Dynamic Changes in Rumen Microbial Diversity and Community Composition Within Rumen Fluid in Response to Various Storage Temperatures and Preservation Times
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Ethics
2.2. Rumen Fluid Acquisition
2.3. Experimental Design
2.4. DNA Extraction, Sequencing, and Data Analysis
2.5. Statistical Analyses
3. Results
3.1. Rumen Bacterial Alpha-Diversity
3.2. Rumen Bacterial Community Composition
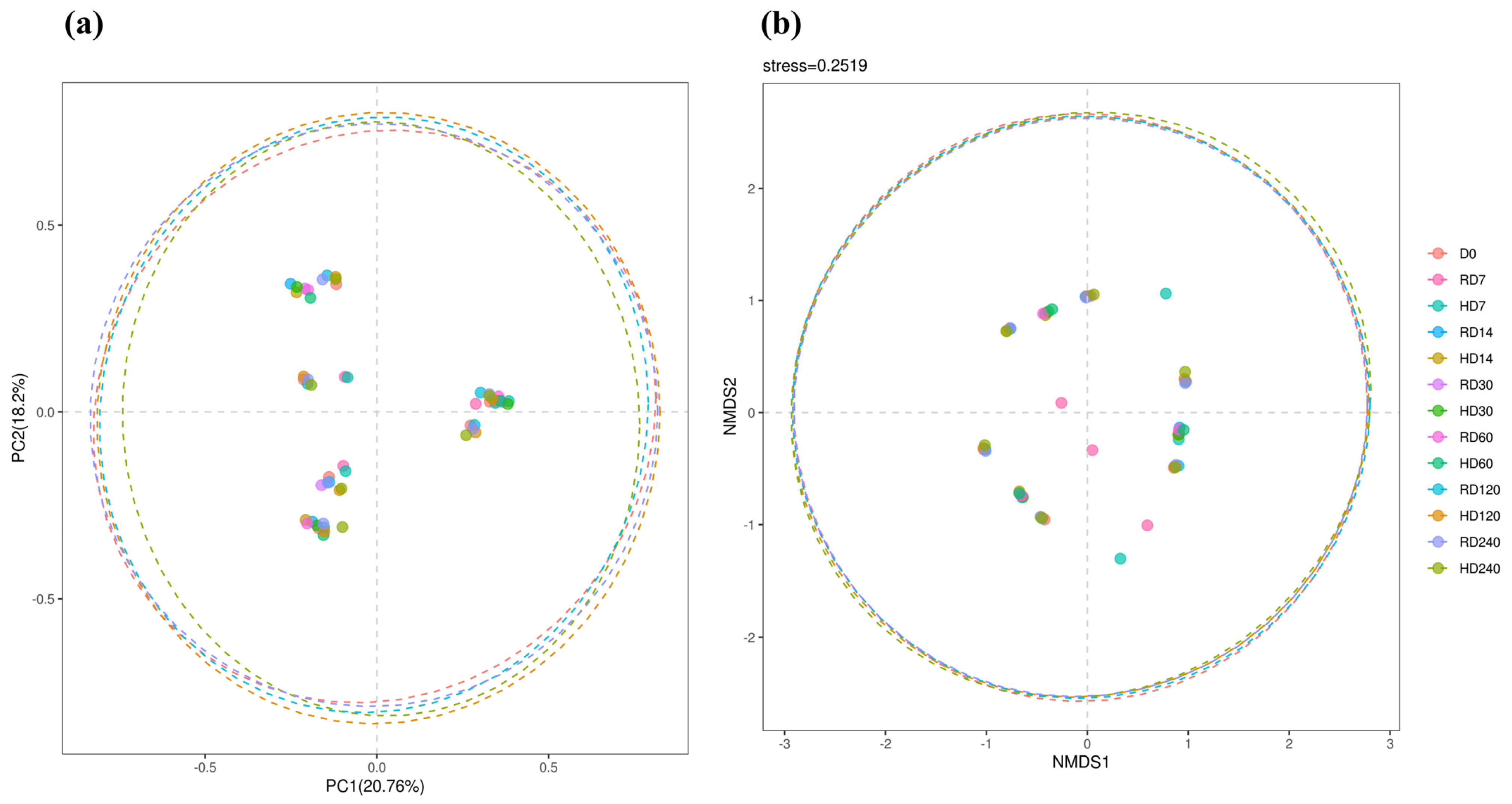
3.3. Rumen Bacterial Beta-Diversity
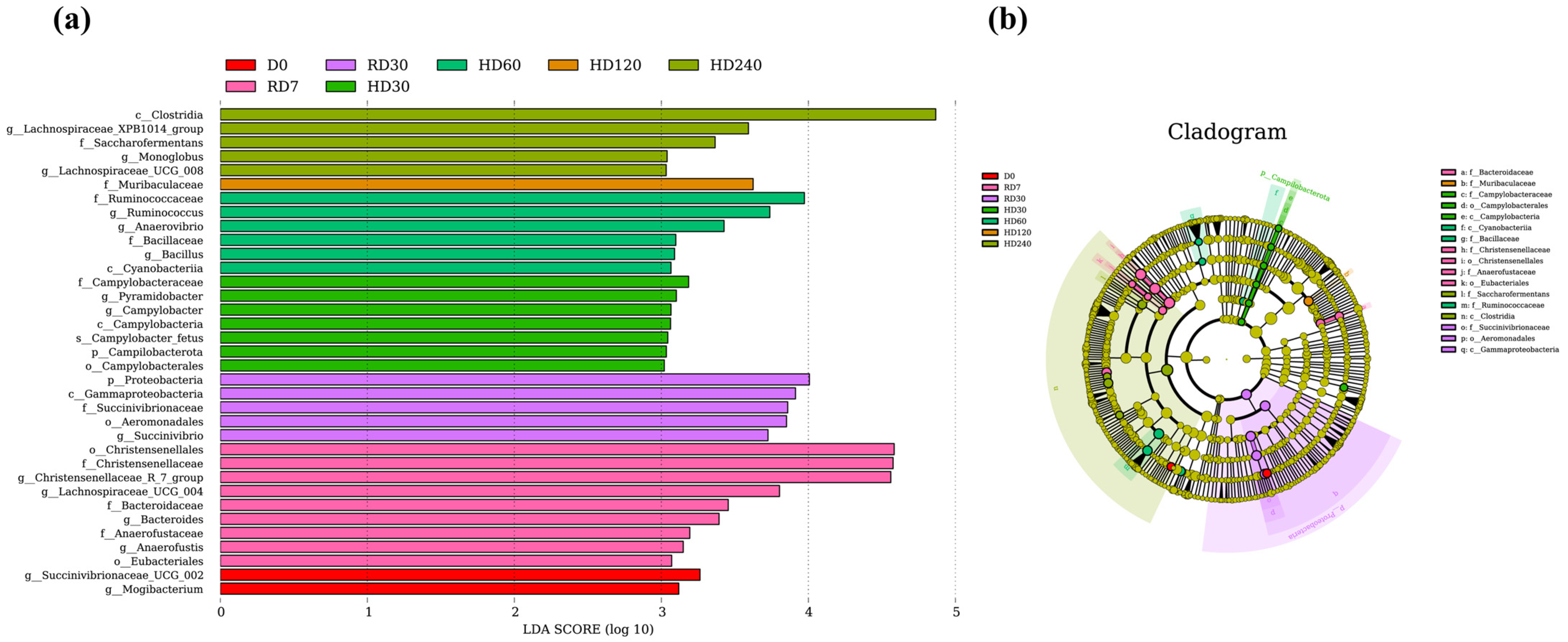
3.4. Biomarker Microbes
3.5. Rumen Bacteria Predicted Metabolic Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NH3-N | Ammonia nitrogen |
| ASVs | Amplicon sequence variants |
| ANOSIM | Analysis of similarities |
| NMDS | Non-metric multidimensional scaling |
| PD | Phylogenetic diversity |
| PCoA | Principal coordinates analysis |
| rRNA | ribosomal RNA |
| SCFAs | Short-chain fatty acids |
References
- Chaucheyras-Durand, F.; Ossa, F. REVIEW: The rumen microbiome: Composition, abundance, diversity, and new investigative tools. Prof. Anim. Sci. 2014, 30, 1–12. [Google Scholar] [CrossRef]
- Keum, G.B.; Pandey, S.; Kim, E.S.; Doo, H.; Kwak, J.; Ryu, S.; Choi, Y.; Kang, J.; Kim, S.; Kim, H.B. Understanding the diversity and roles of the ruminal microbiome. J. Microbiol. 2024, 62, 217–230. [Google Scholar] [CrossRef]
- Lima, J.; Martínez-Álvaro, M.; Mattock, J.; Auffret, M.D.; Duthie, C.A.; Cleveland, M.A.; Dewhurst, R.J.; Watson, M.; Roehe, R. Temporal stability of the rumen microbiome and its longitudinal associations with performance traits in beef cattle. Sci. Rep. 2024, 14, 20772. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wei, Z.; Li, Z.; Ren, J.; Song, Y.; Xu, J.; Liu, A.; Li, X.; Li, M.; Fan, H.; et al. Integrating genome- and transcriptome-wide association studies to uncover the host–microbiome interactions in bovine rumen methanogenesis. iMeta 2024, 3, e234. [Google Scholar] [CrossRef] [PubMed]
- Silva, É.B.; Silva, J.A.; Silva, W.C.; Belo, T.S.; Sousa, C.E.; Santos, M.R.; Neves, K.A.; Rodrigues, T.C.; Camargo-Júnior, R.N.; Lourenço-Júnior, J.D. A Review of the rumen microbiota and the different molecular techniques used to identify microorganisms found in the rumen fluid of ruminants. Animals 2024, 14, 1448. [Google Scholar] [CrossRef] [PubMed]
- Gleason, C.B.; Beckett, L.M.; White, R.R. Rumen fermentation and epithelial gene expression responses to diet ingredients designed to differ in ruminally degradable protein and fiber supplies. Sci. Rep. 2022, 12, 2933. [Google Scholar] [CrossRef]
- Danmaigoro, A.; Muhammad, M.A.; Abubakar, K.; Magiri, R.B.; Bakare, A.G.; Iji, P.A. Morphological and physiological features in small ruminants: An adaptation strategy for survival under changing climatic conditions. Int. J. Biometeorolo. 2024, 68, 1497–1505. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef]
- Dehority Burk, A.; Grubb Jean, A. Effect of short-term chilling of rumen contents on viable bacterial numbers. Appl. Environ. Microbiol. 1980, 39, 376–381. [Google Scholar] [CrossRef]
- Martin, C.C.; Hilgert, A.R.; Guirro, E.C.B.D.P. Influence of food, storage temperature, and time on the extracorporeal viability of ruminal fluid of cattle. Semin. Cienc. Agrar. 2018, 39, 1181–1188. [Google Scholar] [CrossRef]
