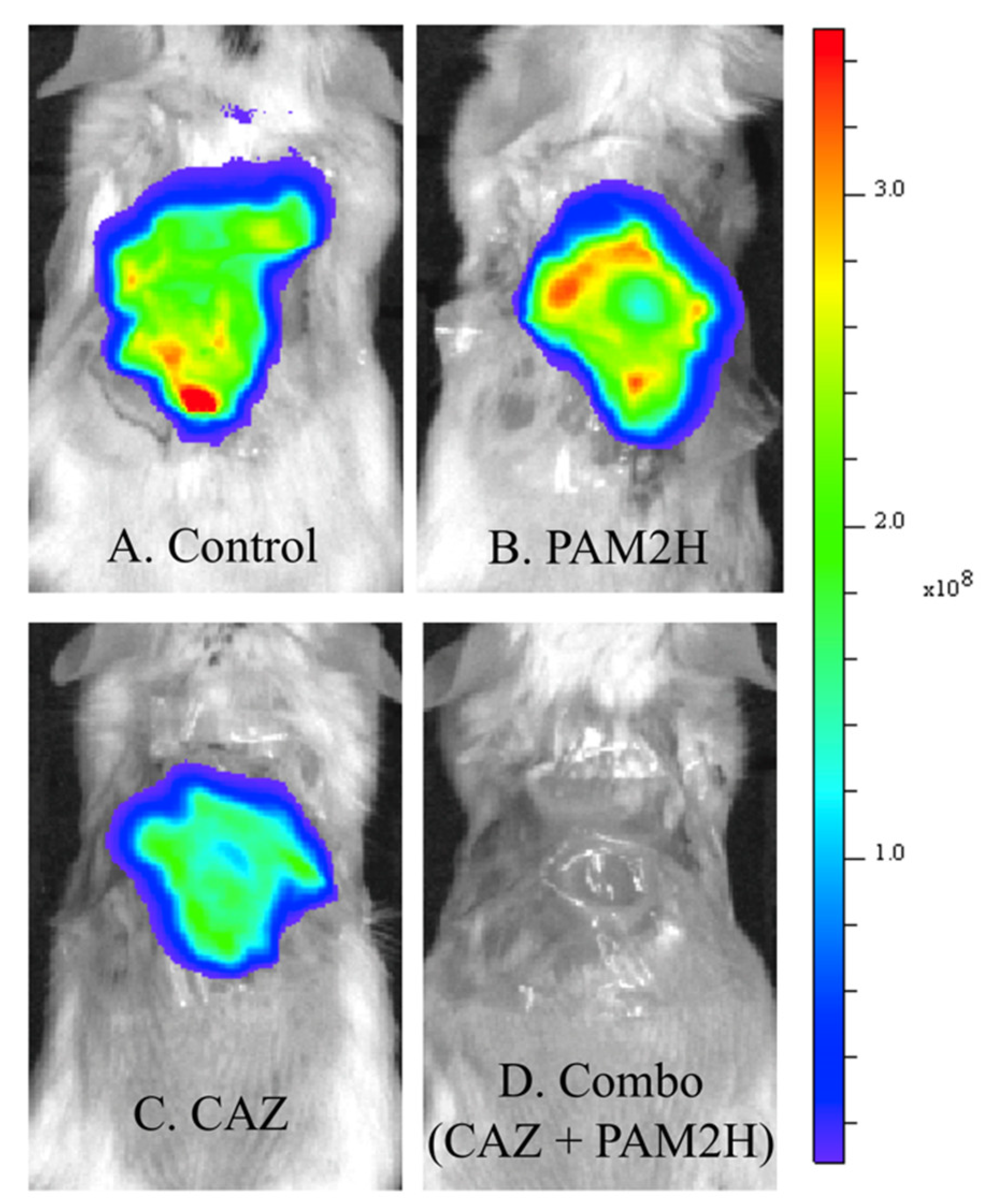
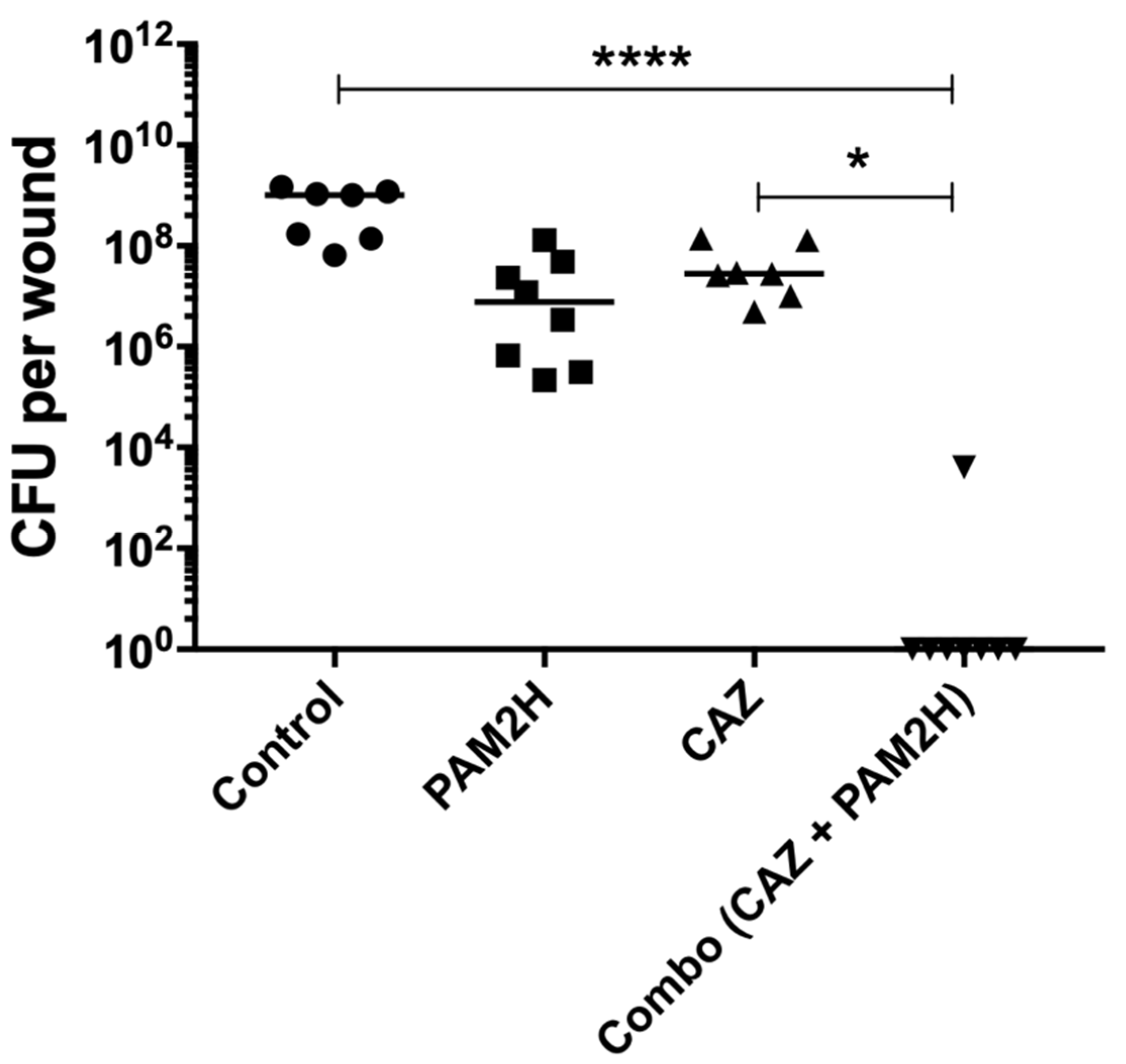
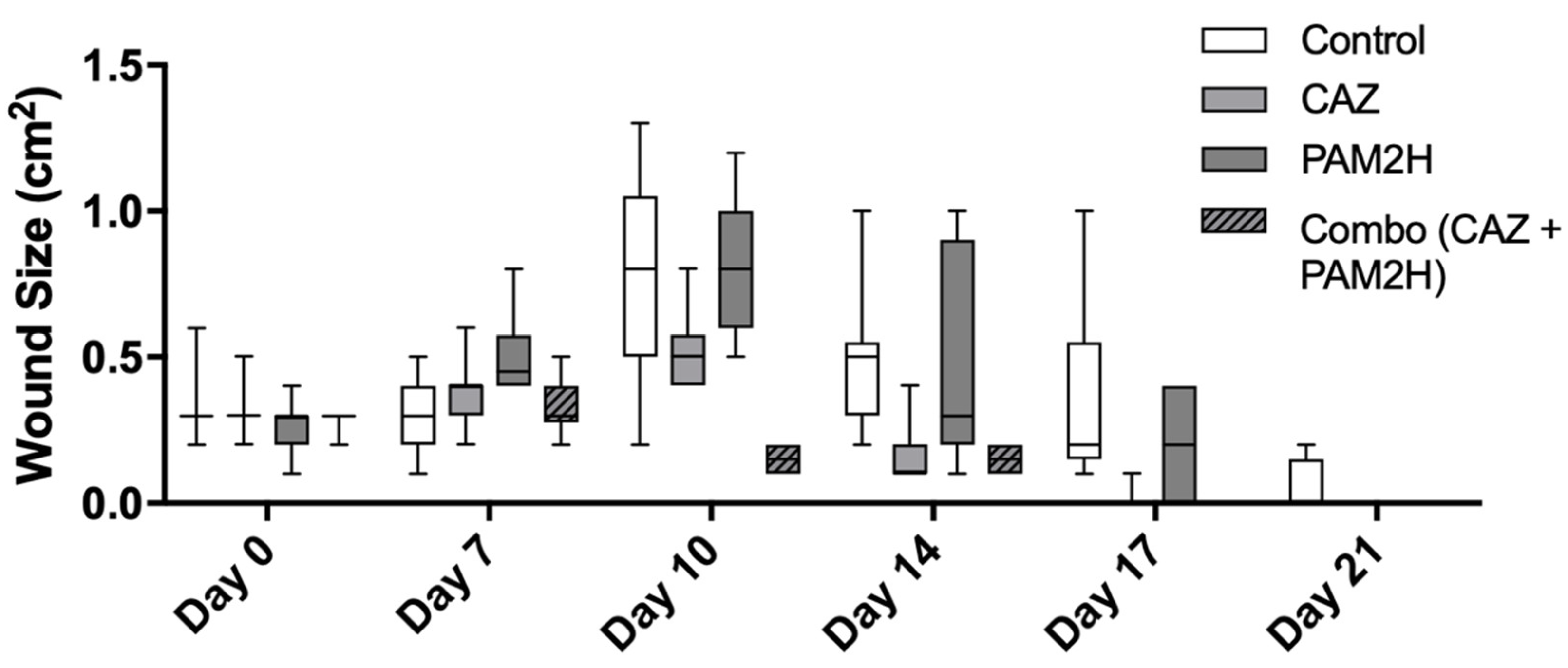
2.2. PAM2H + CAZ Combination Treatment Enhances Efficacy in a Mouse Dorsal Wound Model
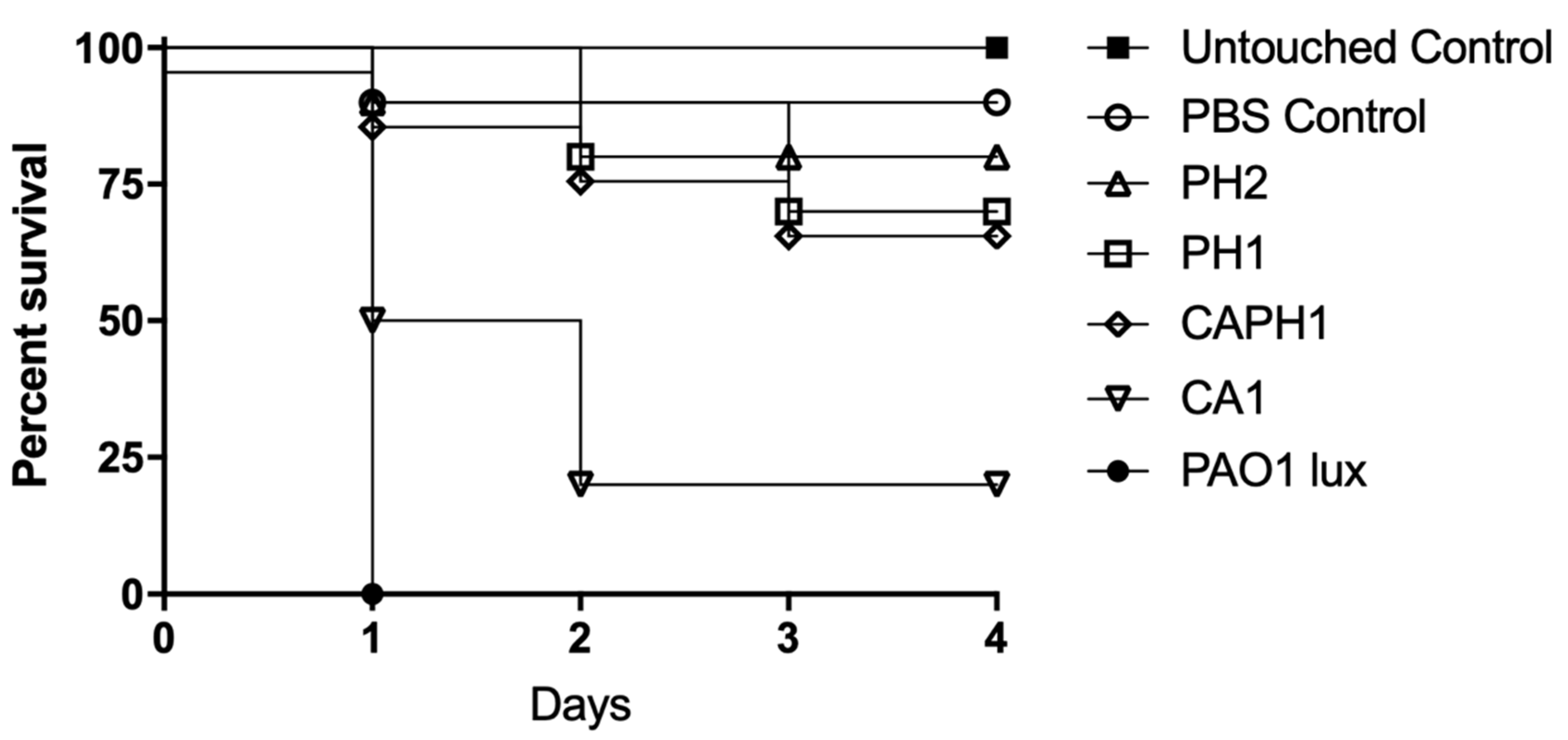
2.3. Recovered Strain Virulence in Wax Worms
2.4. Whole-Genome Sequencing of Recovered PAO1 Strains
3. Materials and Methods
3.1. Bacterial Strains and Bacteriophages
3.2. Bacteriophage Isolation and Cocktail Preparation
3.3. Antibiotics
3.4. Minimum Inhibitory Concentration Assays
3.5. Fractional Inhibitory Concentration Assays
3.6. Assessment of Phage Antibiotic Combination Treatment in a P. aeruginosa Mouse Wound Model
3.7. Assessment of P. aeruginosa Strain Virulence in Galleria mellonella Larvae
3.8. P. aeruginosa DNA Isolation and Whole-Genome Sequencing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lobanovska, M.; Pilla, G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future? Yale J. Boil. Med. 2017, 90, 135–145. [Google Scholar]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Friedman, N.; Temkin, E.; Carmeli, Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016, 22, 416–422. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Tillotson, G.S.; Zinner, S.H. Burden of antimicrobial resistance in an era of decreasing susceptibility. Expert Rev. Anti-Infect. Ther. 2017, 15, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Livermore, D.M. Multiple Mechanisms of Antimicrobial Resistance in Pseudomonas aeruginosa: Our Worst Nightmare? Clin. Infect. Dis. 2002, 34, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016, 24, 862–871. [Google Scholar] [CrossRef]
