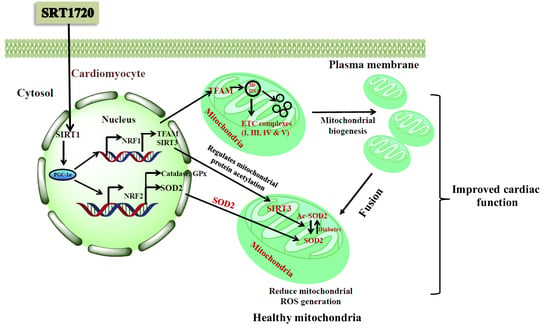
Sirt1 and Sirt3 Activation Improved Cardiac Function of Diabetic Rats via Modulation of Mitochondrial Function
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture
2.1.1. Treatment Conditions for Sirt1 Activation and Inhibition Study
2.1.2. Palmitate Induced Insulin Resistance in H9c2 Cells
2.1.3. Cellular-ROS Measurement
2.1.4. Crude Mitochondrial-ROS Measurement
2.1.5. Mitochondrial Content
2.1.6. Mitochondrial Membrane Potential
2.1.7. Evaluation of Mitochondrial Oxygen Consumption Rate
2.1.8. mRNA Expression Studies by RTPCR
2.1.9. Protein Expression
2.2. In-Vivo Study
2.2.1. Intraperitoneal Glucose Tolerance Test
2.2.2. Electrocardiography
2.2.3. Blood Pressure
2.2.4. Echocardiography
2.2.5. Serum Lipid Profile
2.2.6. Serum Free Fatty Acids
2.2.7. Serum Insulin
2.2.8. mRNA Expression Studies by Real-Time PCR
2.2.9. Protein Expression
2.2.10. Mitochondrial Protein Enrichment
2.2.11. Mitochondrial DNA Isolation
2.2.12. Nuclear Isolation
2.2.13. Histopathology Studies
2.3. Biochemical Assays
2.3.1. Evaluation of Sirt1 Activity
2.3.2. Evaluation of Sirt3 Activity
2.3.3. Antioxidant Parameters
SOD Activity
Catalase
Estimation of GSH
TBARS
2.4. Mitochondrial Enzymatic Assays
2.4.1. Citrate Synthase
2.4.2. β-Hydroxy acyl CoA Dehydrogenase
2.4.3. NADH Dehydrogenase
2.4.4. Succinate Dehydrogenase
2.4.5. Cytochrome-C Oxidase
Statistical Analysis of Data
3. Results
3.1. Sirtuin Activation Reduces Insulin Resistance in High Fructose Diet-Induced Pre-Diabetic Rats and Palmitate-Treated Cardiomyoblast Cells
3.2. Sirtuin Activation Reduces Serum Lipid Profile in High Fructose Diet-Fed Rats
3.3. Sirtuin Activation Improves Structural and Functional Characteristics of Heart in High Fructose Diet-Fed Rats
3.4. Sirtuin Activation Corrected Perturbed ECG Parameters in with High Fructose Diet-Fed Rats
3.5. Sirtuin Activation Reduces Elevated Blood Pressure in High Fructose Diet-Fed Rats
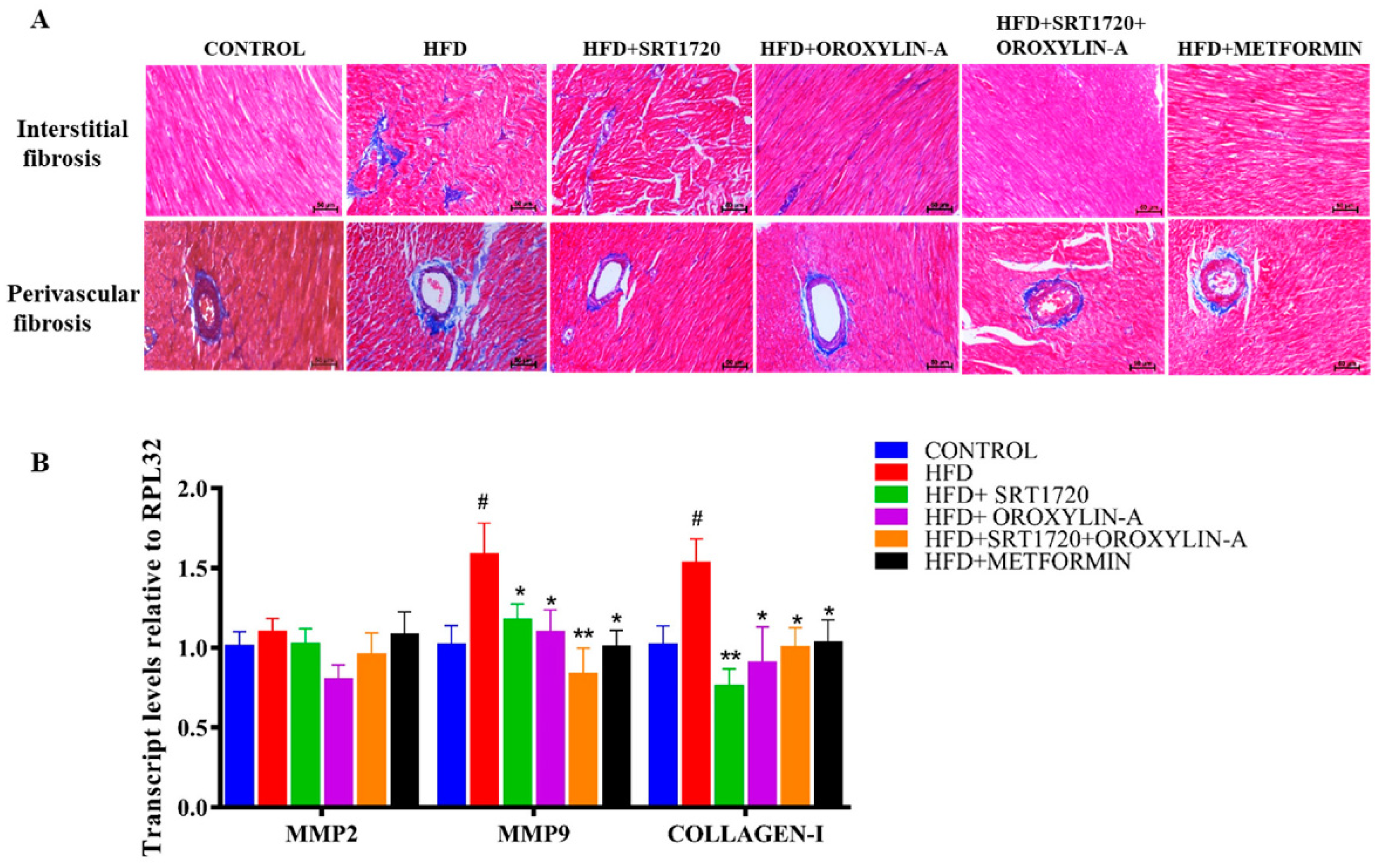
3.6. Sirtuin Activation Reduces Cardiac Fibrosis in High Fructose Diet-Fed Rats
3.7. Sirtuin Activation Reduces Fetal Cardiac Hypertrophic Gene Expression in High Fructose Diet-Fed Rats
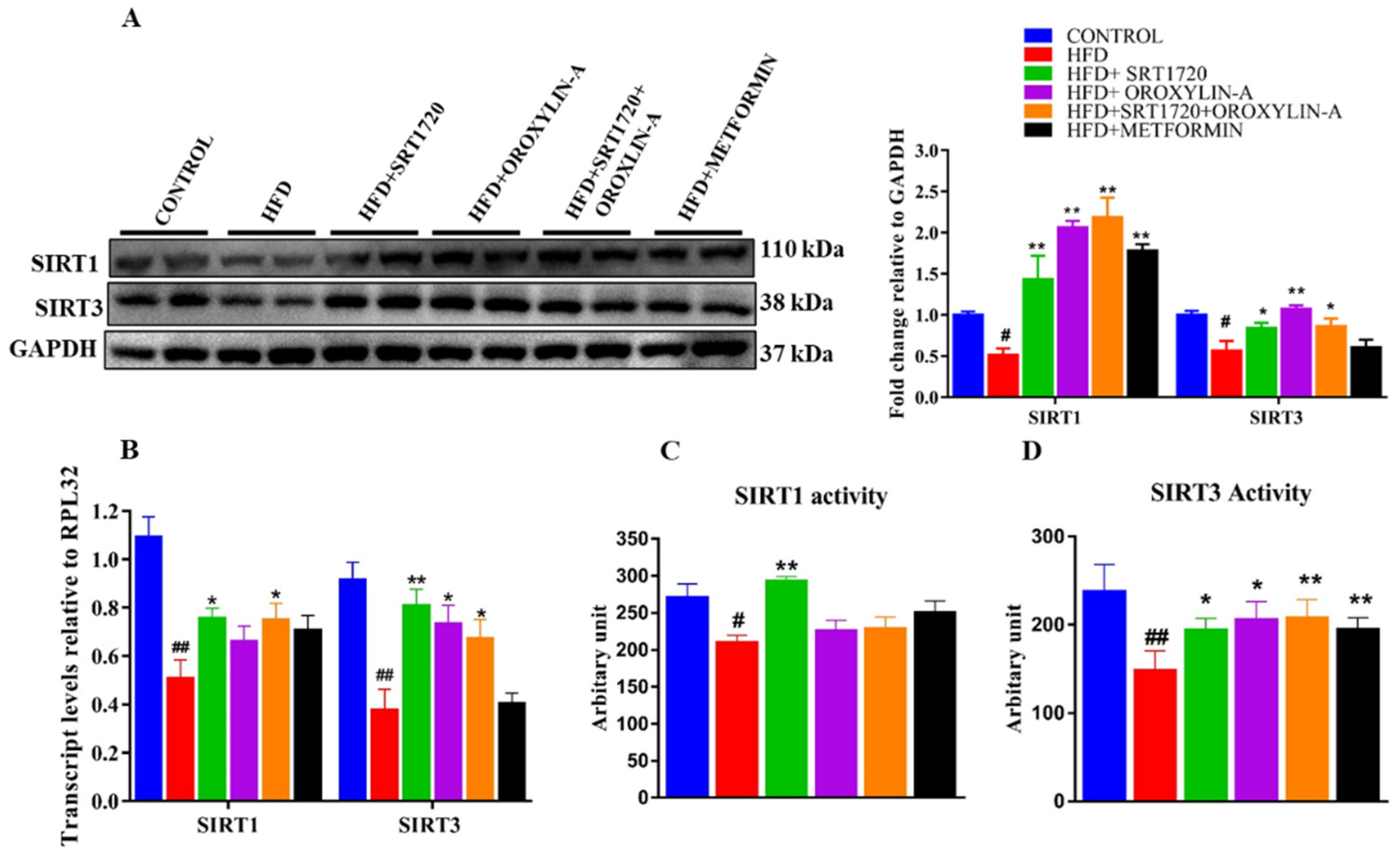
3.8. Sirtuin Activation Increases the Cardiac Expression and Activity of Sirt1 and Sirt3 in High Fructose Diet-Fed Rats