- Takizawa, S.; Baba, Y.; Tada, C.; Fukuda, Y.; Nakai, Y. Preservation of rumen fluid for the pretreatment of waste paper to improve methane production. Waste Manag. 2019, 87, 672–678. [Google Scholar] [CrossRef]
- Belanche, A.; Palma-Hidalgo, J.M.; Nejjam, I.; Serrano, R.; Jiménez, E.; Martín-García, I.; Yáñez-Ruiz, D.R. In vitro assessment of the factors that determine the activity of the rumen microbiota for further applications as inoculum. J. Sci. Food Agric. 2019, 99, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Tunkala, B.Z.; DiGiacomo, K.; Hess, P.S.A.; Dunshea, F.R.; Leury, B.J. Rumen fluid preservation for in vitro gas production systems. Anim. Feed Sci. Technol. 2022, 292, 115405. [Google Scholar] [CrossRef]
- Tunkala, B.Z.; DiGiacomo, K.; Hess, P.S.A.; Dunshea, F.R.; Leury, B.J. Impact of rumen fluid storage on in vitro feed fermentation characteristics. Fermentation 2023, 9, 392. [Google Scholar] [CrossRef]
- Spanghero, M.; Chiaravalli, M.; Colombini, S.; Fabro, C.; Froldi, F.; Mason, F.; Moschini, M.; Sarnataro, C.; Schiavon, S.; Tagliapietra, F. Rumen inoculum collected from cows at slaughter or from a continuous fermenter and preserved in warm, refrigerated, chilled or freeze-dried environments for in vitro tests. Animals 2019, 9, 815. [Google Scholar] [CrossRef]
- Fliegerova, K.; Tapio, I.; Bonin, A.; Mrazek, J.; Callegari, M.L.; Bani, P.; Bayat, A.; Vilkki, J.; Kopečný, J.; Shingfield, K.J.; et al. Effect of DNA extraction and sample preservation method on rumen bacterial population. Anaerobe 2014, 29, 80–84. [Google Scholar] [CrossRef]
- Qiu, Q.; Long, T.; Ouyang, K.; Lei, X.; Qiu, J.; Zhang, J.; Li, Y.; Zhao, X.; Qu, M.; Ouyang, K. Effect of preservation temperature and time on fermentation characteristics, bacterial diversity and community composition of rumen fluid collected from high-grain feeding sheep. Fermentation 2023, 9, 466. [Google Scholar] [CrossRef]
- Poulsen Casper, S.; Kaas Rolf, S.; Aarestrup Frank, M.; Pamp Sünje, J. Standard sample storage conditions have an impact on inferred microbiome composition and antimicrobial resistance patterns. Microbiol. Spectr. 2021, 9, e01387-21. [Google Scholar] [CrossRef]
- Wei, X.; Long, T.; Li, Y.; Ouyang, K.; Qiu, Q. Diet shift may trigger LuxS/AI-2 quorum sensing in rumen bacteria. Bioengineering 2022, 9, 379. [Google Scholar] [CrossRef]
- Lin, M.; Cao, H.; Li, J. Control strategies of ice nucleation, growth, and recrystallization for cryopreservation. Acta Biomater. 2023, 155, 35–56. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The power of microbiome studies: Some considerations on which alpha and beta metrics to use and how to report results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
- Won, M.-Y.; Oyama, L.B.; Courtney, S.J.; Creevey, C.J.; Huws, S.A. Can rumen bacteria communicate to each other? Microbiome 2020, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Qu, M.; Ouyang, K.; Qiu, Q. Effects of storage time and temperature on the fermentation characteristics of rumen fluid from a high-forage diet. Agriculture 2024, 14, 1481. [Google Scholar] [CrossRef]
- Kumar, S.; Najar, I.N.; Sharma, P.; Tamang, S.; Mondal, K.; Das, S.; Sherpa, M.T.; Thakur, N. Temperature—A critical abiotic paradigm that governs bacterial heterogeneity in natural ecological system. Environ. Res. 2023, 234, 116547. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef]
