Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA)
Abstract
1. Introduction
2. Materials and Methods
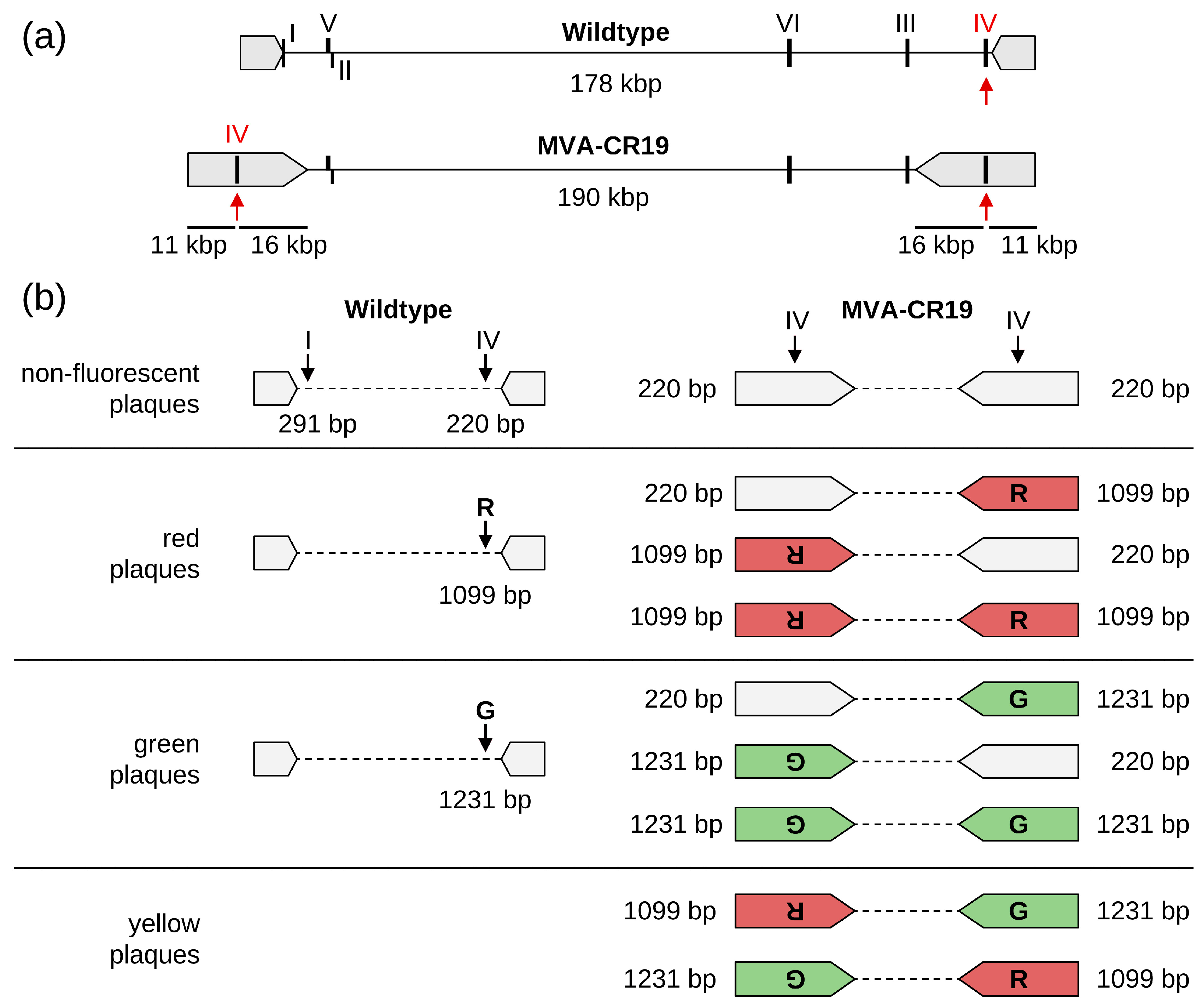
2.1. Design of the Shuttle Plasmids for DS IV
2.2. Generation and Testing of Recombinant MVAs
2.3. CR.pIX and tCR9 Cell Lines
2.4. Quantification of Viruses by Titration and Expression of Fluorescence Markers
3. Results
3.1. Occupancy of DS IV Does Not Interfere with Viability of MVA-CR19
3.2. Consecutive Insertion of EGFP into mCherry-Viruses: An Unusual Recombination Event
3.3. mCherry into EGFP-Expressing MVA-CR19: Confirmation of Copy-Correction at the Terminal Positions
3.4. Insertion of a Selection Marker
3.5. Tetherin Active Also If Activated Late in Replication
3.6. Marker Replacement
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oliveira, G.P.; Rodrigues, R.A.L.; Lima, M.T.; Drumond, B.P.; Abrahão, J.S. Poxvirus Host Range Genes and Virus-Host Spectrum: A Critical Review. Viruses 2017, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, B.; Pérez, P.; Marcos-Villar, L.; Albericio, G.; Astorgano, D.; Álvarez, E.; Sin, L.; Gómez, C.E.; García-Arriaza, J.; Esteban, M. Highly Attenuated Poxvirus-Based Vaccines Against Emerging Viral Diseases. J. Mol. Biol. 2023, 435, 168173. [Google Scholar] [CrossRef] [PubMed]
- Mayr, A.; Hochstein-Mintzel, V.; Stickl, H. Abstammung, Eigenschaften Und Verwendung Des Attenuierten Vaccinia-Stammes MVA. Infection 1975, 3, 6–14. [Google Scholar] [CrossRef]
- Meyer, H.; Sutter, G.; Mayr, A. Mapping of Deletions in the Genome of the Highly Attenuated Vaccinia Virus MVA and Their Influence on Virulence. J. Gen. Virol. 1991, 72 Pt 5, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Meisinger-Henschel, C.; Schmidt, M.; Lukassen, S.; Linke, B.; Krause, L.; Konietzny, S.; Goesmann, A.; Howley, P.; Chaplin, P.; Suter, M.; et al. Genomic Sequence of Chorioallantois Vaccinia Virus Ankara, the Ancestor of Modified Vaccinia Virus Ankara. J. Gen. Virol. 2007, 88, 3249–3259. [Google Scholar] [CrossRef]
- Meisinger-Henschel, C.; Späth, M.; Lukassen, S.; Wolferstätter, M.; Kachelriess, H.; Baur, K.; Dirmeier, U.; Wagner, M.; Chaplin, P.; Suter, M.; et al. Introduction of the Six Major Genomic Deletions of Modified Vaccinia Virus Ankara (MVA) into the Parental Vaccinia Virus Is Not Sufficient to Reproduce an MVA-like Phenotype in Cell Culture and in Mice. J. Virol. 2010, 84, 9907–9919. [Google Scholar] [CrossRef]
- Carroll, M.W.; Moss, B. Host Range and Cytopathogenicity of the Highly Attenuated MVA Strain of Vaccinia Virus: Propagation and Generation of Recombinant Viruses in a Nonhuman Mammalian Cell Line. Virology 1997, 238, 198–211. [Google Scholar] [CrossRef]