3.9. Sirtuin Activation Improves Cardiac Mitochondrial Biogenesis in High Fructose Diet-Fed Rats and Palmitic Acid (PA) Induced Insulin Resistant Cardiomyoblast Cells
3.10. Sirtuin Activation Improves Cardiac Mitochondrial DNA Encoded ETC Complex Gene and Protein Expression, and Their Activity in High Fructose Diet-Fed Rats
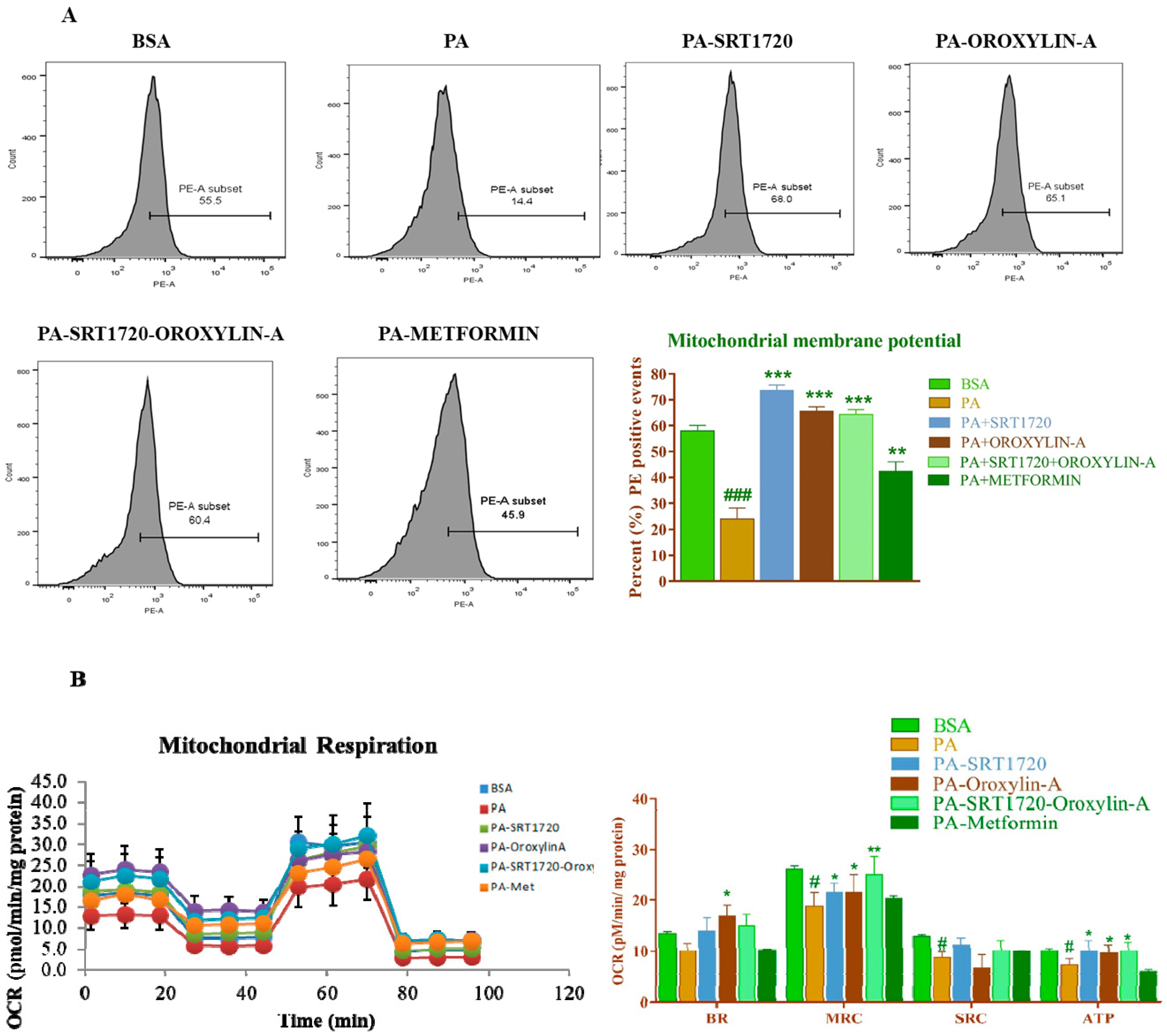
3.11. Sirtuin Modulation Improved Mitochondrial Membrane Potential Andoxygen Consumption Rate in Palmitate-Induced Insulin Resistance Cardiomyoblast Cells
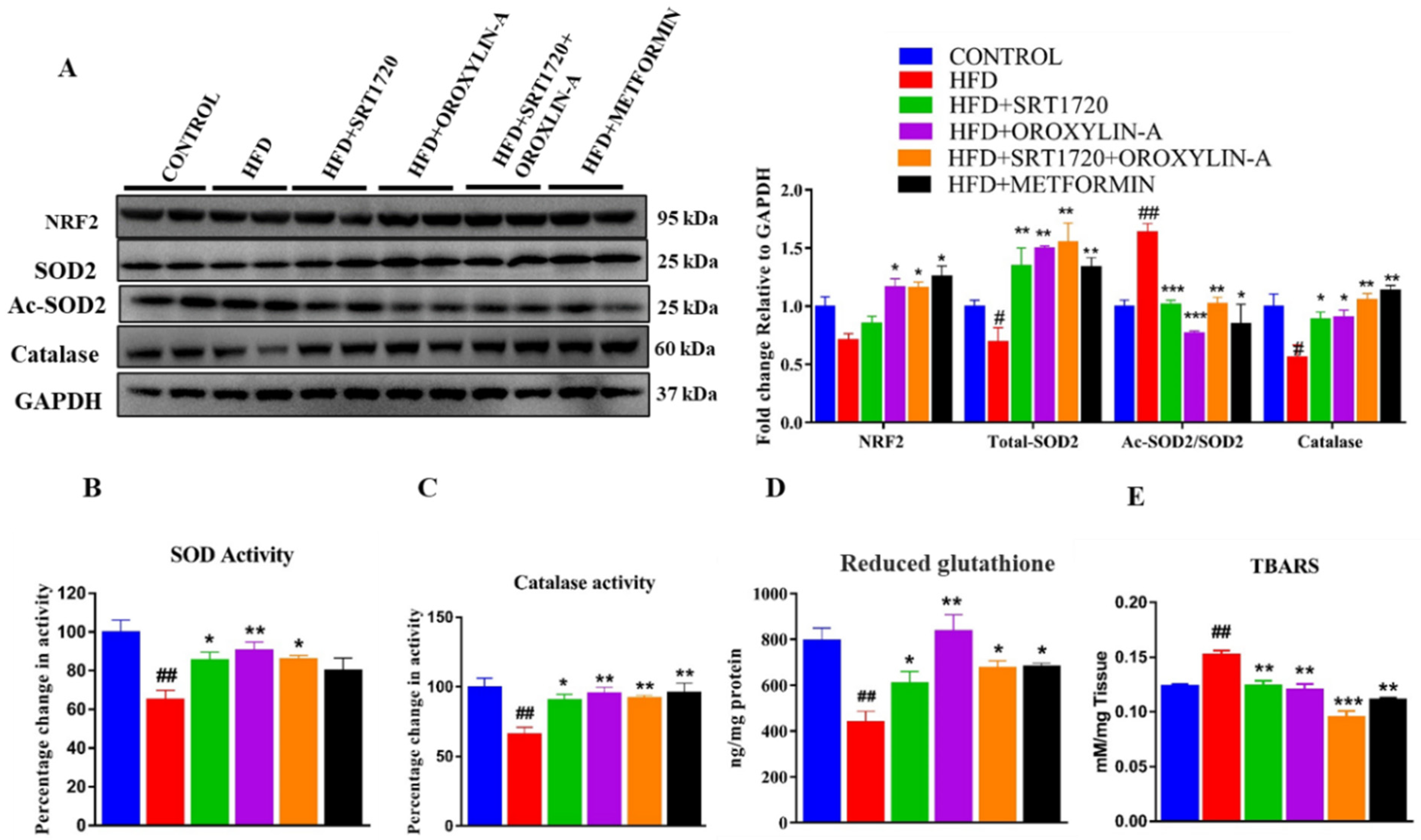
3.12. Sirtuin Activation Improves Cardiac Antioxidant Enzyme Expression and Activity and Reduces Oxidative Damage in High Fructose Diet-Fed Rats
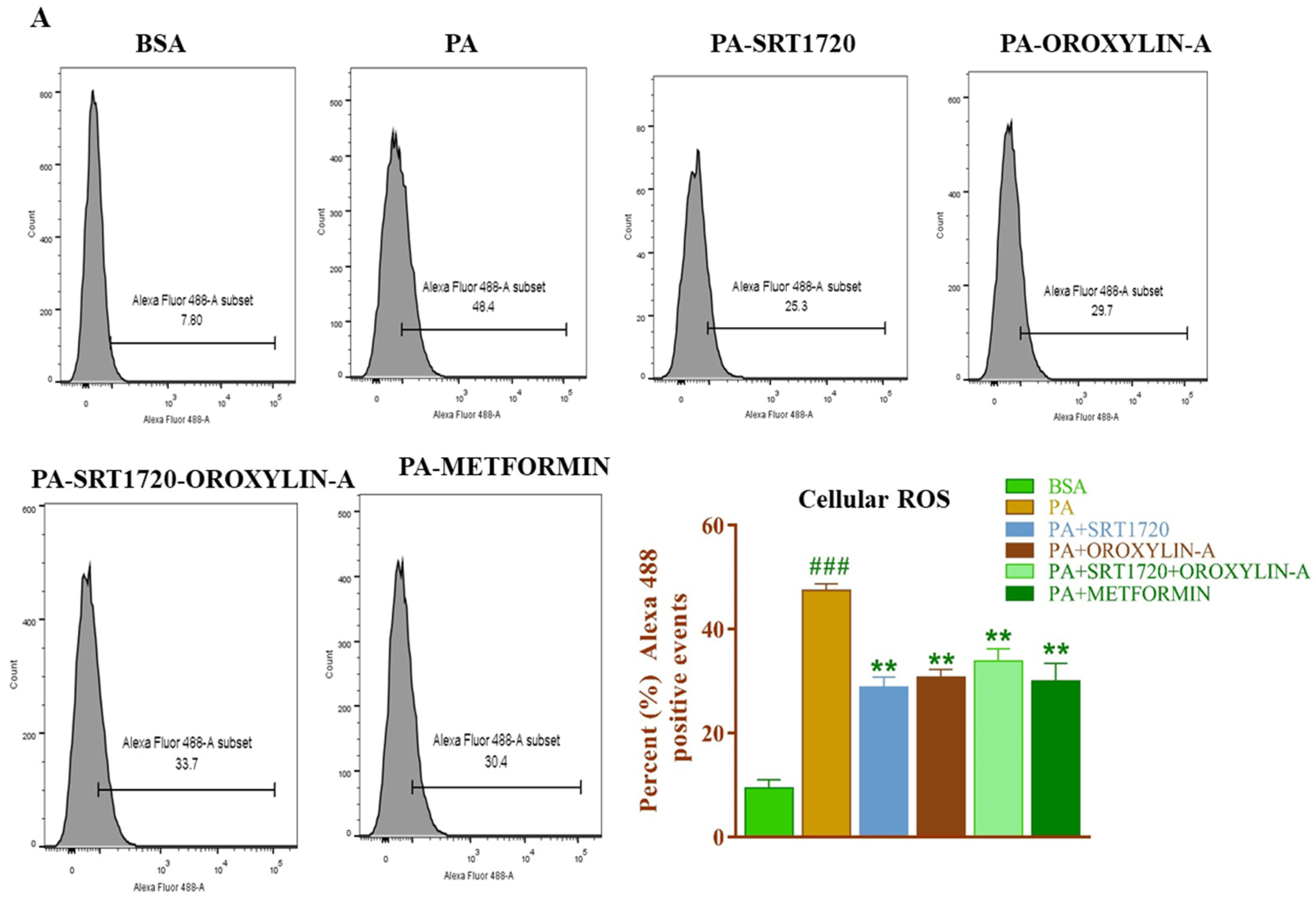
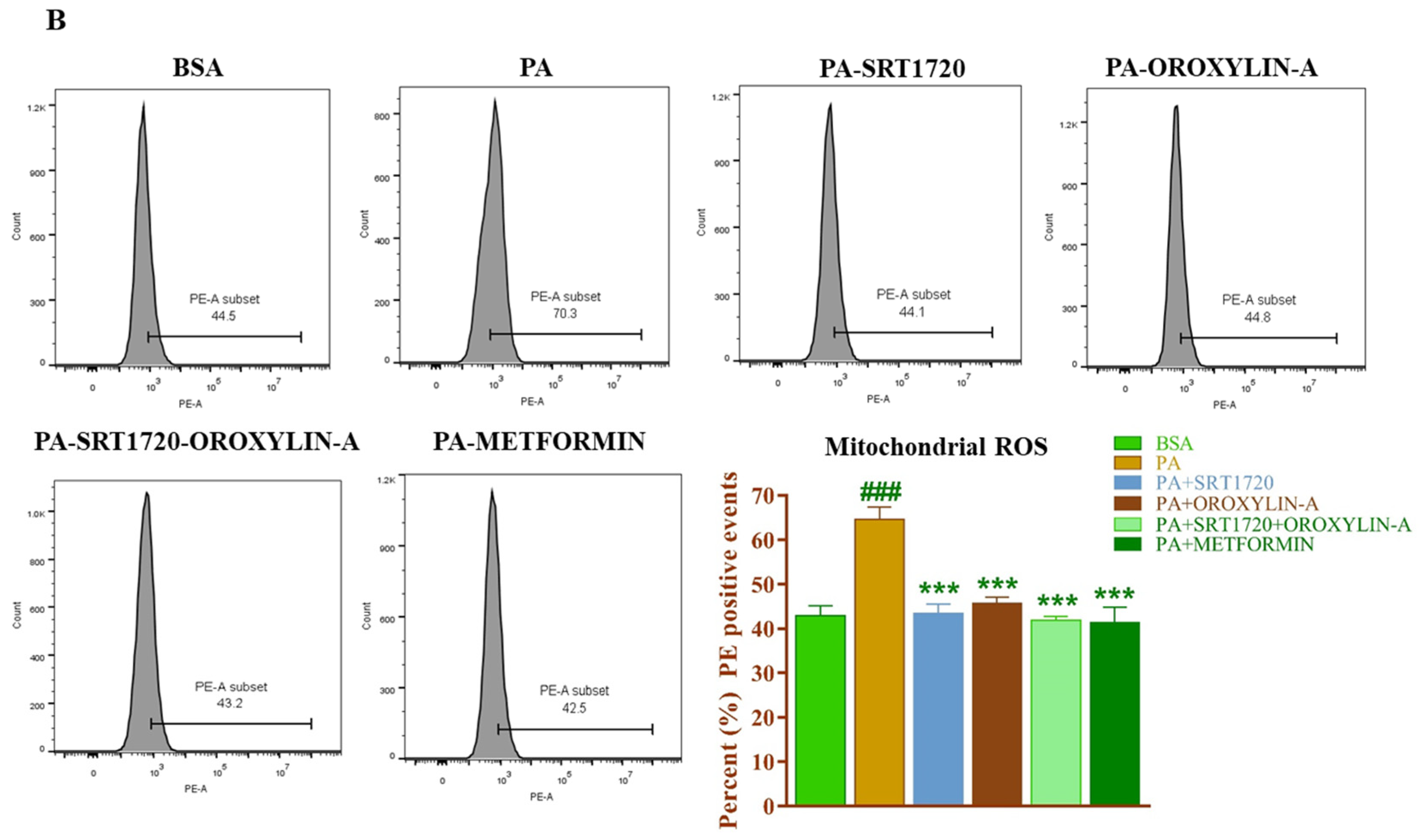
3.13. Effect of Sirtuin Activation on Palmitate-Induced Oxidative Stress in Rat Cardiomyoblast Cells
3.14. Sirtuin Activation Promotes Mitochondrial Fusion in High Fructose Diet-Fed Rats
3.15. Sirt1 Modulation Regulates the Mitochondrial Biogenesis-Related Gene and Protein Expression in Cardiomyoblast Cells
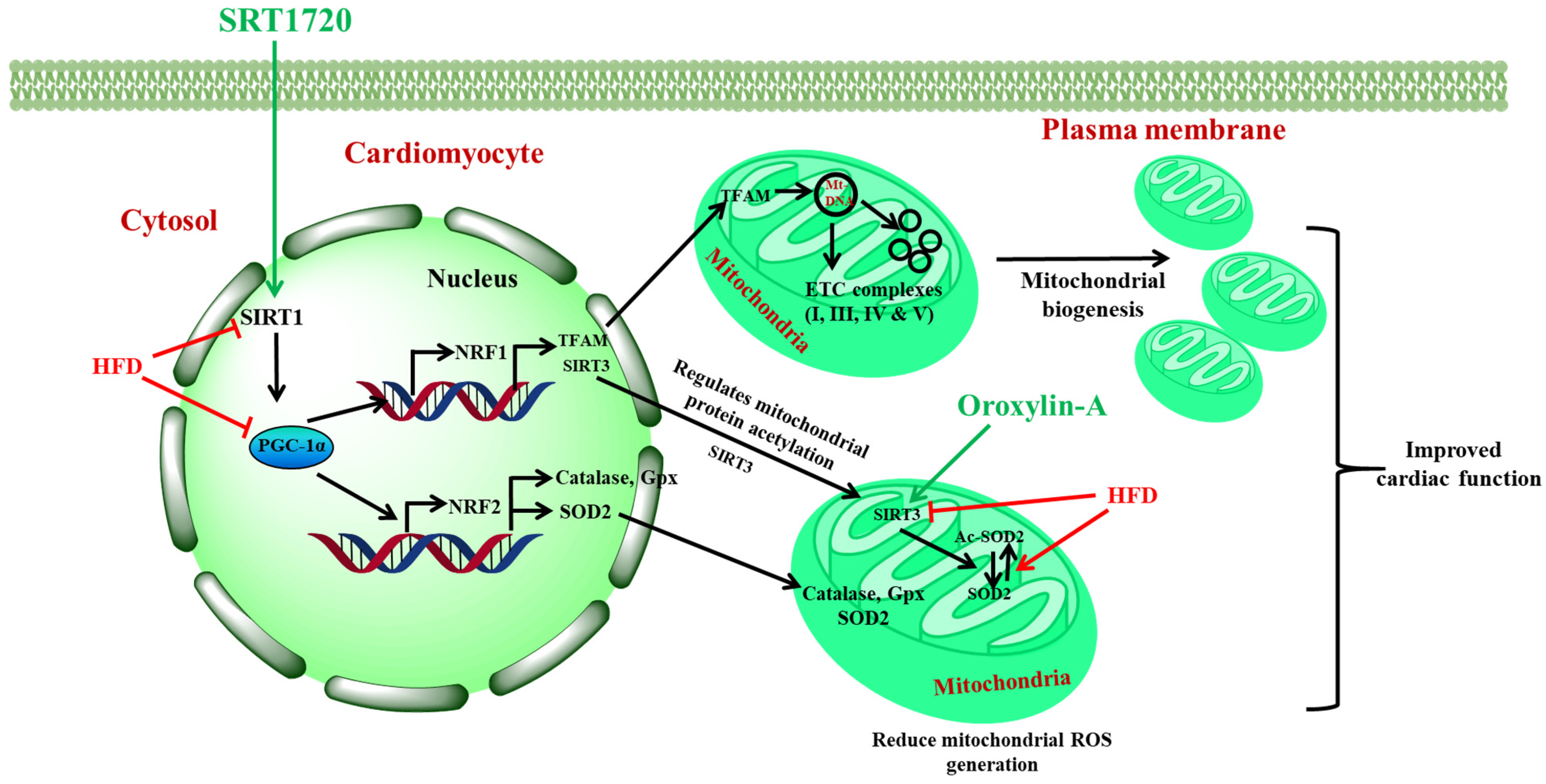
4. Discussion
5. Conclusions
6. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aditya, R.; Kiran, A.R.; Varma, D.S.; Vemuri, R.; Gundamaraju, R. A Review on SIRtuins in Diabetes. Curr. Pharm. Des. 2017, 23, 2299–2307. [Google Scholar] [CrossRef]
- European Society of Cardiology. Global Statistics on Diabetes. Available online: https://www.escardio.org/Education/Diabetes-and-CVD/Recommended-Reading/global-statistics-on-diabetes (accessed on 29 August 2020).
- Joshi, S.R.; Parikh, R.M. India—Diabetes Capital of the World: Now Heading towards Hypertension. J. Assoc. Physicians India 2007, 55, 323–324. [Google Scholar] [PubMed]
- Matheus, A.S.; Tannus, L.R.; Cobas, R.A.; Palma, C.C.; Negrato, C.A.; Gomes, M.B. Impact of Diabetes on Cardiovascular Disease: An Update. Int. J. Hypertens. 2013, 2013, 653789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yandrapalli, S.; Jolly, G.; Horblitt, A.; Sanaani, A.; Aronow, W.S. Cardiovascular Benefits and Safety of Non-Insulin Medications Used in the Treatment of Type 2 Diabetes Mellitus. Postgrad. Med. 2017, 129, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.A.; Hirschey, M.D. Mitochondrial Protein Acetylation Regulates Metabolism. Essays Biochem. 