- Nixon, S.L.; Daly, R.A.; Borton, M.A.; Solden, L.M.; Welch, S.A.; Cole, D.R.; Mouser, P.J.; Wilkins, M.J.; Wrighton, K.C.; Suen, G. Genome-resolved metagenomics extends the environmental distribution of the Verrucomicrobia phylum to the deep terrestrial subsurface. mSphere 2019, 4, 13. [Google Scholar] [CrossRef]
- Tian, R.; Ning, D.; He, Z.; Zhang, P.; Spencer, S.J.; Gao, S.; Shi, W.; Wu, L.; Zhang, Y.; Yang, Y.; et al. Small and mighty: Adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 2020, 8, 51. [Google Scholar] [CrossRef]
- Martinez-Boggio, G.; Monteiro, H.F.; Lima, F.S.; Figueiredo, C.C.; Bisinotto, R.S.; Santos, J.E.P.; Mion, B.; Schenkel, F.S.; Ribeiro, E.S.; Weigel, K.A.; et al. Revealing host genome–microbiome networks underlying feed efficiency in dairy cows. Sci. Rep. 2024, 14, 26060. [Google Scholar] [CrossRef]
- Conte, G.; Dimauro, C.; Daghio, M.; Serra, A.; Mannelli, F.; McAmmond, B.M.; Van Hamme, J.D.; Buccioni, A.; Viti, C.; Mantino, A.; et al. Exploring the relationship between bacterial genera and lipid metabolism in bovine rumen. Animal 2022, 16, 100520. [Google Scholar] [CrossRef]
- Ma, Y.; Deng, X.; Yang, X.; Wang, J.; Li, T.; Hua, G.; Han, D.; Da, L.; Li, R.; Rong, W.; et al. Characteristics of bacterial microbiota in different intestinal segments of Aohan fine-wool sheep. Front. Microbiol. 2022, 13, 874536. [Google Scholar] [CrossRef]
- Luan, J.; Feng, X.; Du, Y.; Yang, D.; Geng, C. Medium-chain fatty acid triglycerides improve feed intake and oxidative stress of finishing bulls by regulating ghrelin concentration and gastrointestinal tract microorganisms and rumen metabolites. Microbiome 2024, 12, 230. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Gao, Y.; Peng, Z.; Ma, X.; You, Y.; Hu, Z.; He, A.; Liao, Y. Isoacids supplementation improves growth performance and feed fiber digestibility associated with ruminal bacterial community in yaks. Front. Microbiol. 2023, 14, 1175880. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J. Production and absorption of volatile fatty acids in the rumen. Livest. Prod. Sci. 1994, 39, 61–69. [Google Scholar] [CrossRef]
- Honerlagen, H.; Reyer, H.; Abou-Soliman, I.; Segelke, D.; Ponsuksili, S.; Trakooljul, N.; Reinsch, N.; Kuhla, B.; Wimmers, K. Microbial signature inferred from genomic breeding selection on milk urea concentration and its relation to proxies of nitrogen-utilization efficiency in Holsteins. J. Dairy Sci. 2023, 106, 4682–4697. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, S.; Spillane, C.; Claffey, N.; Smith, P.E.; O’Rourke, T.; Diskin, M.G.; Waters, S.M. Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front. Microbiol. 2020, 11, 1981. [Google Scholar] [CrossRef]
- McGovern, E.; McGee, M.; Byrne, C.J.; Kenny, D.A.; Kelly, A.K.; Waters, S.M. Investigation into the effect of divergent feed efficiency phenotype on the bovine rumen microbiota across diet and breed. Sci. Rep. 2020, 10, 15317. [Google Scholar] [CrossRef]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
- Zaplana, T.; Miele, S.; Tolonen, A.C. Lachnospiraceae are emerging industrial biocatalysts and biotherapeutics. Front. Bioeng. Biotechnol. 2024, 11, 1324396. [Google Scholar] [CrossRef]