- Summers, W.C. Bacteriophage Therapy. Annu. Rev. Microbiol. 2001, 55, 437–451. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Boas, D.P.A.V.; Sillankorva, S.; Azeredo, J. Phage Therapy: A Step Forward in the Treatment of Pseudomonas aeruginosa Infections. J. Virol. 2015, 89, 7449–7456. [Google Scholar] [CrossRef]
- Hall, A.R.; De Vos, D.; Friman, V.-P.; Pirnay, J.-P.; Buckling, A. Effects of Sequential and Simultaneous Applications of Bacteriophages on Populations of Pseudomonas aeruginosaIn Vitroand in Wax Moth Larvae. Appl. Environ. Microbiol. 2012, 78, 5646–5652. [Google Scholar] [CrossRef]
- Debarbieux, L.; LeDuc, D.; Maura, D.; Morello, E.; Criscuolo, A.; Grossi, O.; Balloy, V.; Touqui, L. Bacteriophages Can Treat and PreventPseudomonas aeruginosaLung Infections. J. Infect. Dis. 2010, 201, 1096–1104. [Google Scholar] [CrossRef]
- Morello, E.; Saussereau, E.; Maura, D.; Huerre, M.; Touqui, L.; Debarbieux, L. Pulmonary Bacteriophage Therapy on Pseudomonas aeruginosa Cystic Fibrosis Strains: First Steps Towards Treatment and Prevention. PLoS ONE 2011, 6, e16963. [Google Scholar] [CrossRef] [PubMed]
- Alemayehu, D.; Casey, P.G.; McAuliffe, O.; Guinane, C.M.; Martin, J.G.; Shanahan, F.; Coffey, A.; Ross, R.P.; Hill, C. Bacteriophages ϕMR299-2 and ϕNH-4 Can Eliminate Pseudomonas aeruginosa in the Murine Lung and on Cystic Fibrosis Lung Airway Cells. mBio 2012, 3, e00029-12. [Google Scholar] [CrossRef]