2012, 52, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Omidian, K.; Rafiei, H.; Bandy, B. Increased Mitochondrial Content and Function by Resveratrol and Select Flavonoids Protects against Benzo[a]Pyrene-Induced Bioenergetic Dysfunction and ROS Generation in a Cell Model of Neoplastic Transformation. Free Radic. Biol. Med. 2020, 152, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Kadiyala, C.; Zheng, L.; Du, Y.; Yohannes, E.; Kao, H.-Y.; Miyagi, M.; Kern, T. Acetylation of Retinal Histones in Diabetes Increases Inflammatory Proteins: Effects of Minocycline and Manipulation of Histone Acetyltransferase (HAT) and Histone Deacetylase (HDAC). J. Biol. Chem. 2012, 287, 25869–25880. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Xu, J.; Li, X.; Liu, Z.; Han, Y.; Xu, X.; Li, X.; Tang, Y.; Liu, Y.; Yu, T.; et al. Sirt3 Modulate Renal Ischemia-Reperfusion Injury through Enhancing Mitochondrial Fusion and Activating the ERK-OPA1 Signaling Pathway. J. Cell. Physiol. 2019, 234, 23495–23506. [Google Scholar] [CrossRef]
- Carnevale, I.; Pellegrini, L.; D’Aquila, P.; Saladini, S.; Lococo, E.; Polletta, L.; Vernucci, E.; Foglio, E.; Coppola, S.; Sansone, L.; et al. SIRT1-SIRT3 Axis Regulates Cellular Response to Oxidative Stress and Etoposide. J. Cell. Physiol. 2017, 232, 1835–1844. [Google Scholar] [CrossRef]
- Kwon, S.; Seok, S.; Yau, P.; Li, X.; Kemper, B.; Kemper, J.K. Obesity and Aging Diminish Sirtuin 1 (SIRT1)-Mediated Deacetylation of SIRT3, Leading to Hyperacetylation and Decreased Activity and Stability of SIRT3. J. Biol. Chem. 2017, 292, 17312–17323. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Dai, S.H.; Li, X.; Luo, P.; Zhu, J.; Wang, Y.H.; Fei, Z.; Jiang, X.F. Sirt1-Sirt3 Axis Regulates Human Blood-Brain Barrier Permeability in Response to Ischemia. Redox Biol. 2018, 14, 229–236. [Google Scholar] [CrossRef] [PubMed]
- De Moura, M.B.; Uppala, R.; Zhang, Y.; Van Houten, B.; Goetzman, E.S. Overexpression of Mitochondrial Sirtuins Alters Glycolysis and Mitochondrial Function in HEK293 Cells. PLoS ONE 2014, 9, e106028. [Google Scholar] [CrossRef] [Green Version]