- McKee, L.A.-O.; La Rosa, S.A.-O.; Westereng, B.A.-O.; Eijsink, V.A.-O.; Pope, P.A.-O.; Larsbrink, J.A.-O. Polysaccharide degradation by the Bacteroidetes: Mechanisms and nomenclature. Environ. Microbiol. Rep. 2021, 13, 559–581. [Google Scholar] [CrossRef]
- Wang, D.; Chen, L.; Tang, G.; Yu, J.; Chen, J.; Li, Z.; Cao, Y.; Lei, X.; Deng, L.; Wu, S.; et al. Multi-omics revealed the long-term effect of ruminal keystone bacteria and the microbial metabolome on lactation performance in adult dairy goats. Microbiome 2023, 11, 215. [Google Scholar] [CrossRef]
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef] [PubMed]
- van der Stel, A.-X.; Wösten, M.M.S.M. Regulation of respiratory pathways in Campylobacterota: A review. Front. Microbiol. 2019, 10, 1719. [Google Scholar] [CrossRef] [PubMed]
- Fassarella, M.; Blaak, E.E.; Penders, J.; Nauta, A.; Smidt, H.; Zoetendal, E.G. Gut microbiome stability and resilience: Elucidating the response to perturbations in order to modulate gut health. Gut 2021, 70, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, Y.; Fu, H.; Jin, S.; He, S.; Wang, Z.; Dong, G.; Li, B.; Guo, S. Does diet or macronutrients intake drive the structure and function of gut microbiota? Front. Microbiol. 2023, 14, 1126189. [Google Scholar] [CrossRef]
- Fayi, T.; Junxi, W. Effects of grazing and barn feeding on biological information of rumen bacterial communities in Pengbo semi-fine wool sheep. Acta Vet. Zootech. Sin. 2019, 50, 2252–2263. [Google Scholar]
- Mandic-Mulec, I.; Stefanic, P.; Elsas, J.D.V. Ecology of Bacillaceae. Microbiol. Spectr. 2015, 3, TBS-0017-2013. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, B.; Zhang, X.; Akbar, M.T.; Wu, T.; Zhang, Y.; Zhi, L.; Shen, Q. Exploration of the Muribaculaceae family in the gut microbiota: Diversity, metabolism, and function. Nutrients 2024, 16, 2660. [Google Scholar] [CrossRef]
- Zu, T.N.K.; Liu, S.; Gerlach, E.S.; Mojadedi, W.; Sund, C.J. Co-feeding glucose with either gluconate or galacturonate during clostridial fermentations provides metabolic fine-tuning capabilities. Sci. Rep. 2021, 11, 29. [Google Scholar] [CrossRef]
- Monteiro, H.F.; Lelis, A.L.J.; Fan, P.; Calvo Agustinho, B.; Lobo, R.R.; Arce-Cordero, J.A.; Dai, X.; Jeong, K.C.; Faciola, A.P. Effects of lactic acid-producing bacteria as direct-fed microbials on the ruminal microbiome. J. Dairy Sci. 2022, 105, 2242–2255. [Google Scholar] [CrossRef]
- Sawane, K.; Hosomi, K.; Park, J.; Ookoshi, K.; Nanri, H.; Nakagata, T.; Chen, Y.-A.; Mohsen, A.; Kawashima, H.; Mizuguchi, K.; et al. Identification of human gut microbiome associated with enterolignan production. Microorganisms 2022, 10, 2169. [Google Scholar] [CrossRef]
- Kim, C.C.; Healey, G.R.; Kelly, W.J.; Patchett, M.L.; Jordens, Z.; Tannock, G.W.; Sims, I.M.; Bell, T.J.; Hedderley, D.; Henrissat, B.; et al. Genomic insights from Monoglobus pectinilyticus: A pectin-degrading specialist bacterium in the human colon. ISME J. 2019, 13, 1437–1456. [Google Scholar] [CrossRef] [PubMed]
- Tomazetto, G.; Hahnke, S.; Wibberg, D.; Pühler, A.; Klocke, M.; Schlüter, A. Proteiniphilum saccharofermentans str. M3/6T isolated from a laboratory biogas reactor is versatile in polysaccharide and oligopeptide utilization as deduced from genome-based metabolic reconstructions. Biotechnol. Rep. 2018, 18, e00254. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Gao, C.; Aziz ur Rahman, M.; Cao, B.; Su, H. Digestive ability, physiological characteristics, and rumen bacterial community of Holstein finishing steers in response to three nutrient density diets as fattening phases advanced. Microorganisms 2020, 8, 335. [Google Scholar] [CrossRef] [PubMed]
