Subclinical Mastitis Related to Streptococcus canis Infection in Dairy Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Herd Description
2.2. Sample Collection and Bacteriological Culture and SCCs
2.3. MALDI-TOF MS Methodology
2.4. Antimicrobial Susceptibilities Test
2.5. Diagnostic PCR Analyses
2.6. Genotyping and Genomic Characterization
3. Results
3.1. First Isolation
3.2. Follow-Up
3.3. Second Isolation
3.4. Biomolecular Analysis
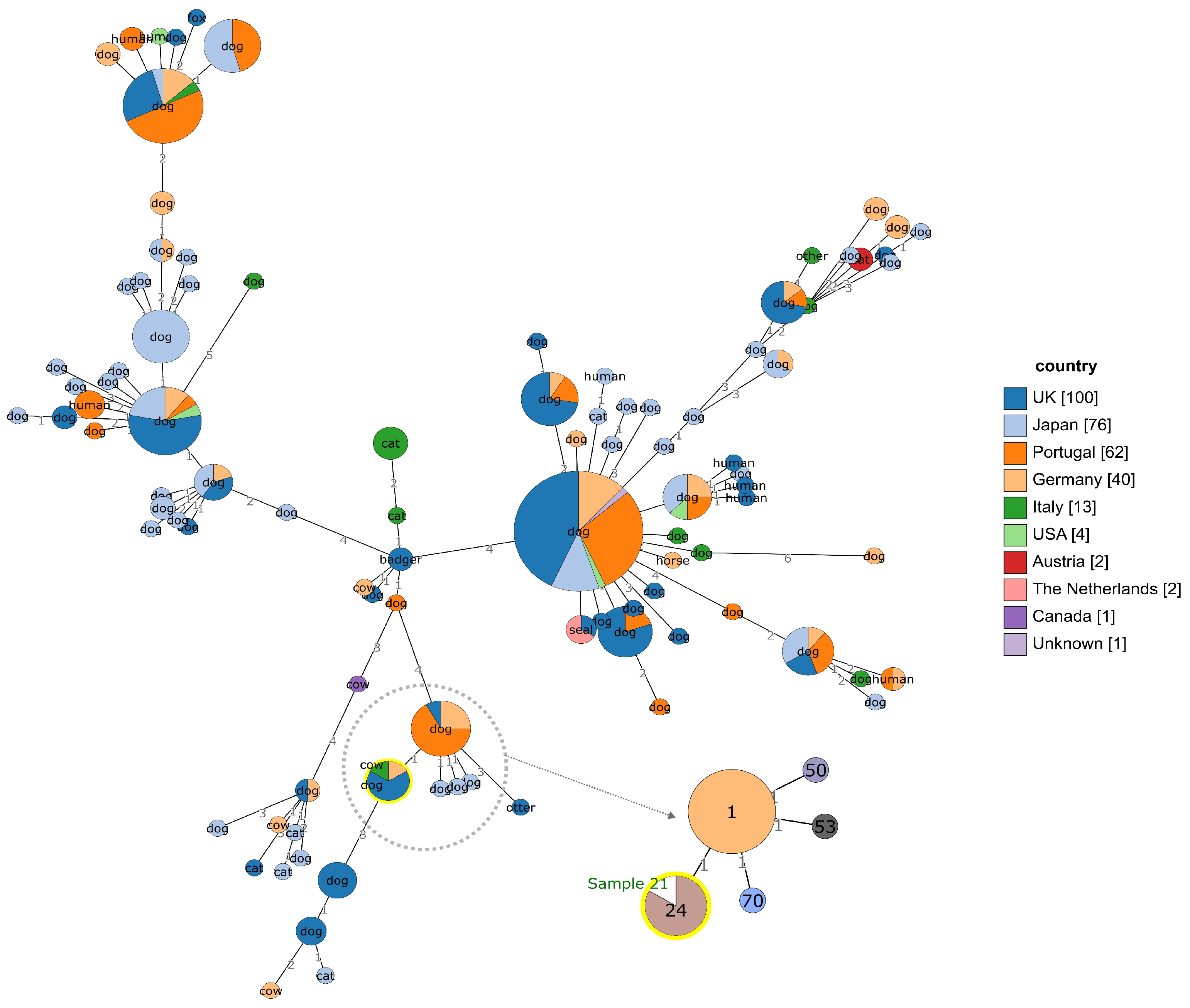
3.5. Genotyping and Genomic Characterization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Devriese, L.A.; Hommez, J.; Kilpper-Balz, R.; Schleifer, K.-H. Streptococcus canis Sp. Nov.: A Species of Group G Streptococci from Animals. Int. J. Syst. Bacteriol. 1986, 36, 422–425. [Google Scholar] [CrossRef]
- Kruger, E.F.; Byrne, B.A.; Pesavento, P.; Hurley, K.F.; Lindsay, L.L.; Sykes, J.E. Relationship between Clinical Manifestations and Pulsed-Field Gel Profiles of Streptococcus canis Isolates from Dogs and Cats. Vet. Microbiol. 2010, 146, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, P.A.; Bannasch, M.J.; Bachmann, R.; Byrne, B.A.; Hurley, K.F. Fatal Streptococcus canis Infections in Intensively Housed Shelter Cats. Vet. Pathol. 2007, 44, 218–221. [Google Scholar] [CrossRef]
- Iglauer, F.; Kunstýr, I.; Mörstedt, R.; Farouq, H.; Wullenweber, M.; Damsch, S. Streptococcus canis Arthritis in a Cat Breeding Colony. J. Exp. Anim. Sci. 1991, 34, 59–65. [Google Scholar]
- Whatmore, A.M.; Engler, K.H.; Gudmundsdottir, G.; Efstratiou, A. Identification of Isolates of Streptococcus canis Infecting Humans. J. Clin. Microbiol. 2001, 39, 4196–4199. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.; Clarridge, J.E.; Young, E.J.; Mizuki, S. The Other Group G Streptococcus: Increased Detection of Streptococcus canis Ulcer Infections in Dog Owners. J. Clin. Microbiol. 2007, 45, 2327–2329. [Google Scholar] [CrossRef]
- Olson, P.S. Streptococcus canis: An Isolate from a Canine Uterus. Vet. Med. Small Anim. Clin. 1975, 70, 933–934. [Google Scholar]
- Corning, B.F.; Murphy, J.C.; Fox, J.G. Group G Streptococcal Lymphadenitis in Rats. J. Clin. Microbiol. 1991, 29, 2720–2723. [Google Scholar] [CrossRef]
- Eberhart, R.J.; Guss, S.B. Group G Streptococci in the Udders of a Pennsylvania Dairy Herd. J. Am. Vet. Med. Assoc. 1970, 157, 1195–1199. [Google Scholar]