- Cheung, K.G.; Cole, L.K.; Xiang, B.; Chen, K.; Ma, X.; Myal, Y.; Hatch, G.M.; Tong, Q.; Dolinsky, V.W. Sirtuin-3 (SIRT3) Protein Attenuates Doxorubicin-Induced Oxidative Stress and Improves Mitochondrial Respiration in H9c2 Cardiomyocytes. J. Biol. Chem. 2015, 290, 10981–10993. [Google Scholar] [CrossRef] [Green Version]
- Keskin, M.; Kurtoglu, S.; Kendirci, M.; Atabek, M.E.; Yazici, C. Homeostasis Model Assessment Is More Reliable Than the Fasting Glucose/Insulin Ratio and Quantitative Insulin Sensitivity Check Index for Assessing Insulin Resistance Among Obese Children and Adolescents. Pediatrics 2005, 115, e500–e503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagul, P.K.; Katare, P.B.; Bugga, P.; Dinda, A.K.; Banerjee, S.K. SIRT-3 Modulation by Resveratrol Improves Mitochondrial Oxidative Phosphorylation in Diabetic Heart through Deacetylation of TFAM. Cells 2018, 7, 235. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating Mitochondrial DAMPs Cause Inflammatory Responses to Injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.K.; McGaffin, K.R.; Huang, X.N.; Ahmad, F. Activation of Cardiac Hypertrophic Signaling Pathways in a Transgenic Mouse with the Human PRKAG2 Thr400Asn Mutation. Biochim. Biophys. Acta 2010, 1802, 284–291. [Google Scholar] [CrossRef] [Green Version]
- Aebi, H.; Wyss, S.R.; Scherz, B.; Skvaril, F. Heterogeneity of Erythrocyte Catalase II. Isolation and Characterization of Normal and Variant Erythrocyte Catalase and Their Subunits. Eur. J. Biochem. 1974, 48, 137–145. [Google Scholar] [CrossRef]
- Ellman, G.L. Tissue Sulfhydryl Groups. Arch Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Mukherjee, S.; Banerjee, S.K.; Maulik, M.; Dinda, A.K.; Talwar, K.K.; Maulik, S.K. Protection against Acute Adriamycin-Induced Cardiotoxicity by Garlic: Role of Endogenous Antioxidants and Inhibition of TNF-Alpha Expression. BMC Pharmacol. 2003, 3, 16. [Google Scholar] [CrossRef] [Green Version]
- Civitarese, A.E.; Carling, S.; Heilbronn, L.K.; Hulver, M.H.; Ukropcova, B.; Deutsch, W.A.; Smith, S.R.; Ravussin, E.; Calerie Pennington Team. Calorie Restriction Increases Muscle Mitochondrial Biogenesis in Healthy Humans. PLoS Med. 2007, 4, e76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Prakash, A.; Dogra, S. Centella Asiatica Attenuates D-Galactose-Induced Cognitive Impairment, Oxidative and Mitochondrial Dysfunction in Mice. Int. J. Alzheimer’s Dis. 2011, 2011, 347569. [Google Scholar] [CrossRef] [Green Version]
