The Repurposing of Cellular Proteins during Enterovirus A71 Infection
Abstract
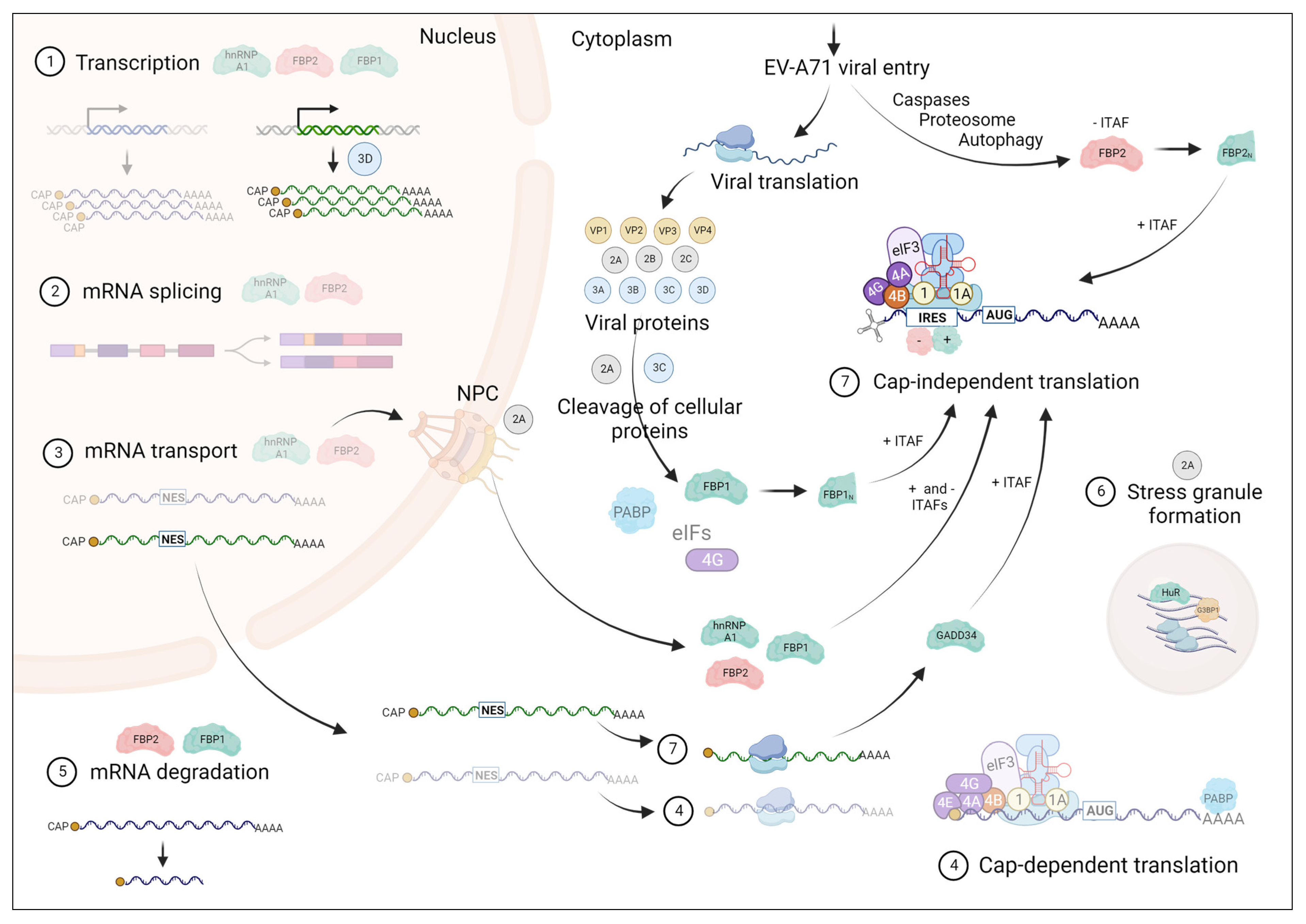
1. Introduction
2. Cellular IRES Trans-Acting Factors (ITAFs) That Regulate EV-A71 Translation
2.1. hnRNP A1
2.2. AUF1 (hnRNP D)
2.3. hnRNP K
2.4. PCBP1 (hnRNP E1)
2.5. PTB (hnRNP I)
2.6. FBP2
N- and C-Terminus Cleaved FBP2
2.7. FBP1
C-Terminus Cleaved FBP1
2.8. Ago2
2.9. HuR
Additive Effects of Ago2 and HuR
2.10. MOV10
2.11. SIRT1
2.12. Sam68
2.13. FUBP3
2.14. GADD34
2.15. DDX3
2.16. APOBEC3G
2.17. Staufen1
2.18. hnRNP H and hnRNP F
2.19. EGR1
2.20. TIA-1 and TIAR
2.21. Additional Cellular Proteins
3. Discussion
4. Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nayak, G.; Bhuyan, S.K.; Bhuyan, R.; Sahu, A.; Kar, D.; Kuanar, A. Global Emergence of Enterovirus 71: A Systematic Review. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 78. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-L.; Chen, C.-M.; Wang, E.-T.; Kuo, H.-W.; Shih, W.-L.; Fang, C.-T.; Liu, D.-P.; Chang, L.-Y. The Secular Trend of Enterovirus A71 after the Implementation of Preventive Measures in Taiwan. BMC Public Health 2022, 22, 1483. [Google Scholar] [CrossRef] [PubMed]
- Enterovirus 71. Available online: https://www.who.int/teams/health-product-policy-and-standards/standards-and-specifications/vaccine-standardization/enterovirus-71 (accessed on 8 November 2023).