- Król, J.; Twardoń, J.; Mrowiec, J.; Podkowik, M.; Dejneka, G.; Debski, B.; Nowicki, T.; Zalewski, W. Short Communication: Streptococcus canis Is Able to Establish a Persistent Udder Infection in a Dairy Herd. J. Dairy Sci. 2015, 98, 7090–7096. [Google Scholar] [CrossRef]
- Wilson, D.J.; Gonzalez, R.N.; Das, H.H. Bovine Mastitis Pathogens in New York and Pennsylvania: Prevalence and Effects on Somatic Cell Count and Milk Production. J. Dairy Sci. 1997, 80, 2592–2598. [Google Scholar] [CrossRef] [PubMed]
- McDonald, T.J.; McDonald, J.S. Streptococci Isolated from Bovine Intramammary Infections. Am. J. Vet. Res. 1976, 37, 377–381. [Google Scholar] [PubMed]
- Tikofsky, L.L.; Zadoks, R.N. Cross-Infection Between Cats and Cows: Origin and Control of Streptococcus canis Mastitis in a Dairy Herd. J. Dairy Sci. 2005, 88, 2707–2713. [Google Scholar] [CrossRef]
- Watts, J.L.; Nickerson, S.C.; Pankey, J.W. A Case Study of Streptococcus Group G Infection in a Dairy Herd. Vet. Microbiol. 1984, 9, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.A.; Stark, D.M. Occurrence and Characterization of Lancefield Group G Streptococci in Bovine Mastitis. Am. J. Vet. Res. 1970, 31, 397–398. [Google Scholar]
- National Mastitis Council. Laboratory Handbook on Bovine Mastitis, 3rd ed.; National Mastitis Council: Verona, WI, USA, 2017. [Google Scholar]
- Barcelos, M.M.; Martins, L.; Grenfell, R.C.; Juliano, L.; Anderson, K.L.; dos Santos, M.V.; Gonçalves, J.L. Comparison of Standard and On-Plate Extraction Protocols for Identification of Mastitis-Causing Bacteria by MALDI-TOF MS. Braz. J. Microbiol. 2019, 50, 849–857. [Google Scholar] [CrossRef]
- CLSI. VET01 Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th Edition, 3rd ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Farina, D.; Bianco, A.; Manzulli, V.; Castellana, S.; Parisi, A.; Caruso, M.; Fraccalvieri, R.; Serrecchia, L.; Rondinone, V.; Pace, L.; et al. Antimicrobial and Phylogenomic Characterization of Bacillus cereus Group Strains Isolated from Different Food Sources in Italy. Antibiotics 2024, 13, 898. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC. A Quality Control Tool for High Throughput Sequence Data [Computer Software]. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 September 2024).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced Multi-Sample Quality Control for High-Throughput Sequencing Data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Nybakken, E.J.; Oppegaard, O.; Gilhuus, M.; Jensen, C.S.; Mylvaganam, H. Identification of Streptococcus Dysgalactiae Using Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry; Refining the Database for Improved Identification. Diagn. Microbiol. Infect. Dis. 2021, 99, 115207. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, M.; Friedman, S.; Saran, A.; Younis, A. An Outbreak of Streptococcus canis Mastitis in a Dairy Herd in Israel. N. Z. Vet. J. 2005, 53, 261–264. [Google Scholar] [CrossRef]
- Hassan, A.A.; Akineden, Ö.; Usleber, E. Identification of Streptococcus canis Isolated from Milk of Dairy Cows with Subclinical Mastitis. J. Clin. Microbiol. 2005, 43, 1234–1238. [Google Scholar] [CrossRef]
- Kovacs, P.; Kis Tamas, E.; Fekete, L.; Szita, G.; Jurkovich, V.; Brydl, E.; Koenyves, L. Outbreak of Streptococcus canis Mastitis in a Hungarian Large Scale Dairy Herd. Magy. Allatorvosok Lapja 2016, 138, 133–139. [Google Scholar]
- Richards, V.P.; Zadoks, R.N.; Pavinski Bitar, P.D.; Lefébure, T.; Lang, P.; Werner, B.; Tikofsky, L.; Moroni, P.; Stanhope, M.J. Genome Characterization and Population Genetic Structure of the Zoonotic Pathogen, Streptococcus canis. BMC Microbiol. 2012, 12, 293. [Google Scholar] [CrossRef]
- Pagnossin, D.; Weir, W.; Smith, A.; Fuentes, M.; Coelho, J.; Oravcova, K. Streptococcus canis Genomic Epidemiology Reveals the Potential for Zoonotic Transfer. Microb. Genom. 2023, 9, mgen000974. [Google Scholar] [CrossRef]
- Fukushima, Y.; Takahashi, T.; Goto, M.; Yoshida, H.; Tsuyuki, Y. Novel Diverse Sequences of the Streptococcus canis M-like Protein (SCM) Gene and Their Prevalence in Diseased Companion Animals: Association of Their Alleles with Sequence Types. J. Infect. Chemother. 2020, 26, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Eibl, C.; Baumgartner, M.; Urbantke, V.; Sigmund, M.; Lichtmannsperger, K.; Wittek, T.; Spergser, J. An Outbreak of Subclinical Mastitis in a Dairy Herd Caused by a Novel Streptococcus canis Sequence Type (ST55). Animals 2021, 11, 550. [Google Scholar] [CrossRef] [PubMed]
- McGuire, A.; Krysa, N.; Mann, S. Hair of the Dog? Periprosthetic Joint Infection with Streptococcus canis. Arthroplast. Today 2021, 8, 53–56. [Google Scholar] [CrossRef]
- Tan, R.; Yee, W.; Cao, D.; Tan, P.; Koh, T. Zoonotic Streptococcus canis Infection in Singapore. Singapore Med. J. 2016, 57, 218. [Google Scholar] [CrossRef] [PubMed]