- Shah, M.S.; Brownlee, M. Molecular and Cellular Mechanisms of Cardiovascular Disorders in Diabetes. Circ. Res. 2016, 118, 1808–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenny, H.C.; Abel, E.D. Heart Failure in Type 2 Diabetes Mellitus. Circ. Res. 2019, 124, 121–141. [Google Scholar] [CrossRef]
- Bagul, P.K.; Deepthi, N.; Sultana, R.; Banerjee, S.K. Resveratrol Ameliorates Cardiac Oxidative Stress in Diabetes through Deacetylation of NFkB-P65 and Histone 3. J. Nutr. Biochem. 2015, 26, 1298–1307. [Google Scholar] [CrossRef]
- Chatterjee, B.; Ghosh, K.; Kanade, S.R. Resveratrol Modulates Epigenetic Regulators of Promoter Histone Methylation and Acetylation That Restores BRCA1, P53, P21CIP1 in Human Breast Cancer Cell Lines. BioFactors 2019, 45, 818–829. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, F.; Ge, X.; Yan, T.; Chen, X.; Shi, X.; Zhai, Q. SIRT1 Improves Insulin Sensitivity under Insulin-Resistant Conditions by Repressing PTP1B. Cell Metab. 2007, 6, 307–319. [Google Scholar] [CrossRef] [Green Version]
- Libri, V.; Brown, A.P.; Gambarota, G.; Haddad, J.; Shields, G.S.; Dawes, H.; Pinato, D.J.; Hoffman, E.; Elliot, P.J.; Vlasuk, G.P.; et al. A Pilot Randomized, Placebo Controlled, Double Blind Phase I Trial of the Novel SIRT1 Activator SRT2104 in Elderly Volunteers. PLoS ONE 2012, 7, e51395. [Google Scholar] [CrossRef]
- Pacholec, M.; Bleasdale, J.E.; Chrunyk, B.; Cunningham, D.; Flynn, D.; Garofalo, R.S.; Griffith, D.; Griffor, M.; Loulakis, P.; Pabst, B.; et al. SRT1720, SRT2183, SRT1460, and Resveratrol Are Not Direct Activators of SIRT1. J. Biol. Chem. 2010, 285, 8340–8351. [Google Scholar] [CrossRef] [Green Version]
- Nishida, K.; Matsushita, T.; Takayama, K.; Tanaka, T.; Miyaji, N.; Ibaraki, K.; Araki, D.; Kanzaki, N.; Matsumoto, T.; Kuroda, R. Intraperitoneal Injection of the SIRT1 Activator SRT1720 Attenuates the Progression of Experimental Osteoarthritis in Mice. Bone Jt. Res. 2018, 7, 252–262. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Chen, H.; Mak, C.; Zaky, A.; Pollock, C.; Saad, S. SRT1720 Attenuates Obesity and Insulin Resistance but Not Liver Damage in the Offspring Due to Maternal and Postnatal High-Fat Diet Consumption. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E196–E203. [Google Scholar] [CrossRef]