- Huang, P.-N.; Shih, S.-R. Update on Enterovirus 71 Infection. Curr. Opin. Virol. 2014, 5, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Ibba, R.; Carta, A.; Madeddu, S.; Caria, P.; Serreli, G.; Piras, S.; Sestito, S.; Loddo, R.; Sanna, G. Inhibition of Enterovirus A71 by a Novel 2-Phenyl-Benzimidazole Derivative. Viruses 2021, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Y.; Zhang, H.; Zheng, Y.; Gou, F.; Yang, X.; Cheng, Y.; McClymont, H.; Li, H.; Liu, X.; et al. Prototypes Virus of Hand, Foot and Mouth Disease Infections and Severe Cases in Gansu, China: A Spatial and Temporal Analysis. BMC Infect. Dis. 2022, 22, 408. [Google Scholar] [CrossRef] [PubMed]
- Wörner, N.; Rodrigo-García, R.; Antón, A.; Castellarnau, E.; Delgado, I.; Vazquez, È.; González, S.; Mayol, L.; Méndez, M.; Solé, E.; et al. Enterovirus-A71 Rhombencephalitis Outbreak in Catalonia: Characteristics, Management and Outcome. Pediatr. Infect. Dis. J. 2021, 40, 628. [Google Scholar] [CrossRef]
- Messacar, K.; Burakoff, A.; Nix, W.A.; Rogers, S.; Oberste, M.S.; Gerber, S.I.; Spence-Davizon, E.; Herlihy, R.; Dominguez, S.R. Notes from the Field: Enterovirus A71 Neurologic Disease in Children—Colorado, 2018. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 1017–1018. [Google Scholar] [CrossRef]
- Nhan, L.N.T.; Hong, N.T.T.; Nhu, L.N.T.; Nguyet, L.A.; Ny, N.T.H.; Thanh, T.T.; Han, D.D.K.; Van, H.M.T.; Thwaites, C.L.; Hien, T.T.; et al. Severe Enterovirus A71 Associated Hand, Foot and Mouth Disease, Vietnam, 2018: Preliminary Report of an Impending Outbreak. Eur. Surveill. 2018, 23, 1800590. [Google Scholar] [CrossRef]
- NIAID Emerging Infectious Diseases/Pathogens|NIH: National Institute of Allergy and Infectious Diseases. Available online: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed on 8 November 2023).
- Li, M.-L.; Shih, S.-R.; Tolbert, B.S.; Brewer, G. Enterovirus A71 Vaccines. Vaccines 2021, 9, 199. [Google Scholar] [CrossRef]
- Yi, E.-J.; Shin, Y.-J.; Kim, J.-H.; Kim, T.-G.; Chang, S.-Y. Enterovirus 71 Infection and Vaccines. Clin. Exp. Vaccine Res. 2017, 6, 4–14. [Google Scholar] [CrossRef]
- Chan, Y.-F.; Sam, I.-C.; AbuBakar, S. Phylogenetic Designation of Enterovirus 71 Genotypes and Subgenotypes Using Complete Genome Sequences. Infect. Genet. Evol. 2010, 10, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Pallansch, M.A. Complete Nucleotide Sequence of Enterovirus 71 Is Distinct from Poliovirus. Virus Res. 1995, 39, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Tolbert, M.; Morgan, C.E.; Pollum, M.; Crespo-Hernández, C.E.; Li, M.-L.; Brewer, G.; Tolbert, B.S. HnRNP A1 Alters the Structure of a Conserved Enterovirus IRES Domain to Stimulate Viral Translation. J. Mol. Biol. 2017, 429, 2841–2858. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.R.; Sarnow, P. Enterovirus 71 Contains a Type I IRES Element That Functions When Eukaryotic Initiation Factor eIF4G Is Cleaved. Virology 2003, 315, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.-C.; Chen, H.-H.; Xu, P.; Wang, R.Y.L. Translation Control of Enterovirus A71 Gene Expression. J. Biomed. Sci. 2020, 27, 22. [Google Scholar] [CrossRef] [PubMed]
- Sommergruber, W.; Ahorn, H.; Klump, H.; Seipelt, J.; Zoephel, A.; Fessl, F.; Krystek, E.; Blaas, D.; Kuechler, E.; Liebig, H.D.; et al. 2A Proteinases of Coxsackie- and Rhinovirus Cleave Peptides Derived from eIF-4γ via a Common Recognition Motif. Virology 1994, 198, 741–745. [Google Scholar] [CrossRef]
- de Breyne, S.; Bonderoff, J.M.; Chumakov, K.M.; Lloyd, R.E.; Hellen, C.U.T. Cleavage of Eukaryotic Initiation Factor eIF5B by Enterovirus 3C Proteases. Virology 2008, 378, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Kuyumcu-Martinez, N.M.; Van Eden, M.E.; Younan, P.; Lloyd, R.E. Cleavage of Poly(A)-Binding Protein by Poliovirus 3C Protease Inhibits Host Cell Translation: A Novel Mechanism for Host Translation Shutoff. Mol. Cell. Biol. 2004, 24, 1779–1790. [Google Scholar] [CrossRef]
- White, J.P.; Reineke, L.C.; Lloyd, R.E. Poliovirus Switches to an eIF2-Independent Mode of Translation during Infection. J. Virol. 2011, 85, 8884–8893. [Google Scholar] [CrossRef]
- Weng, K.-F.; Li, M.-L.; Hung, C.-T.; Shih, S.-R. Enterovirus 71 3C Protease Cleaves a Novel Target CstF-64 and Inhibits Cellular Polyadenylation. PLoS Pathog. 2009, 5, e1000593. [Google Scholar] [CrossRef]