- Drexler, I.; Heller, K.; Wahren, B.; Erfle, V.; Sutter, G. Highly Attenuated Modified Vaccinia Virus Ankara Replicates in Baby Hamster Kidney Cells, a Potential Host for Virus Propagation, but Not in Various Human Transformed and Primary Cells. J. Gen. Virol. 1998, 79 Pt 2, 347–352. [Google Scholar] [CrossRef]
- Blanchard, T.J.; Alcami, A.; Andrea, P.; Smith, G.L. Modified Vaccinia Virus Ankara Undergoes Limited Replication in Human Cells and Lacks Several Immunomodulatory Proteins: Implications for Use as a Human Vaccine. J. Gen. Virol. 1998, 79 Pt 5, 1159–1167. [Google Scholar] [CrossRef]
- Jordan, I.; Horn, D.; Thiele, K.; Haag, L.; Fiddeke, K.; Sandig, V. A Deleted Deletion Site in a New Vector Strain and Exceptional Genomic Stability of Plaque-Purified Modified Vaccinia Ankara (MVA). Virol. Sin. 2020, 35, 212–226. [Google Scholar] [CrossRef]
- Jordan, I.; Horn, D.; John, K.; Sandig, V. A Genotype of Modified Vaccinia Ankara (MVA) That Facilitates Replication in Suspension Cultures in Chemically Defined Medium. Viruses 2013, 5, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, T.G.; Bruno, D.; Martens, C.; Porcella, S.F.; Wolf, Y.I.; Moss, B. Mapping Vaccinia Virus DNA Replication Origins at Nucleotide Level by Deep Sequencing. Proc. Natl. Acad. Sci. USA 2015, 112, 10908–10913. [Google Scholar] [CrossRef] [PubMed]
- Kotwal, G.J.; Moss, B. Analysis of a Large Cluster of Nonessential Genes Deleted from a Vaccinia Virus Terminal Transposition Mutant. Virology 1988, 167, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Favis, N.; Famulski, J.; Evans, D.H. Evolution of and Evolutionary Relationships between Extant Vaccinia Virus Strains. J. Virol. 2015, 89, 1809–1824. [Google Scholar] [CrossRef]
- Evans, D.H. Poxvirus Recombination. Pathogens 2022, 11, 896. [Google Scholar] [CrossRef]
- Brinkmann, A.; Kohl, C.; Pape, K.; Bourquain, D.; Thürmer, A.; Michel, J.; Schaade, L.; Nitsche, A. Extensive ITR Expansion of the 2022 Mpox Virus Genome through Gene Duplication and Gene Loss. Virus Genes 2023, 59, 532–540. [Google Scholar] [CrossRef]
- Brennan, G.; Stoian, A.M.M.; Yu, H.; Rahman, M.J.; Banerjee, S.; Stroup, J.N.; Park, C.; Tazi, L.; Rothenburg, S. Molecular Mechanisms of Poxvirus Evolution. mBio 2023, 14, e0152622. [Google Scholar] [CrossRef]
- Li, M.; Zhang, M.; Ye, Q.; Liu, Y.; Qian, W. Preclinical and Clinical Trials of Oncolytic Vaccinia Virus in Cancer Immunotherapy: A Comprehensive Review. Cancer Biol. Med. 2023, 20, 646–661. [Google Scholar] [CrossRef]