- Tiwari, B.R.; Kim, S.; Rahman, M.; Kim, J. Antibacterial efficacy of lytic Pseudomonas bacteriophage in normal and neutropenic mice models. J. Microbiol. 2011, 49, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell. Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef] [PubMed]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.-P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.-A. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 2016, 215, 703–712. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
- Hagens, S.; Habel, A.; Bläsi, U. Augmentation of the Antimicrobial Efficacy of Antibiotics by Filamentous Phage. Microb. Drug Resist. 2006, 12, 164–168. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Chan, B.K.; E Turner, P.; Kim, S.; Mojibian, H.R.; A Elefteriades, J.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Khawaldeh, A.; Morales, S.; Dillon, B.; Alavidze, Z.; Ginn, A.N.; Thomas, L.; Chapman, S.J.; Dublanchet, A.; Smithyman, A.; Iredell, J.R. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 2011, 60, 1697–1700. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Meletiadis, J.; Pournaras, S.; Roilides, E.; Walsh, T.J. Defining Fractional Inhibitory Concentration Index Cutoffs for Additive Interactions Based on Self-Drug Additive Combinations, Monte Carlo Simulation Analysis, and In Vitro-In Vivo Correlation Data for Antifungal Drug Combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2009, 54, 602–609. [Google Scholar] [CrossRef]