| Item | Storage Time 1 | SEM 2 | p-Value 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D7 | D14 | D30 | D60 | D120 | D240 | Time | Temperature | Interaction | |||
| Chao1 | −80 °C | 673.47 | 460.86 | 852.46 | 678.30 | 480.62 | 1060.26 | 656.52 | ||||
| −20 °C | 673.47 | 495.97 | 920.03 | 722.57 | 416.60 | 765.32 | 713.46 | 103.238 | <0.001 | 0.694 | 0.402 | |
| Average | 673.47 ab | 478.425 b | 886.25 a | 700.44 ab | 448.61 b | 912.79 a | 684.99 ab | |||||
| P0 value 4 | 0.486 | 0.381 | 1.000 | 0.316 | 0.084 | 1.000 | ||||||
| Observed species | −80 °C | 656.33 | 456.00 | 808.67 | 660.67 | 473.33 | 967.33 | 641.50 | ||||
| −20 °C | 656.33 | 489.00 | 860.67 | 671.33 | 416.33 | 741.50 | 685.67 | 92.234 | <0.001 | 0.685 | 0.599 | |
| Average | 656.33 ab | 472.50 b | 834.67 a | 666.00 ab | 444.83 b | 854.42 a | 663.58 ab | |||||
| P0 value | 0.421 | 0.458 | 1.000 | 0.259 | 0.133 | 1.000 | ||||||
| PD whole tree | −80 °C | 55.15 | 45.89 | 59.76 | 53.45 | 43.88 | 66.55 | 54.16 | ||||
| −20 °C | 55.15 | 47.72 | 60.59 | 50.85 | 40.69 | 58.50 | 55.01 | 4.532 | 0.001 | 0.551 | 0.890 | |
| Average | 55.15 abc | 46.81 bc | 60.17 ab | 52.15 abc | 42.28 c | 62.53 a | 54.58 abc | |||||
| P0 value | 0.517 | 0.919 | 0.994 | 0.084 | 0.420 | 1.000 | ||||||
| Shannon index | −80 °C | 7.18 | 7.27 | 7.37 | 7.41 | 7.22 | 7.45 | 7.26 | ||||
| −20 °C | 7.18 | 7.26 | 7.60 | 7.42 | 7.40 | 7.48 | 7.51 | 0.382 | 0.974 | 0.632 | 0.999 | |
| Average | 7.18 | 7.27 | 7.49 | 7.42 | 7.31 | 7.46 | 7.39 | |||||
| P0 value | 1.000 | 0.983 | 0.996 | 1.000 | 0.969 | 0.994 | ||||||
| Simpson index | −80 °C | 0.971 | 0.982 | 0.973 | 0.977 | 0.979 | 0.973 | 0.977 | ||||
| −20 °C | 0.971 | 0.981 | 0.982 | 0.981 | 0.985 | 0.978 | 0.982 | 0.010 | 0.878 | 0.434 | 0.999 | |
| Average | 0.971 | 0.982 | 0.978 | 0.979 | 0.982 | 0.975 | 0.980 | |||||
| P0 value | 0.921 | 0.994 | 0.978 | 0.899 | 0.998 | 0.933 | ||||||
| Item | Storage Time 1 | SEM 2 | p-Value 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D7 | D14 | D30 | D60 | D120 | D240 | Time | Temperature | Interaction | |||
| Bacteroidota | −80 °C | 69.11 | 62.28 | 75.71 | 72.60 | 72.73 | 70.21 | 69.04 | ||||
| −20 °C | 69.11 | 69.24 | 73.97 | 75.80 | 61.85 | 64.04 | 59.59 | 4.228 | 0.137 | 0.265 | 0.352 | |
| Average | 69.11 | 65.76 | 74.84 | 74.20 | 67.29 | 67.13 | 64.31 | |||||
| P0 value 4 | 0.984 | 0.815 | 0.884 | 0.999 | 0.997 | 0.798 | ||||||
| Firmicutes | −80 °C | 25.81 | 33.36 | 21.03 | 22.66 | 23.18 | 25.20 | 26.43 | ||||
| −20 °C | 25.81 | 27.54 | 23.07 | 20.44 | 34.58 | 31.81 | 36.11 | 4.040 | 0.132 | 0.163 | 0.332 | |
| Average | 25.81 | 30.45 | 22.05 | 21.55 | 28.88 | 28.50 | 31.27 | |||||
| P0 value | 0.906 | 0.964 | 0.935 | 0.987 | 0.981 | 0.638 | ||||||
| Proteobacteria | −80 °C | 2.34 | 1.51 | 1.57 | 2.82 | 2.18 | 1.78 | 1.74 | ||||
| −20 °C | 2.34 | 0.74 | 1.25 | 1.69 | 1.16 | 0.93 | 0.76 | 0.462 | 0.031 | 0.006 | 0.817 | |
| Average | 2.34 | 1.13 | 1.41 | 2.25 | 1.67 | 1.35 | 1.25 | |||||
| P0 value | 0.132 | 0.400 | 1.000 | 0.758 | 0.133 | 0.072 | ||||||
| Verrucomicrobiota | −80 °C | 0.70 | 0.62 | 0.15 | 0.35 | 0.22 | 0.76 | 0.48 | ||||