- D’Alise, A.M.; Leoni, G.; Cotugno, G.; Siani, L.; Vitale, R.; Ruzza, V.; Garzia, I.; Antonucci, L.; Micarelli, E.; Venafra, V.; et al. Phase I Trial of Viral Vector-Based Personalized Vaccination Elicits Robust Neoantigen-Specific Antitumor T-Cell Responses. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2024, 30, 2412–2423. [Google Scholar] [CrossRef]
- Bendjama, K.; Quemeneur, E. Modified Vaccinia Virus Ankara-Based Vaccines in the Era of Personalized Immunotherapy of Cancer. Hum. Vaccines Immunother. 2017, 13, 1997–2003. [Google Scholar] [CrossRef] [PubMed]
- Altenburg, A.F.; Kreijtz, J.H.C.M.; de Vries, R.D.; Song, F.; Fux, R.; Rimmelzwaan, G.F.; Sutter, G.; Volz, A. Modified Vaccinia Virus Ankara (MVA) as Production Platform for Vaccines against Influenza and Other Viral Respiratory Diseases. Viruses 2014, 6, 2735–2761. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bruno, D.P.; Martens, C.A.; Porcella, S.F.; Moss, B. Genome-Wide Analysis of the 5’ and 3’ Ends of Vaccinia Virus Early mRNAs Delineates Regulatory Sequences of Annotated and Anomalous Transcripts. J. Virol. 2011, 85, 5897–5909. [Google Scholar] [CrossRef] [PubMed]
- Yuen, L.; Moss, B. Oligonucleotide Sequence Signaling Transcriptional Termination of Vaccinia Virus Early Genes. Proc. Natl. Acad. Sci. USA 1987, 84, 6417–6421. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Svensjö, T.; Winkler, T.; Lu, M.; Eriksson, C.; Eriksson, E. Tetracycline Repressor, tetR, Rather than the tetR-Mammalian Cell Transcription Factor Fusion Derivatives, Regulates Inducible Gene Expression in Mammalian Cells. Hum. Gene Ther. 1998, 9, 1939–1950. [Google Scholar] [CrossRef]
- Neckermann, P.; Mohr, M.; Billmeier, M.; Karlas, A.; Boilesen, D.R.; Thirion, C.; Holst, P.J.; Jordan, I.; Sandig, V.; Asbach, B.; et al. Transgene Expression Knock-down in Recombinant Modified Vaccinia Virus Ankara Vectors Improves Genetic Stability and Sustained Transgene Maintenance across Multiple Passages. Front. Immunol. 2024, 15, 1338492. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Sisler, J.R.; Moss, B. Compact, Synthetic, Vaccinia Virus Early/Late Promoter for Protein Expression. BioTechniques 1997, 23, 1094–1097. [Google Scholar] [CrossRef]
- Jordan, I.; Vos, A.; Beilfuss, S.; Neubert, A.; Breul, S.; Sandig, V. An Avian Cell Line Designed for Production of Highly Attenuated Viruses. Vaccine 2009, 27, 748–756. [Google Scholar] [CrossRef]
- Jordan, I.; Northoff, S.; Thiele, M.; Hartmann, S.; Horn, D.; Höwing, K.; Bernhardt, H.; Oehmke, S.; von Horsten, H.; Rebeski, D.; et al. A Chemically Defined Production Process for Highly Attenuated Poxviruses. Biol. J. Int. Assoc. Biol. Stand. 2011, 39, 50–58. [Google Scholar] [CrossRef]