- Buehrle, D.J.; Shields, R.K.; Clarke, L.G.; Potoski, B.A.; Clancy, C.J.; Nguyen, M.H. Carbapenem-Resistant Pseudomonas aeruginosa Bacteremia: Risk Factors for Mortality and Microbiologic Treatment Failure. Antimicrob. Agents Chemother. 2016, 61. [Google Scholar] [CrossRef]
- Nair, A.B.; Jacob, S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.G.; Black, C.C.; Pavlicek, R.L.; Honnold, C.L.; Wise, M.C.; Alamneh, Y.A.; Moon, J.K.; Kessler, J.L.; Si, Y.; Williams, R.; et al. Validation of a Novel Murine Wound Model of Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2013, 58, 1332–1342. [Google Scholar] [CrossRef]
- Rampioni, G.; Pillai, C.R.; Longo, F.; Bondì, R.; Baldelli, V.; Messina, M.; Imperi, F.; Visca, P.; Leoni, L. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Choi, K.-H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Farlow, J.; Freyberger, H.R.; He, Y.; Ward, A.M.; Rutvisuttinunt, W.; Li, T.; Campbell, R.; Jacobs, A.C.; Nikolich, M.P.; Filippov, A.A. Complete Genome Sequences of 10 Phages Lytic against Multidrug-Resistant Pseudomonas aeruginosa. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 11th ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Vipra, A.; Desai, S.N.; Junjappa, R.P.; Roy, P.; Poonacha, N.; Ravinder, P.; Sriram, B.; Padmanabhan, S. Determining the Minimum Inhibitory Concentration of Bacteriophages: Potential Advantages. Adv. Microbiol. 2013, 3, 181–190. [Google Scholar] [CrossRef]