| −20 °C | 0.70 | 0.53 | 0.13 | 0.44 | 0.27 | 0.71 | 0.32 | 0.210 | 0.038 | 0.814 | 0.997 | |
| Average | 0.70 a | 0.58 ab | 0.14 b | 0.39 ab | 0.24 ab | 0.74 a | 0.40 ab | |||||
| P0 value | 0.996 | 0.118 | 0.751 | 0.307 | 1.000 | 0.572 | ||||||
| Desulfobacterota | −80 °C | 0.29 | 0.36 | 0.32 | 0.25 | 0.31 | 0.34 | 0.44 | ||||
| −20 °C | 0.29 | 0.34 | 0.38 | 0.24 | 0.63 | 0.59 | 0.60 | 0.146 | 0.353 | 0.181 | 0.852 | |
| Average | 0.29 | 0.35 | 0.35 | 0.25 | 0.47 | 0.46 | 0.52 | |||||
| P0 value | 0.999 | 1.000 | 1.000 | 0.872 | 0.751 | 0.445 | ||||||
| Spirochaetota | −80 °C | 0.46 | 0.41 | 0.36 | 0.52 | 0.37 | 0.43 | 0.25 | ||||
| −20 °C | 0.46 | 0.43 | 0.43 | 0.34 | 0.20 | 0.51 | 0.34 | 0.101 | 0.244 | 0.815 | 0.743 | |
| Average | 0.46 | 0.42 | 0.40 | 0.43 | 0.29 | 0.47 | 0.29 | |||||
| P0 value | 0.999 | 0.994 | 1.000 | 0.589 | 1.000 | 0.390 | ||||||
| Synergistota | −80 °C | 0.36 | 0.54 | 0.33 | 0.21 | 0.32 | 0.31 | 0.50 | ||||
| −20 °C | 0.36 | 0.35 | 0.21 | 0.21 | 0.22 | 0.27 | 0.47 | 0.136 | 0.327 | 0.360 | 0.993 | |
| Average | 0.36 | 0.45 | 0.27 | 0.21 | 0.27 | 0.29 | 0.49 | |||||
| P0 value | 0.996 | 0.991 | 0.910 | 0.990 | 0.994 | 0.925 | ||||||
| Actinobacteriota | −80 °C | 0.17 | 0.18 | 0.15 | 0.05 | 0.17 | 0.25 | 0.33 | ||||
| −20 °C | 0.17 | 0.25 | 0.16 | 0.25 | 0.52 | 0.43 | 0.82 | 0.219 | 0.279 | 0.127 | 0.860 | |
| Average | 0.17 | 0.22 | 0.16 | 0.15 | 0.34 | 0.34 | 0.58 | |||||
| P0 value | 1.000 | 1.000 | 1.000 | 0.983 | 0.954 | 0.262 | ||||||
| Patescibacteria | −80 °C | 0.11 | 0.16 | 0.08 | 0.14 | 0.16 | 0.18 | 0.30 | ||||
| −20 °C | 0.11 | 0.15 | 0.11 | 0.16 | 0.26 | 0.25 | 0.50 | 0.065 | <0.001 | 0.099 | 0.546 | |
| Average | 0.11 b | 0.16 b | 0.10 b | 0.15 b | 0.21 ab | 0.21 ab | 0.40 a | |||||
| P0 value | 0.991 | 1.000 | 0.996 | 0.712 | 0.430 | <0.001 | ||||||
| Cyanobacteria | −80 °C | 0.22 | 0.13 | 0.13 | 0.20 | 0.11 | 0.25 | 0.19 | ||||
| −20 °C | 0.22 | 0.07 | 0.15 | 0.26 | 0.14 | 0.24 | 0.26 | 0.075 | 0.307 | 0.722 | 0.981 | |
| Average | 0.22 | 0.10 | 0.14 | 0.23 | 0.12 | 0.24 | 0.22 | |||||
| P0 value | 0.624 | 0.924 | 1.000 | 0.818 | 1.000 | 1.000 | ||||||
| Item | Storage Time 1 | SEM 2 | p-Value 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D7 | D14 | D30 | D60 | D120 | D240 | Time | Temperature | Interaction | |||
| Prevotella | −80 °C | 43.03 | 34.87 | 54.33 | 45.54 | 48.22 | 41.86 | 39.60 | ||||
| −20 °C | 43.03 | 40.71 | 51.76 | 46.16 | 34.59 | 36.61 | 30.05 | 7.7617 | 0.355 | 0.409 | 0.919 | |
| Average | 43.03 | 37.79 | 53.04 | 45.85 | 41.40 | 39.23 | 34.83 | |||||
| P0 value 4 | 0.993 | 0.847 | 1.000 | 1.000 | 0.996 | 0.845 | ||||||
| Rikenellaceae RC9 gut group | −80 °C | 6.97 | 8.30 | 6.89 | 9.64 | 7.96 | 7.64 | 10.46 | ||||
| −20 °C | 6.97 | 9.06 | 8.13 | 10.45 | 10.96 | 9.64 | 12.89 | 1.6991 | 0.048 | 0.119 | 0.968 | |
| Average | 6.97 b | 8.68 ab | 7.51 ab | 10.04 ab | 9.46 ab | 8.64 ab | 11.67 a | |||||
| P0 value | 0.948 | 1.000 | 0.539 | 0.757 | 0.885 | 0.021 | ||||||
| Prevotellaceae UCG-003 | −80 °C | 4.82 | 4.25 | 3.08 | 3.81 | 3.60 | 5.83 | 5.86 | ||||
| −20 °C | 4.82 | 4.54 | 2.97 | 4.71 | 3.51 | 3.99 | 4.13 | 1.6956 | 0.895 | 0.690 | 0.970 | |
| Average | 4.82 | 4.39 | 3.02 | 4.26 | 3.55 | 4.91 | 4.99 | |||||
| P0 value | 1.000 | 0.933 | 1.000 | 0.988 | 1.000 | 1.000 | ||||||