- Lei, X.; Xiao, X.; Xue, Q.; Jin, Q.; He, B.; Wang, J. Cleavage of Interferon Regulatory Factor 7 by Enterovirus 71 3C Suppresses Cellular Responses. J. Virol. 2013, 87, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Han, N.; Xiao, X.; Jin, Q.; He, B.; Wang, J. Enterovirus 71 3C Inhibits Cytokine Expression through Cleavage of the TAK1/TAB1/TAB2/TAB3 Complex. J. Virol. 2014, 88, 9830–9841. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-L.; Lin, J.-Y.; Chen, B.-S.; Weng, K.-F.; Shih, S.-R.; Calderon, J.D.; Tolbert, B.S.; Brewer, G. EV71 3C Protease Induces Apoptosis by Cleavage of hnRNP A1 to Promote Apaf-1 Translation. PLoS ONE 2019, 14, e0221048. [Google Scholar] [CrossRef]
- Sweeney, T.R.; Abaeva, I.S.; Pestova, T.V.; Hellen, C.U.T. The Mechanism of Translation Initiation on Type 1 Picornavirus IRESs. EMBO J. 2014, 33, 76–92. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.W.; Wu, J.; Wang, X.; Guo, H.; Sun, S. Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses. mBio 2023, 14, e0035823. [Google Scholar] [CrossRef]
- López-Ulloa, B.; Fuentes, Y.; Pizarro-Ortega, M.S.; López-Lastra, M. RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation. Viruses 2022, 14, 188. [Google Scholar] [CrossRef]
- Martínez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Lozano, G.; Diaz-Toledano, R. Picornavirus IRES Elements: RNA Structure and Host Protein Interactions. Virus Res. 2015, 206, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Salas, E. The Impact of RNA Structure on Picornavirus IRES Activity. Trends Microbiol. 2008, 16, 230–237. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Embarek, A.M. Insights into Structural and Mechanistic Features of Viral IRES Elements. Front. Microbiol. 2018, 8, 2629. [Google Scholar] [CrossRef]
- Pestova, T.V.; Hellen, C.U.; Wimmer, E. A Conserved AUG Triplet in the 5’ Nontranslated Region of Poliovirus Can Function as an Initiation Codon in Vitro and in Vivo. Virology 1994, 204, 729–737. [Google Scholar] [CrossRef]
- Lulla, V.; Dinan, A.M.; Hosmillo, M.; Chaudhry, Y.; Sherry, L.; Irigoyen, N.; Nayak, K.M.; Stonehouse, N.J.; Zilbauer, M.; Goodfellow, I.; et al. An Upstream Protein-Coding Region in Enteroviruses Modulates Virus Infection in Gut Epithelial Cells. Nat. Microbiol. 2019, 4, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, Y.; Liu, G.; Jiang, Y.; Shen, S.; Bi, R.; Huang, H.; Cheng, T.; Wang, C.; Wei, W. A Second Open Reading Frame in Human Enterovirus Determines Viral Replication in Intestinal Epithelial Cells. Nat. Commun. 2019, 10, 4066. [Google Scholar] [CrossRef]
- Lizcano-Perret, B.; Michiels, T. Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses. Viruses 2021, 13, 1210. [Google Scholar] [CrossRef]
- Wang, C.; Sun, M.; Yuan, X.; Ji, L.; Jin, Y.; Cardona, C.J.; Xing, Z. Enterovirus 71 Suppresses Interferon Responses by Blocking Janus Kinase (JAK)/Signal Transducer and Activator of Transcription (STAT) Signaling through Inducing Karyopherin-A1 Degradation. J. Biol. Chem. 2017, 292, 10262–10274. [Google Scholar] [CrossRef] [PubMed]
- Shiroki, K.; Isoyama, T.; Kuge, S.; Ishii, T.; Ohmi, S.; Hata, S.; Suzuki, K.; Takasaki, Y.; Nomoto, A. Intracellular Redistribution of Truncated La Protein Produced by Poliovirus 3Cpro-Mediated Cleavage. J. Virol. 1999, 73, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, D.; Wang, M.; Cheng, A.; Zhu, Y.; Mao, S.; Ou, X.; Zhao, X.; Huang, J.; Gao, Q.; et al. Multiple Functions of Heterogeneous Nuclear Ribonucleoproteins in the Positive Single-Stranded RNA Virus Life Cycle. Front. Immunol. 2022, 13, 989298. [Google Scholar] [CrossRef]
- Mayeda, A.; Krainer, A.R. Regulation of Alternative Pre-mRNA Splicing by hnRNP A1 and Splicing Factor SF2. Cell 1992, 68, 365–375. [Google Scholar] [CrossRef]
- Jean-Philippe, J.; Paz, S.; Caputi, M. hnRNP A1: The Swiss Army Knife of Gene Expression. Int. J. Mol. Sci. 2013, 14, 18999–19024. [Google Scholar] [CrossRef]
- Michael, W.M.; Choi, M.; Dreyfuss, G. A Nuclear Export Signal in hnRNP A1: A Signal-Mediated, Temperature-Dependent Nuclear Protein Export Pathway. Cell 1995, 83, 415–422. [Google Scholar] [CrossRef]
- Henics, T.; Sanfridson, A.; Hamilton, B.J.; Nagy, E.; Rigby, W.F. Enhanced Stability of Interleukin-2 mRNA in MLA 144 Cells. Possible Role of Cytoplasmic AU-Rich Sequence-Binding Proteins. J. Biol. Chem. 1994, 269, 5377–5383. [Google Scholar] [CrossRef]