- Sopirala, M.M.; Mangino, J.E.; Gebreyes, W.A.; Biller, B.; Bannerman, T.; Balada-Llasat, J.-M.; Pancholi, P. Synergy Testing by Etest, Microdilution Checkerboard, and Time-Kill Methods for Pan-Drug-Resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4678–4683. [Google Scholar] [CrossRef]
- Agún, S.; Fernández, L.; González-Menéndez, E.; Martínez, B.; Rodríguez, A.; García, P. Study of the Interactions between Bacteriophage phiIPLA-RODI and Four Chemical Disinfectants for the Elimination of Staphylococcus aureus Contamination. Viruses 2018, 10, 103. [Google Scholar] [CrossRef]
- Regeimbal, J.M.; Jacobs, A.C.; Corey, B.W.; Henry, M.S.; Thompson, M.G.; Pavlicek, R.L.; Quinones, J.; Hannah, R.M.; Ghebremedhin, M.; Crane, N.J.; et al. Personalized Therapeutic Cocktail of Wild Environmental Phages Rescues Mice from Acinetobacter baumannii Wound Infections. Antimicrob. Agents Chemother. 2016, 60, 5806–5816. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Peleg, A.Y.; Jara, S.; Monga, D.; Eliopoulos, G.M.; Moellering, R.C.; Mylonakis, E. Galleria mellonella as a Model System to Study Acinetobacter baumannii Pathogenesis and Therapeutics. Antimicrob. Agents Chemother. 2009, 53, 2605–2609. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. (v. 38. 63). BBDuk. Jt Genome Inst. Available online: https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbduk-guide/ (accessed on 25 June 2020).