| Christensenellaceae R-7 group | −80 °C | 6.58 | 8.27 | 1.59 | 1.58 | 1.65 | 3.52 | 3.36 | ||||
| −20 °C | 6.58 | 7.06 | 1.67 | 0.71 | 5.16 | 4.75 | 5.82 | 1.1726 | <0.001 | 0.250 | 0.430 | |
| Average | 6.58 ab | 7.67 a | 1.63 c | 1.15 c | 3.40 bc | 4.14 abc | 4.59 abc | |||||
| P0 value | 0.965 | 0.002 | <0.001 | 0.112 | 0.157 | 0.369 | ||||||
| Prevotellaceae UCG-001 | −80 °C | 3.71 | 2.44 | 3.79 | 2.16 | 3.67 | 3.81 | 3.44 | ||||
| −20 °C | 3.71 | 2.06 | 3.12 | 2.87 | 2.07 | 2.55 | 2.06 | 1.6449 | 0.973 | 0.467 | 0.992 | |
| Average | 3.71 | 2.25 | 3.45 | 2.51 | 2.87 | 3.18 | 2.75 | |||||
| P0 value | 0.972 | 1.000 | 0.990 | 0.998 | 1.000 | 0.991 | ||||||
| Succiniclasticum | −80 °C | 1.83 | 2.80 | 2.92 | 1.69 | 2.72 | 2.86 | 3.15 | ||||
| −20 °C | 1.83 | 1.82 | 2.63 | 2.47 | 2.32 | 2.63 | 2.95 | 0.7633 | 0.552 | 0.652 | 0.979 | |
| Average | 1.83 | 2.31 | 2.77 | 2.08 | 2.52 | 2.75 | 3.05 | |||||
| P0 value | 0.996 | 0.873 | 1.000 | 0.969 | 0.754 | 0.450 | ||||||
| Selenomonas | −80 °C | 1.35 | 1.80 | 2.48 | 3.84 | 3.38 | 2.60 | 2.17 | ||||
| −20 °C | 1.35 | 1.33 | 2.70 | 1.69 | 2.13 | 1.90 | 1.48 | 0.8247 | 0.432 | 0.114 | 0.876 | |
| Average | 1.35 | 1.57 | 2.59 | 2.77 | 2.76 | 2.25 | 1.83 | |||||
| P0 value | 1.000 | 0.735 | 0.600 | 0.608 | 0.827 | 0.991 | ||||||
| NK4A214 group | −80 °C | 2.51 | 2.97 | 1.28 | 1.36 | 1.26 | 1.21 | 1.90 | ||||
| −20 °C | 2.51 | 2.49 | 1.13 | 1.23 | 2.46 | 2.17 | 3.10 | 0.8231 | 0.390 | 0.411 | 0.881 | |
| Average | 2.51 | 2.73 | 1.21 | 1.29 | 1.86 | 1.69 | 2.50 | |||||
| P0 value | 1.000 | 0.685 | 0.749 | 0.985 | 0.880 | 1.000 | ||||||
| Veillonellaceae UCG-001 | −80 °C | 0.98 | 1.30 | 1.82 | 1.34 | 1.74 | 1.96 | 2.15 | ||||
| −20 °C | 0.98 | 0.92 | 1.70 | 1.80 | 1.81 | 1.87 | 2.65 | 0.4267 | 0.006 | 0.792 | 0.942 | |
| Average | 0.98 b | 1.11 ab | 1.76 ab | 1.57 ab | 1.77 ab | 1.91 ab | 2.40 a | |||||
| P0 value | 1.000 | 0.525 | 0.796 | 0.497 | 0.118 | 0.003 | ||||||
| Item | Storage Time 1 | SEM 2 | p-Value 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D7 | D14 | D30 | D60 | D120 | D240 | Time | Temperature | Interaction | |||
| Metabolism of cofactors and vitamins | −80 °C | 14.87 | 14.70 | 15.07 | 14.87 | 14.96 | 14.80 | 14.77 | ||||
| −20 °C | 14.87 | 14.81 | 14.97 | 15.02 | 14.33 | 14.44 | 14.12 | 0.239 | 0.160 | 0.111 | 0.427 | |
| Average | 14.87 | 14.75 | 15.02 | 14.95 | 14.64 | 14.62 | 14.44 | |||||
| P0 value 4 | 0.999 | 0.996 | 1.000 | 0.958 | 0.851 | 0.313 | ||||||
| Carbohydrate metabolism | −80 °C | 14.03 | 13.82 | 14.07 | 14.01 | 14.09 | 13.83 | 13.90 | ||||
| −20 °C | 14.03 | 13.93 | 14.11 | 14.08 | 14.05 | 13.96 | 13.83 | 0.152 | 0.530 | 0.678 | 0.985 | |
| Average | 14.03 | 13.87 | 14.09 | 14.05 | 14.07 | 13.89 | 13.87 | |||||
| P0 value | 0.946 | 0.999 | 1.000 | 1.000 | 0.935 | 0.854 | ||||||
| Amino acid metabolism | −80 °C | 13.05 | 13.01 | 13.03 | 13.03 | 13.08 | 12.92 | 13.02 | ||||
| −20 °C | 13.05 | 13.00 | 13.07 | 13.08 | 13.10 | 13.03 | 13.00 | 0.110 | 0.954 | 0.637 | 0.992 | |
| Average | 13.05 | 13.01 | 13.05 | 13.06 | 13.09 | 12.98 | 13.01 | |||||
| P0 value | 1.000 | 1.000 | 1.000 | 1.000 | 0.981 | 1.000 | ||||||
| Metabolism of terpenoids and polyketides | −80 °C | 8.64 | 8.83 | 8.15 | 8.49 | 8.34 | 8.70 | 8.53 | ||||
| −20 °C | 8.64 | 8.63 | 8.27 | 8.42 | 8.77 | 8.74 | 9.03 | 0.217 | 0.176 | 0.324 | 0.610 | |