- Darling, A.J.; Boose, J.A.; Spaltro, J. Virus Assay Methods: Accuracy and Validation. Biol. J. Int. Assoc. Biol. Stand. 1998, 26, 105–110. [Google Scholar] [CrossRef]
- Dobson, B.M.; Tscharke, D.C. Redundancy Complicates the Definition of Essential Genes for Vaccinia Virus. J. Gen. Virol. 2015, 96, 3326–3337. [Google Scholar] [CrossRef] [PubMed]
- Tapia, F.; Jordan, I.; Genzel, Y.; Reichl, U. Efficient and Stable Production of Modified Vaccinia Ankara Virus in Two-Stage Semi-Continuous and in Continuous Stirred Tank Cultivation Systems. PLoS ONE 2017, 12, e0182553. [Google Scholar] [CrossRef] [PubMed]
- Perez-Caballero, D.; Zang, T.; Ebrahimi, A.; McNatt, M.W.; Gregory, D.A.; Johnson, M.C.; Bieniasz, P.D. Tetherin Inhibits HIV-1 Release by Directly Tethering Virions to Cells. Cell 2009, 139, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, L.S.; Shors, S.T.; Murphy, B.R.; Moss, B. Development of a Replication-Deficient Recombinant Vaccinia Virus Vaccine Effective against Parainfluenza Virus 3 Infection in an Animal Model. Vaccine 1996, 14, 1451–1458. [Google Scholar] [CrossRef] [PubMed]
- Shockett, P.E.; Schatz, D.G. Diverse Strategies for Tetracycline-Regulated Inducible Gene Expression. Proc. Natl. Acad. Sci. USA 1996, 93, 5173–5176. [Google Scholar] [CrossRef]
- Kieser, Q.; Noyce, R.S.; Shenouda, M.; Lin, Y.-C.J.; Evans, D.H. Cytoplasmic Factories, Virus Assembly, and DNA Replication Kinetics Collectively Constrain the Formation of Poxvirus Recombinants. PLoS ONE 2020, 15, e0228028. [Google Scholar] [CrossRef]
- Payne, L.G.; Kristenson, K. Mechanism of Vaccinia Virus Release and Its Specific Inhibition by N1-Isonicotinoyl-N2-3-Methyl-4-Chlorobenzoylhydrazine. J. Virol. 1979, 32, 614–622. [Google Scholar] [CrossRef]
- Massung, R.F.; Knight, J.C.; Esposito, J.J. Topography of Variola Smallpox Virus Inverted Terminal Repeats. Virology 1995, 211, 350–355. [Google Scholar] [CrossRef]
- Greseth, M.D.; Traktman, P. The Life Cycle of the Vaccinia Virus Genome. Annu. Rev. Virol. 2022, 9, 239–259. [Google Scholar] [CrossRef]
- McFadden, G.; Dales, S. Biogenesis of Poxviruses: Mirror-Image Deletions in Vaccinia Virus DNA. Cell 1979, 18, 101–108. [Google Scholar] [CrossRef]
- Mackett, M.; Smith, G.L.; Moss, B. General Method for Production and Selection of Infectious Vaccinia Virus Recombinants Expressing Foreign Genes. J. Virol. 1984, 49, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Kremer, M.; Volz, A.; Kreijtz, J.H.C.M.; Fux, R.; Lehmann, M.H.; Sutter, G. Easy and Efficient Protocols for Working with Recombinant Vaccinia Virus MVA. Methods Mol. Biol. Clifton N. J. 2012, 890, 59–92. [Google Scholar] [CrossRef]