- Miller, J.R.; Koren, S.; Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 2010, 95, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Mather, A.E.; Sánchez-Busó, L.; Page, A.J.; Parkhill, J.; Keane, J.A.; Harris, S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 2017, 3, e000131. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Nbsp; MLST. Available online: https://github.com/tseemann/mlst (accessed on 25 June 2020).
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Snippy. Rapid Haploid Variant Calling and Core Genome Alignment. Available online: https://github.com/tseemann/snippy (accessed on 25 June 2020).
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]

| Isolate No. | Source | MIC (µg/mL) | MIC (PFU/mL) | |||
|---|---|---|---|---|---|---|
| CAZ | MEM | GEN | CIP | PAM2H | ||
| MRSN321 | Wound | 32 | 16 | 2 | 0.025 | 1 × 100 |
| MRSN994 | Respiratory | 32 | 32 | 4 | 4 | 1 × 106 |
| MRSN2108 | Tissue | 16 | 64 | 2 | 8 | >1 × 109 |
| MRSN5498 | Tissue | 16 | 32 | 256 | 8 | 1 × 108 |
| MRSN5508 | Fluid | 16 | 32 | 128 | 8 | 1 × 108 |
| MRSN5519 | Wound | 32 | 32 | 128 | 32 | 1 × 106 |
| MRSN6695 | Urine | 128 | 32 | 8 | 2 | >1 × 109 |
| MRSN8915 | Urine | 4 | 32 | 16 | 64 | 1 × 104 |
| MRSN11538 | Wound | 8 | 32 | 128 | 4 | 1 × 109 |
| MRSN12282 | Respiratory | 32 | 64 | 128 | 64 | 1 × 102 |
| MRSN15678 | Wound | 16 | 16 | 128 | 32 | 1 × 106 |
| MRSN16345 | Urine | 16 | 8 | 1 | 32 | 1 × 100 |
| MRSN23861 | Respiratory | 16 | 256 | 2 | 32 | 1 × 107 |
| MRSN409937 | Fluid | 128 | 16 | 2 | 8 | 1 × 100 |
| PAO1::lux * | Laboratory | 2 | 1 | 32 | 1 | 1 × 100 |
| PAO1 | Laboratory | 1 | 2 | 1 | 1 | 1 × 100 |
| Antibiotic | CLSI MIC Breakpoints µg/mL | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Ceftazidime (CAZ) | ≤8 | 16 | ≥32 |
| Ciprofloxacin (CIP) | ≤0.5 | 1 | ≥2 |
| Gentamicin (GEN) | ≤4 | 8 | ≥16 |
| Meropenem (MEM) | ≤2 | 4 | ≥8 |
| Isolate No. | Ceftazidime (CAZ) | Meropenem (MEM) | Gentamicin (GEN) | Ciprofloxacin (CIP) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL) | MIC in the Presence of PAM2H (µg/mL) | Amount of PAM2H (PFU/mL) | MIC (µg/mL) | MIC in the Presence of PAM2H (µg/mL) | Amount of PAM2H (PFU/mL) | MIC (µg/mL) | MIC in the Presence of PAM2H (µg/mL) | Amount of PAM2H (PFU/mL) | MIC (µg/mL) | MIC in Presence of PAM2H (µg/mL) | Amount of PAM2H (PFU/mL) | |
| PA321 | 32 | 0.0625 | 1 × 100 | 16 | 0.0625 | 1 × 101 | 2 | 0.007813 | 1 × 101 | 0.25 | 0.000976563 | 1 × 100 |
| PA994 | 32 | 0.125 | 1 × 107 | 32 | 0.125 | 1 × 107 | 4 | 0.015625 | 1 × 107 | 4 | 0.015625 | 1 × 107 |