- Wei, L.; Zhou, Y.; Dai, Q.; Qiao, C.; Zhao, L.; Hui, H.; Lu, N.; Guo, Q.L. Oroxylin A Induces Dissociation of Hexokinase II from the Mitochondria and Inhibits Glycolysis by SIRT3-Mediated Deacetylation of Cyclophilin D in Breast Carcinoma. Cell Death Dis. 2013, 4, e601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Zhou, Y.; Qiao, C.; Ni, T.; Li, Z.; You, Q.; Guo, Q.; Lu, N. Oroxylin A Inhibits Glycolysis-Dependent Proliferation of Human Breast Cancer via Promoting SIRT3-Mediated SOD2 Transcription and HIF1alpha Destabilization. Cell Death Dis. 2015, 6, e1714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhang, J.; Lin, Y.; Lei, Q.; Guan, K.L.; Zhao, S.; Xiong, Y. Tumour Suppressor SIRT3 Deacetylates and Activates Manganese Superoxide Dismutase to Scavenge ROS. EMBO Rep. 2011, 12, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Minor, R.K.; Baur, J.A.; Gomes, A.P.; Ward, T.M.; Csiszar, A.; Mercken, E.M.; Abdelmohsen, K.; Shin, Y.K.; Canto, C.; Scheibye-Knudsen, M.; et al. SRT1720 Improves Survival and Healthspan of Obese Mice. Sci. Rep. 2011, 1, 70. [Google Scholar] [CrossRef] [Green Version]
- Jing, E.; Emanuelli, B.; Hirschey, M.D.; Boucher, J.; Lee, K.Y.; Lombard, D.; Verdin, E.M.; Kahn, C.R. Sirtuin-3 (Sirt3) Regulates Skeletal Muscle Metabolism and Insulin Signaling via Altered Mitochondrial Oxidation and Reactive Oxygen Species Production. Proc. Natl. Acad. Sci. USA 2011, 108, 14608–14613. [Google Scholar] [CrossRef] [Green Version]
- Lantier, L.; Williams, A.S.; Williams, I.M.; Yang, K.K.; Bracy, D.P.; Goelzer, M.; James, F.D.; Gius, D.; Wasserman, D.H. SIRT3 Is Crucial for Maintaining Skeletal Muscle Insulin Action and Protects Against Severe Insulin Resistance in High-Fat-Fed Mice. Diabetes 2015, 64, 3081–3092. [Google Scholar] [CrossRef] [Green Version]
- VanHoose, L.; Sawers, Y.; Loganathan, R.; Vacek, J.L.; Stehno-Bittel, L.; Novikova, L.; Al-Jarrah, M.; Smirnova, I.V. Electrocardiographic Changes with the Onset of Diabetes and the Impact of Aerobic Exercise Training in the Zucker Diabetic Fatty (ZDF) Rat. Cardiovasc. Diabetol. 2010, 9, 56. [Google Scholar] [CrossRef] [Green Version]