- Kugler, F.; Drexler, I.; Protzer, U.; Hoffmann, D.; Moeini, H. Generation of Recombinant MVA-Norovirus: A Comparison Study of Bacterial Artificial Chromosome- and Marker-Based Systems. Virol. J. 2019, 16, 100. [Google Scholar] [CrossRef] [PubMed]
- Antoine, G.; Scheiflinger, F.; Dorner, F.; Falkner, F.G. The Complete Genomic Sequence of the Modified Vaccinia Ankara Strain: Comparison with Other Orthopoxviruses. Virology 1998, 244, 365–396. [Google Scholar] [CrossRef] [PubMed]
- Vallée, G.; Norris, P.; Paszkowski, P.; Noyce, R.S.; Evans, D.H. Vaccinia Virus Gene Acquisition through Nonhomologous Recombination. J. Virol. 2021, 95, e0031821. [Google Scholar] [CrossRef]
- Upton, C.; Slack, S.; Hunter, A.L.; Ehlers, A.; Roper, R.L. Poxvirus Orthologous Clusters: Toward Defining the Minimum Essential Poxvirus Genome. J. Virol. 2003, 77, 7590–7600. [Google Scholar] [CrossRef]
- Qin, L.; Upton, C.; Hazes, B.; Evans, D.H. Genomic Analysis of the Vaccinia Virus Strain Variants Found in Dryvax Vaccine. J. Virol. 2011, 85, 13049–13060. [Google Scholar] [CrossRef]
- Elde, N.C.; Child, S.J.; Eickbush, M.T.; Kitzman, J.O.; Rogers, K.S.; Shendure, J.; Geballe, A.P.; Malik, H.S. Poxviruses Deploy Genomic Accordions to Adapt Rapidly against Host Antiviral Defenses. Cell 2012, 150, 831–841. [Google Scholar] [CrossRef]
- Sliva, K.; Resch, T.; Kraus, B.; Goffinet, C.; Keppler, O.T.; Schnierle, B.S. The Cellular Antiviral Restriction Factor Tetherin Does Not Inhibit Poxviral Replication. J. Virol. 2012, 86, 1893–1896. [Google Scholar] [CrossRef]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.-F. Lessons from the Host Defences of Bats, a Unique Viral Reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef]
- Hayward, J.A.; Tachedjian, M.; Johnson, A.; Irving, A.T.; Gordon, T.B.; Cui, J.; Nicolas, A.; Smith, I.; Boyd, V.; Marsh, G.A.; et al. Unique Evolution of Antiviral Tetherin in Bats. J. Virol. 2022, 96, e0115222. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cooper, T.; Howley, P.M.; Hayball, J.D. From Crescent to Mature Virion: Vaccinia Virus Assembly and Maturation. Viruses 2014, 6, 3787–3808. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, M.; Samolej, J.; Evans, R.J.; Yakimovich, A.; White, I.J.; Kriston-Vizi, J.; Martin-Serrano, J.; Sundquist, W.I.; Frickel, E.-M.; Mercer, J. Vaccinia Virus Hijacks ESCRT-Mediated Multivesicular Body Formation for Virus Egress. Life Sci. Alliance 2021, 4, e202000910. [Google Scholar] [CrossRef] [PubMed]