- Hamilton, B.J.; Burns, C.M.; Nichols, R.C.; Rigby, W.F. Modulation of AUUUA Response Element Binding by Heterogeneous Nuclear Ribonucleoprotein A1 in Human T Lymphocytes. The Roles of Cytoplasmic Location, Transcription, and Phosphorylation. J. Biol. Chem. 1997, 272, 28732–28741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-S.; Manche, L.; Xu, R.-M.; Krainer, A.R. hnRNP A1 Associates with Telomere Ends and Stimulates Telomerase Activity. RNA 2006, 12, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Cáceres, J.F. The Multifunctional RNA-Binding Protein hnRNP A1 Is Required for Processing of miR-18a. Nat. Struct. Mol. Biol. 2007, 14, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Mili, S.; Shu, H.J.; Zhao, Y.; Piñol-Roma, S. Distinct RNP Complexes of Shuttling hnRNP Proteins with Pre-mRNA and mRNA: Candidate Intermediates in Formation and Export of mRNA. Mol. Cell. Biol. 2001, 21, 7307–7319. [Google Scholar] [CrossRef] [PubMed]
- Piñol-Roma, S.; Dreyfuss, G. Shuttling of Pre-mRNA Binding Proteins between Nucleus and Cytoplasm. Nature 1992, 355, 730–732. [Google Scholar] [CrossRef]
- Levengood, J.D.; Tolbert, B.S. Idiosyncrasies of hnRNP A1-RNA Recognition: Can Binding Mode Influence Function. Semin. Cell Dev. Biol. 2019, 86, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.E.; Meagher, J.L.; Levengood, J.D.; Delproposto, J.; Rollins, C.; Stuckey, J.A.; Tolbert, B.S. The First Crystal Structure of the UP1 Domain of hnRNP A1 Bound to RNA Reveals a New Look for an Old RNA Binding Protein. J. Mol. Biol. 2015, 427, 3241–3257. [Google Scholar] [CrossRef] [PubMed]
- Kooshapur, H.; Choudhury, N.R.; Simon, B.; Mühlbauer, M.; Jussupow, A.; Fernandez, N.; Jones, A.N.; Dallmann, A.; Gabel, F.; Camilloni, C.; et al. Structural Basis for Terminal Loop Recognition and Stimulation of Pri-miRNA-18a Processing by hnRNP A1. Nat. Commun. 2018, 9, 2479. [Google Scholar] [CrossRef]
- Barraud, P.; Allain, F.H.-T. Solution Structure of the Two RNA Recognition Motifs of hnRNP A1 Using Segmental Isotope Labeling: How the Relative Orientation between RRMs Influences the Nucleic Acid Binding Topology. J. Biomol. NMR 2013, 55, 119–138. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Shih, S.-R.; Pan, M.; Li, C.; Lue, C.-F.; Stollar, V.; Li, M.-L. hnRNP A1 Interacts with the 5′ Untranslated Regions of Enterovirus 71 and Sindbis Virus RNA and Is Required for Viral Replication. J. Virol. 2009, 83, 6106–6114. [Google Scholar] [CrossRef]
- Levengood, J.D.; Tolbert, M.; Li, M.-L.; Tolbert, B.S. High-Affinity Interaction of hnRNP A1 with Conserved RNA Structural Elements Is Required for Translation and Replication of Enterovirus 71. RNA Biol. 2013, 10, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-Y.; Li, M.-L.; Huang, P.-N.; Chien, K.-Y.; Horng, J.-T.; Shih, S.-R. Heterogeneous Nuclear Ribonuclear Protein K Interacts with the Enterovirus 71 5’ Untranslated Region and Participates in Virus Replication. J. Gen. Virol. 2008, 89 Pt 10, 2540–2549. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Li, M.-L.; Shih, S.-R. Far Upstream Element Binding Protein 2 Interacts with Enterovirus 71 Internal Ribosomal Entry Site and Negatively Regulates Viral Translation. Nucleic Acids Res. 2009, 37, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Davila-Calderon, J.; Li, M.-L.; Penumutchu, S.R.; Haddad, C.; Malcolm, L.; Hargrove, A.E.; Brewer, G.; Tolbert, B.S. Enterovirus Evolution Reveals the Mechanism of an RNA-Targeted Antiviral and Determinants of Viral Replication. bioRxiv 2023. [Google Scholar] [CrossRef]
- Fialcowitz, E.J.; Brewer, B.Y.; Keenan, B.P.; Wilson, G.M. A Hairpin-like Structure within an AU-Rich mRNA-Destabilizing Element Regulates Trans-Factor Binding Selectivity and mRNA Decay Kinetics. J. Biol. Chem. 2005, 280, 22406–22417. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Brewer, G. The Regulation of mRNA Stability in Mammalian Cells: 2.0. Gene 2012, 500, 10. [Google Scholar] [CrossRef]
- Pont, A.R.; Sadri, N.; Hsiao, S.J.; Smith, S.; Schneider, R.J. mRNA Decay Factor AUF1 Maintains Normal Aging, Telomere Maintenance, and Suppression of Senescence by Activation of Telomerase Transcription. Mol. Cell 2012, 47, 5–15. [Google Scholar] [CrossRef]
- Laroia, G.; Cuesta, R.; Brewer, G.; Schneider, R.J. Control of mRNA Decay by Heat Shock-Ubiquitin-Proteasome Pathway. Science 1999, 284, 499–502. [Google Scholar] [CrossRef]
- Lu, J.-Y.; Bergman, N.; Sadri, N.; Schneider, R.J. Assembly of AUF1 with eIF4G–Poly(A) Binding Protein Complex Suggests a Translation Function in AU-Rich mRNA Decay. RNA 2006, 12, 883–893. [Google Scholar] [CrossRef]
- Sinsimer, K.S.; Gratacós, F.M.; Knapinska, A.M.; Lu, J.; Krause, C.D.; Wierzbowski, A.V.; Maher, L.R.; Scrudato, S.; Rivera, Y.M.; Gupta, S.; et al. Chaperone Hsp27, a Novel Subunit of AUF1 Protein Complexes, Functions in AU-Rich Element-Mediated mRNA Decay. Mol. Cell. Biol. 2008, 28, 5223–5237. [Google Scholar] [CrossRef] [PubMed]