- Grigorescu, E.D.; Lacatusu, C.M.; Floria, M.; Mihai, B.M.; Cretu, I.; Sorodoc, L. Left Ventricular Diastolic Dysfunction in Type 2 Diabetes-Progress and Perspectives. Diagnostics 2019, 9, 121. [Google Scholar] [CrossRef] [Green Version]
- Davarpasand, T.; Hosseinsabet, A.; Omidi, F.; Mehrabi-Pari, S. Interaction Effect of Diabetes and Acute Myocardial Infarction on the Left Atrial Function as Evaluated by 2-D Speckle-Tracking Echocardiography. Ultrasound Med. Biol. 2020, 46, 1490–1503. [Google Scholar] [CrossRef]
- Bagul, P.K.; Dinda, A.K.; Banerjee, S.K. Effect of Resveratrol on Sirtuins Expression and Cardiac Complications in Diabetes. Biochem. Biophys. Res. Commun. 2015, 468, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Thaung, H.P.A.; Baldi, J.C.; Wang, H.-Y.; Hughes, G.; Cook, R.F.; Bussey, C.T.; Sheard, P.W.; Bahn, A.; Jones, P.P.; Schwenke, D.O.; et al. Increased Efferent Cardiac Sympathetic Nerve Activity and Defective Intrinsic Heart Rate Regulation in Type 2 Diabetes. Diabetes 2015, 64, 2944–2956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luongo, T.S.; Lambert, J.P.; Gross, P.; Nwokedi, M.; Lombardi, A.A.; Shanmughapriya, S.; Carpenter, A.C.; Kolmetzky, D.; Gao, E.; van Berlo, J.H.; et al. The Mitochondrial Na+/Ca2+ Exchanger Is Essential for Ca2+ Homeostasis and Viability. Nature 2017, 545, 93–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, R.; Srodulski, S.; Peng, X.; Margulies, K.B.; Despa, F.; Despa, S. Intracellular Na+ Concentration ([Na+]i) Is Elevated in Diabetic Hearts Due to Enhanced Na+-Glucose Cotransport. J. Am. Heart Assoc. 2015, 4, e002183. [Google Scholar] [CrossRef] [Green Version]
- Pierce, G.N.; Dhalla, N.S. Sarcolemmal Na+-K+-ATPase Activity in Diabetic Rat Heart. Am. J. Physiol. 1983, 245, C241–C247. [Google Scholar] [CrossRef]
- Szűcs, G.; Sója, A.; Péter, M.; Sárközy, M.; Bruszel, B.; Siska, A.; Földesi, I.; Szabó, Z.; Janáky, T.; Vígh, L.; et al. Prediabetes Induced by Fructose-Enriched Diet Influences Cardiac Lipidome and Proteome and Leads to Deterioration of Cardiac Function Prior to the Development of Excessive Oxidative Stress and Cell Damage. Available online: https://www.hindawi.com/journals/omcl/2019/3218275/ (accessed on 11 December 2020).