| PA2108 | 16 | 0.0625 | 1 × 104 | 64 | 0.25 | 1 × 103 | 2 | 0.007813 | 1 × 103 | 8 | 0.03125 | 1 × 103 |
| PA5498 | 16 | 2 | 1 × 105 | 32 | 0.25 | 1 × 107 | 256 | 0.5 | 1 × 107 | 8 | 0.03125 | 1 × 107 |
| PA5508 | 16 | 0.0625 | 1 × 105 | 32 | 0.125 | 1 × 102 | 128 | 0.5 | 1 × 102 | 8 | 0.03125 | 1 × 106 |
| PA5519 | 32 | 0.125 | 1 × 105 | 32 | 0.125 | 1 × 106 | 128 | 0.5 | 1 × 106 | 32 | 0.125 | 1 × 105 |
| PA6695 | 128 | 128 | 1 × 102 | 32 | 32 | 1 × 102 | 8 | 8 | 1 × 102 | 2 | 1 | 1 × 102 |
| PA8915 | 4 | 0.015625 | 1 × 100 | 32 | 16 | 1 × 100 | 16 | 16 | 1 × 100 | 64 | 0.25 | 1 × 100 |
| PA11538 | 8 | 8 | 1 × 107 | 32 | 16 | 1 × 102 | 128 | 16 | 1 × 103 | 4 | 0.25 | 1 × 107 |
| PA12282 | 32 | 0.125 | 1 × 101 | 64 | 16 | 1 × 100 | 128 | 2 | 1 × 100 | 64 | 0.25 | 1 × 100 |
| PA15678 | 16 | 0.0625 | 1 × 105 | 16 | 2 | 1 × 104 | 128 | 8 | 1 × 105 | 32 | 0.125 | 1 × 105 |
| PA16345 | 16 | 0.0625 | 1 × 100 | 8 | 0.125 | 1 × 101 | 1 | 0.003906 | 1 × 100 | 32 | 0.125 | 1 × 100 |
| PA23861 | 16 | 16 | 1 × 103 | 256 | 256 | 1 × 102 | 2 | 0.007813 | 1 × 107 | 32 | 32 | 1 × 108 |
| PA409937 | 128 | 0.5 | 1 × 100 | 16 | 2 | 1 × 101 | 2 | 0.015625 | 1 × 101 | 8 | 0.03125 | 1 × 100 |
| PAO1::lux | 2 | 0.007813 | 1 × 100 | 1 | 0.003906 | 1 × 100 | 32 | 0.125 | 1 × 100 | 1 | 0.003906 | 1 × 100 |
| PAO1 | 1 | 0.003906 | 1 × 100 | 2 | 0.03125 | 1 × 100 | 1 | 0.003906 | 1 × 100 | 1 | 0.003906 | 1 × 100 |
| FICAB | ||||
|---|---|---|---|---|
| Isolate No. | CAZ | MEM | GEN | CIP |
| MRSN321 | 0.002 | 0.004 | 0.004 | 0.039 |
| MRSN994 | 0.001 | 0.004 | 0.004 | 0.004 |
| MRSN2108 | 0.004 | 0.004 | 0.004 | 0.004 |
| MRSN5498 | 0.125 | 0.008 | 0.002 | 0.004 |
| MRSN5508 | 0.004 | 0.004 | 0.004 | 0.004 |
| MRSN5519 | 0.004 | 0.004 | 0.004 | 0.004 |
| MRSN6695 | 1 | 1 | 1 | 0.5 |
| MRSN8915 | 0.004 | 0.5 | 1 | 0 |
| MRSN11538 | 1 | 0.5 | 0.125 | 0.063 |
| MRSN12282 | 0.004 | 0.25 | 0.016 | 0.004 |
| MRSN15678 | 0.004 | 0.125 | 0.063 | 0.004 |
| MRSN16345 | 0.004 | 0.016 | 0.004 | 0.004 |
| MRSN23861 | 1 | 1 | 0.004 | 1 |
| MRSN409937 | 0.004 | 0.125 | 0.008 | 0.004 |
| PAO1::lux | 0.004 | 0.004 | 0.004 | 0.004 |
| PAO1 | 0.004 | 0.016 | 0.004 | 0.004 |
| Product 1 | Mutation 2 | Impact 3 | Known Phage Receptor 4 | PAO1::lux Mutant Isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH1 | PH2 | PH3 | PH4 | CA1 | CA2 | CA3 | CA4 | CAPH1 | CAPH2 | ||||
| type 4 fimbrial biogenesis outer membrane protein PilQ | SNP (A > C) | Thr605Pro | Yes | + | |||||||||
| B-band O-antigen polymerase | Insertion (A) | Thr46 (fs) | Yes | + | |||||||||