- Davila-Calderon, J.; Patwardhan, N.N.; Chiu, L.-Y.; Sugarman, A.; Cai, Z.; Penutmutchu, S.R.; Li, M.-L.; Brewer, G.; Hargrove, A.E.; Tolbert, B.S. IRES-Targeting Small Molecule Inhibits Enterovirus 71 Replication via Allosteric Stabilization of a Ternary Complex. Nat. Commun. 2020, 11, 4775. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-Y.; Li, M.-L.; Brewer, G. mRNA Decay Factor AUF1 Binds the Internal Ribosomal Entry Site of Enterovirus 71 and Inhibits Virus Replication. PLoS ONE 2014, 9, e103827. [Google Scholar] [CrossRef] [PubMed]
- Cathcart, A.L.; Rozovics, J.M.; Semler, B.L. Cellular mRNA Decay Protein AUF1 Negatively Regulates Enterovirus and Human Rhinovirus Infections. J. Virol. 2013, 87, 10423–10434. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-Y.; Brewer, G.; Li, M.-L. HuR and Ago2 Bind the Internal Ribosome Entry Site of Enterovirus 71 and Promote Virus Translation and Replication. PLoS ONE 2015, 10, e0140291. [Google Scholar] [CrossRef] [PubMed]
- Stains, J.P.; Lecanda, F.; Towler, D.A.; Civitelli, R. Heterogeneous Nuclear Ribonucleoprotein K Represses Transcription from a Cytosine/Thymidine-Rich Element in the Osteocalcin Promoter. Biochem. J. 2005, 385 Pt 2, 613–623. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cao, W.; Razanau, A.; Feng, D.; Lobo, V.G.; Xie, J. Control of Alternative Splicing by Forskolin through hnRNP K during Neuronal Differentiation. Nucleic Acids Res. 2012, 40, 8059–8071. [Google Scholar] [CrossRef]
- Fan, X.; Xiong, H.; Wei, J.; Gao, X.; Feng, Y.; Liu, X.; Zhang, G.; He, Q.-Y.; Xu, J.; Liu, L. Cytoplasmic hnRNPK Interacts with GSK3β and Is Essential for the Osteoclast Differentiation. Sci. Rep. 2015, 5, 17732. [Google Scholar] [CrossRef]
- Fukuda, T.; Naiki, T.; Saito, M.; Irie, K. hnRNP K Interacts with RNA Binding Motif Protein 42 and Functions in the Maintenance of Cellular ATP Level during Stress Conditions. Genes Cells 2009, 14, 113–128. [Google Scholar] [CrossRef]
- Habelhah, H.; Shah, K.; Huang, L.; Ostareck-Lederer, A.; Burlingame, A.L.; Shokat, K.M.; Hentze, M.W.; Ronai, Z. ERK Phosphorylation Drives Cytoplasmic Accumulation of hnRNP-K and Inhibition of mRNA Translation. Nat. Cell Biol. 2001, 3, 325–330. [Google Scholar] [CrossRef]
- Bomsztyk, K.; Denisenko, O.; Ostrowski, J. hnRNP K: One Protein Multiple Processes. Bioessays 2004, 26, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Leffers, H.; Dejgaard, K.; Celis, J.E. Characterisation of Two Major Cellular Poly(rC)-Binding Human Proteins, Each Containing Three K-Homologous (KH) Domains. Eur. J. Biochem. 1995, 230, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Dong, X.; Li, Y.; Zhang, Q.; Kim, C.; Song, Y.; Kang, L.; Liu, Y.; Wu, K.; Wu, J. PolyC-Binding Protein 1 Interacts with 5′-Untranslated Region of Enterovirus 71 RNA in Membrane-Associated Complex to Facilitate Viral Replication. PLoS ONE 2014, 9, e87491. [Google Scholar] [CrossRef] [PubMed]
- Thyagarajan, A.; Szaro, B.G. Phylogenetically Conserved Binding of Specific K Homology Domain Proteins to the 3’-Untranslated Region of the Vertebrate Middle Neurofilament mRNA. J. Biol. Chem. 2004, 279, 49680–49688. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Rayala, S.K.; Gururaj, A.E.; Talukder, A.H.; O’Malley, B.W.; Kumar, R. Signaling-Dependent and Coordinated Regulation of Transcription, Splicing, and Translation Resides in a Single Coregulator, PCBP1. Proc. Natl. Acad. Sci. USA 2007, 104, 5866–5871. [Google Scholar] [CrossRef] [PubMed]
- Lian, W.-X.; Yin, R.-H.; Kong, X.-Z.; Zhang, T.; Huang, X.-H.; Zheng, W.-W.; Yang, Y.; Zhan, Y.-Q.; Xu, W.-X.; Yu, M.; et al. THAP11, a Novel Binding Protein of PCBP1, Negatively Regulates CD44 Alternative Splicing and Cell Invasion in a Human Hepatoma Cell Line. FEBS Lett. 2012, 586, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Huang, X.-H.; Dong, L.; Hu, D.; Ge, C.; Zhan, Y.-Q.; Xu, W.-X.; Yu, M.; Li, W.; Wang, X.; et al. PCBP-1 Regulates Alternative Splicing of the CD44 Gene and Inhibits Invasion in Human Hepatoma Cell Line HepG2 Cells. Mol. Cancer 2010, 9, 72. [Google Scholar] [CrossRef]
- Malik, A.K.; Flock, K.E.; Godavarthi, C.L.; Loh, H.H.; Ko, J.L. Molecular Basis Underlying the Poly C Binding Protein 1 as a Regulator of the Proximal Promoter of Mouse Mu-Opioid Receptor Gene. Brain Res. 2006, 1112, 33–45. [Google Scholar] [CrossRef]