- Park, J.H.; Ku, H.J.; Kim, J.K.; Park, J.-W.; Lee, J.H. Amelioration of High Fructose-Induced Cardiac Hypertrophy by Naringin. Sci. Rep. 2018, 8, 9464. [Google Scholar] [CrossRef] [Green Version]
- Reaven, G.M.; Ho, H.; Hoffman, B.B. Attenuation of Fructose-Induced Hypertension in Rats by Exercise Training. Hypertension 1988, 12, 129–132. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Amlal, H.; Haas, P.J.; Dringenberg, U.; Fussell, S.; Barone, S.L.; Engelhardt, R.; Zuo, J.; Seidler, U.; Soleimani, M. Fructose-Induced Hypertension: Essential Role of Chloride and Fructose Absorbing Transporters PAT1 and Glut5. Kidney Int. 2008, 74, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Shamhart, P.E.; Luther, D.J.; Adapala, R.K.; Bryant, J.E.; Petersen, K.A.; Meszaros, J.G.; Thodeti, C.K. Hyperglycemia Enhances Function and Differentiation of Adult Rat Cardiac Fibroblasts. Can. J. Physiol. Pharmacol. 2014, 92, 598–604. [Google Scholar] [CrossRef] [Green Version]
- Patti, M.E.; Corvera, S. The Role of Mitochondria in the Pathogenesis of Type 2 Diabetes. Endocr. Rev. 2010, 31, 364–395. [Google Scholar] [CrossRef] [Green Version]
- Montaigne, D.; Marechal, X.; Coisne, A.; Debry, N.; Modine, T.; Fayad, G.; Potelle, C.; El Arid, J.M.; Mouton, S.; Sebti, Y.; et al. Myocardial Contractile Dysfunction Is Associated with Impaired Mitochondrial Function and Dynamics in Type 2 Diabetic but Not in Obese Patients. Circulation 2014, 130, 554–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, M.R.; Bagul, P.K.; Katare, P.B.; Anwar Mohammed, S.; Padiya, R.; Banerjee, S.K. Garlic Activates SIRT-3 to Prevent Cardiac Oxidative Stress and Mitochondrial Dysfunction in Diabetes. Life Sci. 2016, 164, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Khalek, W.A.; Cortade, F.; Ollendorff, V.; Lapasset, L.; Tintignac, L.; Chabi, B.; Wrutniak-Cabello, C. SIRT3, a Mitochondrial NAD+-Dependent Deacetylase, Is Involved in the Regulation of Myoblast Differentiation. PLoS ONE 2014, 9, e114388. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Wang, R.; Xue, Y.; Liu, X.; Zhang, H.-B.; Chen, Y.; Fang, F.; Chang, Y. Sirtuin 3, a New Target of PGC-1α, Plays an Important Role in the Suppression of ROS and Mitochondrial Biogenesis. PLoS ONE 2010, 5, e11707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemire, B. WormBook: The Online Review of C. elegans Biology [Internet]. 14 September 2005. Available online: https://www.ncbi.nlm.nih.gov/books/NBK19694/ (accessed on 29 August 2020).
- Fex, M.; Nicholas, L.M.; Vishnu, N.; Medina, A.; Sharoyko, V.V.; Nicholls, D.G.; Spégel, P.; Mulder, H. The Pathogenetic Role of β-Cell Mitochondria in Type 2 Diabetes. J. Endocrinol. 2018, 236, R145–R159. [Google Scholar] [CrossRef] [Green Version]
- Galloway, C.A.; Yoon, Y. Mitochondrial Dynamics in Diabetic Cardiomyopathy. Antioxid. Redox Signal. 2015, 22, 1545–1562. [Google Scholar] [CrossRef] [Green Version]
- Zorzano, A.; Liesa, M.; Palacin, M. Mitochondrial Dynamics as a Bridge between Mitochondrial Dysfunction and Insulin Resistance. Arch. Physiol. Biochem. 2009, 115, 1–12. [Google Scholar] [CrossRef]
- Rehman, K.; Akash, M.S.H. Mechanism of Generation of Oxidative Stress and Pathophysiology of Type 2 Diabetes Mellitus: How Are They Interlinked? J. Cell. Biochem. 2017, 118, 3577–3585. [Google Scholar] [CrossRef]
- Mendes, F.; Martins, D. The Impact of Exercise on Inflammation, Oxidative Stress and Remodelling In Cardiac Muscle. Arq. Bras. Cardiol. 2020, 114, 106–108. [Google Scholar] [CrossRef]
- Jukarainen, S.; Heinonen, S.; Rämö, J.T.; Rinnankoski-Tuikka, R.; Rappou, E.; Tummers, M.; Muniandy, M.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; et al. Obesity Is Associated With Low NAD(+)/SIRT Pathway Expression in Adipose Tissue of BMI-Discordant Monozygotic Twins. J. Clin. Endocrinol. Metab. 2016, 101, 275–283. [Google Scholar] [CrossRef] [Green Version]





| Parameter | Control | HFD | HFD + SRT1720 | HFD + Oroxylin-A | HFD + SRT1720 + Oroxylin-A | HFD + Metformin |
|---|---|---|---|---|---|---|
| Triglycerides. (mg/dL) | 81.5 ± 17.85 | 158 ± 24.2 ## | 70.5 ± 10.3 *** | 52.3 ± 10.3 *** | 75.5 ± 31.6 *** | 54.0 ± 12.6 *** |
| Cholesterol (mg/dL) | 77.5 ± 4.46 | 96.3 ± 4.0 ## | 84.6 ± 7.03 * | 9.47 ± 9.7 | 86.8 ± 12.6 * | 97 ± 14.7 |
| LDL (mg/dL) | 16 ± 2.92 | 23.4 ± 2.7 # | 18.44 ± 2.0 * | 19.8 ± 2.4 | 18.6 ± 2.3 * | 19.2 ± 1.79 |
| HDL (mg/dL) | 78.8. ±4.39 | 78.1 ± 1.6 | 86 ± 3.2 | 91.8 ± 3.8 * | 79.8 ± 3.8 | 93.8 ± 5.06 * |
| Free fatty acid (mM/L) | 0.84 ± 0.12 | 1.38 ± 0.15 ## | 0.97 ± 0.12 * | 0.88 ± 0.07 * | 1.08 ± 0.19 * | 0.81 ± 0.19 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paramesha, B.; Anwar, M.S.; Meghwani, H.; Maulik, S.K.; Arava, S.K.; Banerjee, S.K. Sirt1 and Sirt3 Activation Improved Cardiac Function of Diabetic Rats via Modulation of Mitochondrial Function. Antioxidants 2021, 10, 338. https://doi.org/10.3390/antiox10030338
Paramesha B, Anwar MS, Meghwani H, Maulik SK, Arava SK, Banerjee SK. Sirt1 and Sirt3 Activation Improved Cardiac Function of Diabetic Rats via Modulation of Mitochondrial Function. Antioxidants. 2021; 10(3):338. https://doi.org/10.3390/antiox10030338
Chicago/Turabian StyleParamesha, Bugga, Mohammed Soheb Anwar, Himanshu Meghwani, Subir Kumar Maulik, Sudheer Kumar Arava, and Sanjay K Banerjee. 2021. "Sirt1 and Sirt3 Activation Improved Cardiac Function of Diabetic Rats via Modulation of Mitochondrial Function" Antioxidants 10, no. 3: 338. https://doi.org/10.3390/antiox10030338