| Average | 8.64 | 8.73 | 8.21 | 8.45 | 8.55 | 8.72 | 8.78 | |||||
| P0 value | 0.999 | 0.425 | 0.977 | 1.000 | 0.999 | 0.980 | ||||||
| Metabolism of other amino acids | −80 °C | 6.72 | 6.75 | 6.68 | 6.72 | 6.94 | 6.86 | 6.83 | ||||
| −20 °C | 6.72 | 6.69 | 6.71 | 6.71 | 6.73 | 6.86 | 7.04 | 0.118 | 0.191 | 0.941 | 0.723 | |
| Average | 6.72 | 6.72 | 6.70 | 6.71 | 6.84 | 6.86 | 6.94 | |||||
| P0 value | 1.000 | 1.000 | 1.000 | 0.958 | 0.775 | 0.285 | ||||||
| Replication and repair | −80 °C | 6.51 | 6.54 | 6.47 | 6.47 | 6.47 | 6.42 | 6.45 | ||||
| −20 °C | 6.51 | 6.59 | 6.50 | 6.52 | 6.47 | 6.46 | 6.41 | 0.052 | 0.145 | 0.532 | 0.949 | |
| Average | 6.51 | 6.57 | 6.49 | 6.50 | 6.47 | 6.44 | 6.43 | |||||
| P0 value | 0.888 | 1.000 | 1.000 | 0.994 | 0.652 | 0.562 | ||||||
| Glycan biosynthesis and metabolism | −80 °C | 6.21 | 5.98 | 6.46 | 6.25 | 6.33 | 6.18 | 6.08 | ||||
| −20 °C | 6.21 | 6.19 | 6.37 | 6.38 | 5.70 | 5.82 | 5.52 | 0.250 | 0.195 | 0.175 | 0.521 | |
| Average | 6.21 | 6.08 | 6.41 | 6.31 | 6.01 | 6.00 | 5.80 | |||||
| P0 value | 0.999 | 0.980 | 0.999 | 0.987 | 0.948 | 0.425 | ||||||
| Energy metabolism | −80 °C | 5.69 | 5.69 | 6.05 | 6.04 | 6.06 | 6.01 | 6.04 | ||||
| −20 °C | 5.69 | 5.94 | 6.06 | 5.71 | 6.01 | 5.99 | 5.94 | 0.099 | 0.001 | 0.531 | 0.329 | |
| Average | 5.69 b | 5.81 ab | 6.05 a | 5.88 ab | 6.03 a | 6.00 a | 5.99 ab | |||||
| P0 value | 0.853 | 0.009 | 0.464 | 0.014 | 0.005 | 0.008 | ||||||
| Lipid metabolism | −80 °C | 4.03 | 4.28 | 3.45 | 3.99 | 3.59 | 4.05 | 3.88 | ||||
| −20 °C | 4.03 | 4.08 | 3.84 | 3.97 | 4.24 | 4.06 | 4.30 | 0.209 | 0.382 | 0.120 | 0.434 | |
| Average | 4.03 | 4.18 | 3.65 | 3.98 | 3.92 | 4.05 | 4.09 | |||||
| P0 value | 0.990 | 0.525 | 1.000 | 0.998 | 1.000 | 1.000 | ||||||
| Translation | −80 °C | 3.52 | 3.58 | 3.48 | 3.49 | 3.50 | 3.48 | 3.51 | ||||
| −20 °C | 3.52 | 3.57 | 3.49 | 3.52 | 3.52 | 3.51 | 3.50 | 0.022 | 0.011 | 0.337 | 0.957 | |
| Average | 3.52 ab | 3.58 a | 3.49 b | 3.50 b | 3.51 ab | 3.49 b | 3.50 b | |||||
| P0 value | 0.217 | 0.613 | 0.970 | 0.991 | 0.629 | 0.885 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Cheng, J.; Xie, Y.; Ouyang, K.; Qu, M.; Pan, K.; Qiu, Q. Dynamic Changes in Rumen Microbial Diversity and Community Composition Within Rumen Fluid in Response to Various Storage Temperatures and Preservation Times. Vet. Sci. 2025, 12, 234. https://doi.org/10.3390/vetsci12030234
Liu C, Cheng J, Xie Y, Ouyang K, Qu M, Pan K, Qiu Q. Dynamic Changes in Rumen Microbial Diversity and Community Composition Within Rumen Fluid in Response to Various Storage Temperatures and Preservation Times. Veterinary Sciences. 2025; 12(3):234. https://doi.org/10.3390/vetsci12030234
Chicago/Turabian StyleLiu, Chang, Jin Cheng, Yunong Xie, Kehui Ouyang, Mingren Qu, Ke Pan, and Qinghua Qiu. 2025. "Dynamic Changes in Rumen Microbial Diversity and Community Composition Within Rumen Fluid in Response to Various Storage Temperatures and Preservation Times" Veterinary Sciences 12, no. 3: 234. https://doi.org/10.3390/vetsci12030234
APA StyleLiu, C., Cheng, J., Xie, Y., Ouyang, K., Qu, M., Pan, K., & Qiu, Q. (2025). Dynamic Changes in Rumen Microbial Diversity and Community Composition Within Rumen Fluid in Response to Various Storage Temperatures and Preservation Times. Veterinary Sciences, 12(3), 234. https://doi.org/10.3390/vetsci12030234