- Han, S.P.; Tang, Y.H.; Smith, R. Functional Diversity of the hnRNPs: Past, Present and Perspectives. Biochem. J. 2010, 430, 379–392. [Google Scholar] [CrossRef]
- Patton, J.G.; Mayer, S.A.; Tempst, P.; Nadal-Ginard, B. Characterization and Molecular Cloning of Polypyrimidine Tract-Binding Protein: A Component of a Complex Necessary for Pre-mRNA Splicing. Genes Dev. 1991, 5, 1237–1251. [Google Scholar] [CrossRef]
- Hamid, F.M.; Makeyev, E.V. Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5’ Splice Site Choice. PLoS Genet. 2014, 10, e1004771. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Masuda, A.; Ohe, K.; Ito, M.; Hutchinson, D.O.; Mayeda, A.; Engel, A.G.; Ohno, K. HnRNP L and hnRNP LL Antagonistically Modulate PTB-Mediated Splicing Suppression of CHRNA1 Pre-mRNA. Sci. Rep. 2013, 3, 2931. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, M.; Raffalli-Mathieu, F.; Lang, M.A. Identification of a Regulatory Cis-Element within the 3’-Untranslated Region of the Murine Inducible Nitric Oxide Synthase (iNOS) mRNA; Interaction with Heterogeneous Nuclear Ribonucleoproteins I and L and Role in the iNOS Gene Expression. Mol. Immunol. 2007, 44, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.; Yaman, I.; Gaccioli, F.; Zeenko, V.V.; Wang, C.; Caprara, M.G.; Venema, R.C.; Komar, A.A.; Snider, M.D.; Hatzoglou, M. The hnRNA-Binding Proteins hnRNP L and PTB Are Required for Efficient Translation of the Cat-1 Arginine/Lysine Transporter mRNA during Amino Acid Starvation. Mol. Cell. Biol. 2009, 29, 2899–2912. [Google Scholar] [CrossRef] [PubMed]
- Jafarifar, F.; Yao, P.; Eswarappa, S.M.; Fox, P.L. Repression of VEGFA by CA-Rich Element-Binding microRNAs Is Modulated by hnRNP L. EMBO J. 2011, 30, 1324–1334. [Google Scholar] [CrossRef]
- Xi, J.; Ye, F.; Wang, G.; Han, W.; Wei, Z.; Yin, B.; Yuan, J.; Qiang, B.; Peng, X. Polypyrimidine Tract-Binding Protein Regulates Enterovirus 71 Translation Through Interaction with the Internal Ribosomal Entry Site. Virol. Sin. 2019, 34, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A New Regulatory Protein, KSRP, Mediates Exon Inclusion through an Intronic Splicing Enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Davis-Smyth, T.; Duncan, R.C.; Zheng, T.; Michelotti, G.; Levens, D. The Far Upstream Element-Binding Proteins Comprise an Ancient Family of Single-Strand DNA-Binding Transactivators. J. Biol. Chem. 1996, 271, 31679–31687. [Google Scholar] [CrossRef]
- Lellek, H.; Kirsten, R.; Diehl, I.; Apostel, F.; Buck, F.; Greeve, J. Purification and Molecular Cloning of a Novel Essential Component of the Apolipoprotein B mRNA Editing Enzyme-Complex. J. Biol. Chem. 2000, 275, 19848–19856. [Google Scholar] [CrossRef]
- Gherzi, R.; Chen, C.-Y.; Trabucchi, M.; Ramos, A.; Briata, P. The Role of KSRP in mRNA Decay and microRNA Precursor Maturation. Wiley Interdiscip. Rev. RNA 2010, 1, 230–239. [Google Scholar] [CrossRef]
- Trabucchi, M.; Briata, P.; Filipowicz, W.; Rosenfeld, M.G.; Ramos, A.; Gherzi, R. How to Control miRNA Maturation? RNA Biol. 2009, 6, 536–540. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Briata, P.; Chen, C.-Y.; Ramos, A.; Gherzi, R. Functional and Molecular Insights into KSRP Function in mRNA Decay. Biochim. Biophys. Acta 2013, 1829, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Kung, Y.-A.; Weng, K.-F.; Lin, J.-Y.; Horng, J.-T.; Shih, S.-R. Enterovirus 71 Infection Cleaves a Negative Regulator for Viral Internal Ribosomal Entry Site-Driven Translation. J. Virol. 2013, 87, 3828–3838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, Q.M. Far Upstream Element Binding Protein 1: A Commander of Transcription, Translation and Beyond. Oncogene 2013, 32, 2907–2916. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.; Bazar, L.; Michelotti, G.; Tomonaga, T.; Krutzsch, H.; Avigan, M.; Levens, D. A Sequence-Specific, Single-Strand Binding Protein Activates the Far Upstream Element of c-Myc and Defines a New DNA-Binding Motif. Genes Dev. 1994, 8, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Olanich, M.E.; Moss, B.L.; Piwnica-Worms, D.; Townsend, R.R.; Weber, J.D. Identification of FUSE-Binding Protein 1 as a Regulatory mRNA-Binding Protein That Represses Nucleophosmin Translation. Oncogene 2011, 30, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Harris, D.; Pandey, V.N. The FUSE Binding Protein Is a Cellular Factor Required for Efficient Replication of Hepatitis C Virus. J. Virol. 2008, 82, 5761–5773. [Google Scholar] [CrossRef][Green Version]