- Lacave, G.; Coutard, A.; Troché, G.; Augusto, S.; Pons, S.; Zuber, B.; Laurent, V.; Amara, M.; Couzon, B.; Bédos, J.-P.; et al. Endocarditis Caused by Streptococcus canis: An Emerging Zoonosis? Infection 2016, 44, 111–114. [Google Scholar] [CrossRef]
- Pagnossin, D.; Smith, A.; Oravcová, K.; Weir, W. Streptococcus canis, the Underdog of the Genus. Vet. Microbiol. 2022, 273, 109524. [Google Scholar] [CrossRef]
- Bechtold, V.; Petzl, W.; Huber-Schlenstedt, R.; Gangl, A.; Sorge, U.S. Antimicrobial Resistance of Streptococcus Dysgalactiae, Streptococcus Agalactiae, and Streptococcus canis in Quarter Milk Samples from Bavaria, Southern Germany, between 2012 and 2022. J. Dairy Sci. 2024, 107, 8452–8463. [Google Scholar] [CrossRef]
| T0 | T1 | ||||||
|---|---|---|---|---|---|---|---|
| CMT | SCC | ISOL. | CMT | SCC | ISOL. | WGS | |
| Sample 1, 3, 5, 6, 8 | P | N | N | NT | NT | NT | NT |
| Sample 2, 7 | P | P | P | N | N | N | T0 |
| Sample 4 | P | P | P | P | P | P | T0 |
| Sample 10, 16, 17, 24, 25, 28 | N | NT | NT | P | N | N | NT |
| Sample 13 | N | NT | NT | P | P | P | NT |
| Sample 21 | N | NT | NT | P | P | P | T1 |
| Sample 9, 11, 12, 14, 15, 18, 19, 20, 22, 23, 26, 27, 29, 30, 31 | N | NT | NT | N | N | N | NT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sposato, A.; Del Sambro, L.; Castellana, S.; Catalano, E.; Galgano, M.; Castellana, A.; Caffò, A.; Manzulli, V.; Caruso, M.; Marino, L.; et al. Subclinical Mastitis Related to Streptococcus canis Infection in Dairy Cattle. Vet. Sci. 2025, 12, 286. https://doi.org/10.3390/vetsci12030286
Sposato A, Del Sambro L, Castellana S, Catalano E, Galgano M, Castellana A, Caffò A, Manzulli V, Caruso M, Marino L, et al. Subclinical Mastitis Related to Streptococcus canis Infection in Dairy Cattle. Veterinary Sciences. 2025; 12(3):286. https://doi.org/10.3390/vetsci12030286
Chicago/Turabian StyleSposato, Alessio, Laura Del Sambro, Stefano Castellana, Elisabetta Catalano, Michela Galgano, Antonella Castellana, Annamaria Caffò, Viviana Manzulli, Marta Caruso, Leonardo Marino, and et al. 2025. "Subclinical Mastitis Related to Streptococcus canis Infection in Dairy Cattle" Veterinary Sciences 12, no. 3: 286. https://doi.org/10.3390/vetsci12030286
APA StyleSposato, A., Del Sambro, L., Castellana, S., Catalano, E., Galgano, M., Castellana, A., Caffò, A., Manzulli, V., Caruso, M., Marino, L., Milano, A., & Addante, L. (2025). Subclinical Mastitis Related to Streptococcus canis Infection in Dairy Cattle. Veterinary Sciences, 12(3), 286. https://doi.org/10.3390/vetsci12030286