- Tolonen, N.; Doglio, L.; Schleich, S.; Krijnse Locker, J. Vaccinia Virus DNA Replication Occurs in Endoplasmic Reticulum-Enclosed Cytoplasmic Mini-Nuclei. Mol. Biol. Cell 2001, 12, 2031–2046. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.J.; Evans, D.H. Vaccinia Virus Particles Mix Inefficiently, and in a Way That Would Restrict Viral Recombination, in Coinfected Cells. J. Virol. 2010, 84, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, A.S.; Maruri-Avidal, L.; Bisht, H.; Hansen, B.T.; Schwartz, C.L.; Fischer, E.R.; Meng, X.; Xiang, Y.; Moss, B. Enigmatic Origin of the Poxvirus Membrane from the Endoplasmic Reticulum Shown by 3D Imaging of Vaccinia Virus Assembly Mutants. Proc. Natl. Acad. Sci. USA 2017, 114, E11001–E11009. [Google Scholar] [CrossRef]
- Hyun, J. Poxvirus under the Eyes of Electron Microscope. Appl. Microsc. 2022, 52, 11. [Google Scholar] [CrossRef]
- Di Lullo, G.; Soprana, E.; Panigada, M.; Palini, A.; Erfle, V.; Staib, C.; Sutter, G.; Siccardi, A.G. Marker Gene Swapping Facilitates Recombinant Modified Vaccinia Virus Ankara Production by Host-Range Selection. J. Virol. Methods 2009, 156, 37–43. [Google Scholar] [CrossRef]
- Staib, C.; Drexler, I.; Ohlmann, M.; Wintersperger, S.; Erfle, V.; Sutter, G. Transient Host Range Selection for Genetic Engineering of Modified Vaccinia Virus Ankara. BioTechniques 2000, 28, 1137–1142, 1144–1146, 1148. [Google Scholar] [CrossRef]
- Antoshkina, I.V.; Glazkova, D.V.; Urusov, F.A.; Bogoslovskaya, E.V.; Shipulin, G.A. Comparison of Recombinant MVA Selection Methods Based on F13L, D4R and K1L Genes. Viruses 2022, 14, 528. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abidi, S.; Elhazaz Fernandez, A.; Seehase, N.; Hanisch, L.; Karlas, A.; Sandig, V.; Jordan, I. Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA). Vaccines 2024, 12, 1377. https://doi.org/10.3390/vaccines12121377
Abidi S, Elhazaz Fernandez A, Seehase N, Hanisch L, Karlas A, Sandig V, Jordan I. Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA). Vaccines. 2024; 12(12):1377. https://doi.org/10.3390/vaccines12121377
Chicago/Turabian StyleAbidi, Sirine, Aurora Elhazaz Fernandez, Nicole Seehase, Lina Hanisch, Alexander Karlas, Volker Sandig, and Ingo Jordan. 2024. "Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA)" Vaccines 12, no. 12: 1377. https://doi.org/10.3390/vaccines12121377
APA StyleAbidi, S., Elhazaz Fernandez, A., Seehase, N., Hanisch, L., Karlas, A., Sandig, V., & Jordan, I. (2024). Expression of an Efficient Selection Marker Out of a Duplicated Site in the ITRs of a Modified Vaccinia Virus Ankara (MVA). Vaccines, 12(12), 1377. https://doi.org/10.3390/vaccines12121377