| B-band O-antigen polymerase | SNP (A > G) | Tyr249Cys | Yes | + | |||||||||
| type 4 fimbrial biogenesis protein PilY1 | Deletion (AGACCAGCTT) | Gln520 (fs) | Yes | + | |||||||||
| type 4 fimbrial biogenesis protein PilB | Deletion (A) | His414 (fs) | Yes | + | |||||||||
| type 4 fimbrial biogenesis protein PilB | Deletion (CGGA) | Arg258 (fs) | Yes | + | |||||||||
| glucose-6-phosphate isomerase | Insertion (A) | Thr219 (fs) | No | + | |||||||||
| oxidoreductase | SNP (T > A) | Ser133Thr | No | + | + | + | + | + | + | + | + | + | + |
| Phage Name | Family | Genus |
|---|---|---|
| EPa5 | Siphoviridae | Abidjanvirus |
| EPa11 | Myoviridae | Pbunavirus |
| EPa15 | Myoviridae | Pbunavirus |
| EPa22 | Myoviridae | Pbunavirus |
| EPa43 | Siphoviridae | Abidjanvirus |
| Treatment Groups | Treatment Location | |
|---|---|---|
| Topical (25 µL) | Intraperitoneal (5 µL/g) | |
| 1 × per day | 2 × per day | |
| Phage-Treated Group | PAM2H cocktail | PBS |
| Ceftazidime-Treated Group | PBS | Ceftazidime (CAZ) |
| Combination-Treated Group | PAM2H cocktail | Ceftazidime (CAZ) |
| Control-Treated Group | PBS | PBS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engeman, E.; Freyberger, H.R.; Corey, B.W.; Ward, A.M.; He, Y.; Nikolich, M.P.; Filippov, A.A.; Tyner, S.D.; Jacobs, A.C. Synergistic Killing and Re-Sensitization of Pseudomonas aeruginosa to Antibiotics by Phage-Antibiotic Combination Treatment. Pharmaceuticals 2021, 14, 184. https://doi.org/10.3390/ph14030184
Engeman E, Freyberger HR, Corey BW, Ward AM, He Y, Nikolich MP, Filippov AA, Tyner SD, Jacobs AC. Synergistic Killing and Re-Sensitization of Pseudomonas aeruginosa to Antibiotics by Phage-Antibiotic Combination Treatment. Pharmaceuticals. 2021; 14(3):184. https://doi.org/10.3390/ph14030184
Chicago/Turabian StyleEngeman, Emily, Helen R. Freyberger, Brendan W. Corey, Amanda M. Ward, Yunxiu He, Mikeljon P. Nikolich, Andrey A. Filippov, Stuart D. Tyner, and Anna C. Jacobs. 2021. "Synergistic Killing and Re-Sensitization of Pseudomonas aeruginosa to Antibiotics by Phage-Antibiotic Combination Treatment" Pharmaceuticals 14, no. 3: 184. https://doi.org/10.3390/ph14030184
APA StyleEngeman, E., Freyberger, H. R., Corey, B. W., Ward, A. M., He, Y., Nikolich, M. P., Filippov, A. A., Tyner, S. D., & Jacobs, A. C. (2021). Synergistic Killing and Re-Sensitization of Pseudomonas aeruginosa to Antibiotics by Phage-Antibiotic Combination Treatment. Pharmaceuticals, 14(3), 184. https://doi.org/10.3390/ph14030184