- Chien, H.-L.; Liao, C.-L.; Lin, Y.-L. FUSE Binding Protein 1 Interacts with Untranslated Regions of Japanese Encephalitis Virus RNA and Negatively Regulates Viral Replication. J. Virol. 2011, 85, 4698–4706. [Google Scholar] [CrossRef]
- Huang, P.-N.; Lin, J.-Y.; Locker, N.; Kung, Y.-A.; Hung, C.-T.; Lin, J.-Y.; Huang, H.-I.; Li, M.-L.; Shih, S.-R. Far Upstream Element Binding Protein 1 Binds the Internal Ribosomal Entry Site of Enterovirus 71 and Enhances Viral Translation and Viral Growth. Nucleic Acids Res. 2011, 39, 9633–9648. [Google Scholar] [CrossRef]
- Hung, C.-T.; Kung, Y.-A.; Li, M.-L.; Brewer, G.; Lee, K.-M.; Liu, S.-T.; Shih, S.-R. Additive Promotion of Viral Internal Ribosome Entry Site-Mediated Translation by Far Upstream Element-Binding Protein 1 and an Enterovirus 71-Induced Cleavage Product. PLoS Pathog. 2016, 12, e1005959. [Google Scholar] [CrossRef]
- Höck, J.; Meister, G. The Argonaute Protein Family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; MacRae, I.J. The Crystal Structure of Human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed]
- Pantazopoulou, V.I.; Delis, A.D.; Georgiou, S.; Pagakis, S.N.; Filippa, V.; Dragona, E.; Kloukina, I.; Chatzitheodoridis, E.; Trebicka, J.; Velentzas, A.D.; et al. AGO2 Localizes to Cytokinetic Protrusions in a P38-Dependent Manner and Is Needed for Accurate Cell Division. Commun. Biol. 2021, 4, 726. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Steitz, J.A. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.M.; Steitz, J.A. HuR and mRNA Stability. Cell. Mol. Life Sci. 2001, 58, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, S.; Jens, M.; Theil, K.; Schwanhäusser, B.; Selbach, M.; Landthaler, M.; Rajewsky, N. Transcriptome-Wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol. Cell 2011, 43, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.C.; Steitz, J.A. Overexpression of HuR, a Nuclear-Cytoplasmic Shuttling Protein, Increases the in Vivo Stability of ARE-Containing mRNAs. EMBO J. 1998, 17, 3448–3460. [Google Scholar] [CrossRef] [PubMed]
- Uren, P.J.; Burns, S.C.; Ruan, J.; Singh, K.K.; Smith, A.D.; Penalva, L.O.F. Genomic Analyses of the RNA-Binding Protein Hu Antigen R (HuR) Identify a Complex Network of Target Genes and Novel Characteristics of Its Binding Sites. J. Biol. Chem. 2011, 286, 37063–37066. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Zhang, J.; Ma, J.; Xu, X.; Wang, Y.; Zhou, Z.; Jiang, D.; Shen, S.; Ding, Y.; et al. Silencing of HuR Inhibits Osteosarcoma Cell Epithelial-Mesenchymal Transition via AGO2 in Association With Long Non-Coding RNA XIST. Front. Oncol. 2021, 11, 601982. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, B.; Huang, H.; Zhao, Z. Enterovirus 71 Induces Anti-Viral Stress Granule-like Structures in RD Cells. Biochem. Biophys. Res. Commun. 2016, 476, 212–217. [Google Scholar] [CrossRef]
- Wang, H.; Chang, L.; Wang, X.; Su, A.; Feng, C.; Fu, Y.; Chen, D.; Zheng, N.; Wu, Z. MOV10 Interacts with Enterovirus 71 Genomic 5’UTR and Modulates Viral Replication. Biochem. Biophys. Res. Commun. 2016, 479, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 Is a 5’ to 3’ RNA Helicase Contributing to UPF1 mRNA Target Degradation by Translocation along 3’ UTRs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Lührmann, R.; Tuschl, T. Identification of Novel Argonaute-Associated Proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Abudu, A.; Wang, X.; Dang, Y.; Zhou, T.; Xiang, S.-H.; Zheng, Y.-H. Identification of Molecular Determinants from Moloney Leukemia Virus 10 Homolog (MOV10) Protein for Virion Packaging and Anti-HIV-1 Activity. J. Biol. Chem. 2012, 287, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Islam, R. Mammalian Sirt1: Insights on Its Biological Functions. Cell Commun. Signal. 2011, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Schoonjans, K.; Auwerx, J. Sirtuin Functions in Health and Disease. Mol. Endocrinol. 2007, 21, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-S.; Ott, M. The Ups and Downs of SIRT1. Trends Biochem. Sci. 2008, 33, 517–525. [Google Scholar] [CrossRef]
- Pagans, S.; Pedal, A.; North, B.J.; Kaehlcke, K.; Marshall, B.L.; Dorr, A.; Hetzer-Egger, C.; Henklein, P.; Frye, R.; McBurney, M.W.; et al. SIRT1 Regulates HIV Transcription via Tat Deacetylation. PLoS Biol. 2005, 3, e41. [Google Scholar] [CrossRef]
- Ren, J.-H.; Tao, Y.; Zhang, Z.-Z.; Chen, W.-X.; Cai, X.-F.; Chen, K.; Ko, B.C.B.; Song, C.-L.; Ran, L.-K.; Li, W.-Y.; et al. Sirtuin 1 Regulates Hepatitis B Virus Transcription and Replication by Targeting Transcription Factor AP-1. J. Virol. 2014, 88, 2442–2451. [Google Scholar] [CrossRef]
- Han, Y.; Wang, L.; Cui, J.; Song, Y.; Luo, Z.; Chen, J.; Xiong, Y.; Zhang, Q.; Liu, F.; Ho, W.; et al. SIRT1 Inhibits EV71 Genome Replication and RNA Translation by Interfering with the Viral Polymerase and 5′UTR RNA. J. Cell Sci. 2016, 129, 4534–4547. [Google Scholar] [CrossRef]
- Matter, N.; Herrlich, P.; König, H. Signal-Dependent Regulation of Splicing via Phosphorylation of Sam68. Nature 2002, 420, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Paronetto, M.P.; Cappellari, M.; Busà, R.; Pedrotti, S.; Vitali, R.; Comstock, C.; Hyslop, T.; Knudsen, K.E.; Sette, C. Alternative Splicing of the Cyclin D1 Proto-Oncogene Is Regulated by the RNA-Binding Protein Sam68. Cancer Res. 2010, 70, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Jiménez, F.; Sánchez-Margalet, V. Role of Sam68 in Post-Transcriptional Gene Regulation. Int. J. Mol. Sci. 2013, 14, 23402–23419. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, S.; Totty, N.F.; Hsuan, J.J.; Courtneidge, S.A. A Target for Src in Mitosis. Nature 1994, 368, 871–874. [Google Scholar] [CrossRef]
- Macias, M.J.; Wiesner, S.; Sudol, M. WW and SH3 Domains, Two Different Scaffolds to Recognize Proline-Rich Ligands. FEBS Lett. 2002, 513, 30–37. [Google Scholar] [CrossRef]
- Galarneau, A.; Richard, S. The STAR RNA Binding Proteins GLD-1, QKI, SAM68 and SLM-2 Bind Bipartite RNA Motifs. BMC Mol. Biol. 2009, 10, 47. [Google Scholar] [CrossRef]
- Lin, Q.; Taylor, S.J.; Shalloway, D. Specificity and Determinants of Sam68 RNA Binding: Implications for the biological function of k homology domains. J. Biol. Chem. 1997, 272, 27274–27280. [Google Scholar] [CrossRef]
- Lawrence, P.; Schafer, E.A.; Rieder, E. The Nuclear Protein Sam68 Is Cleaved by the FMDV 3C Protease Redistributing Sam68 to the Cytoplasm during FMDV Infection of Host Cells. Virology 2012, 425, 40–52. [Google Scholar] [CrossRef]
- McBride, A.E.; Schlegel, A.; Kirkegaard, K. Human Protein Sam68 Relocalization and Interaction with Poliovirus RNA Polymerase in Infected Cells. Proc. Natl. Acad. Sci. USA 1996, 93, 2296–2301. [Google Scholar] [CrossRef]
- Zhang, H.; Song, L.; Cong, H.; Tien, P. Nuclear Protein Sam68 Interacts with the Enterovirus 71 Internal Ribosome Entry Site and Positively Regulates Viral Protein Translation. J. Virol. 2015, 89, 10031–10043. [Google Scholar] [CrossRef]
- Xu, P.; Tong, W.; Chen, Y.-M. FUSE Binding Protein FUBP3 Is a Potent Regulator in Japanese Encephalitis Virus Infection. Virol. J. 2021, 18, 224. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Anandram, S.; Ross, C.; Srivastava, S. FUBP3 Regulates Chronic Myeloid Leukaemia Progression through PRC2 Complex Regulated PAK1-ERK Signalling. J. Cell. Mol. Med. 2023, 27, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.-J.; Lin, C.-W.; Lai, C.-C.; Lan, Y.-C.; Lai, C.-H.; Hung, C.-H.; Hsueh, K.-C.; Lin, T.-H.; Chang, H.C.; Wan, L.; et al. Kaempferol Inhibits Enterovirus 71 Replication and Internal Ribosome Entry Site (IRES) Activity through FUBP and HNRP Proteins. Food Chem. 2011, 128, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-I.; Chang, Y.-Y.; Lin, J.-Y.; Kuo, R.-L.; Liu, H.-P.; Shih, S.-R.; Wu, C.-C. Interactome Analysis of the EV71 5′ Untranslated Region in Differentiated Neuronal Cells SH-SY5Y and Regulatory Role of FBP3 in Viral Replication. Proteomics 2016, 16, 2351–2362. [Google Scholar] [CrossRef] [PubMed]
- Brush, M.H.; Weiser, D.C.; Shenolikar, S. Growth Arrest and DNA Damage-Inducible Protein GADD34 Targets Protein Phosphatase 1α to the Endoplasmic Reticulum and Promotes Dephosphorylation of the α Subunit of Eukaryotic Translation Initiation Factor 2. Mol. Cell. Biol. 2003, 23, 1292–1303. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Jiang, X.; Xin, H.; Zheng, X.; Xue, X.; Chen, J.-L.; Qi, B. GADD34-Mediated Dephosphorylation of eIF2α Facilitates Pseudorabies Virus Replication by Maintaining de Novo Protein Synthesis. Vet. Res. 2021, 52, 148. [Google Scholar] [CrossRef] [PubMed]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The Integrated Stress Response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef] [PubMed]
- Dalet, A.; Argüello, R.J.; Combes, A.; Spinelli, L.; Jaeger, S.; Fallet, M.; Vu Manh, T.-P.; Mendes, A.; Perego, J.; Reverendo, M.; et al. Protein Synthesis Inhibition and GADD34 Control IFN-β Heterogeneous Expression in Response to dsRNA. EMBO J. 2017, 36, 761–782. [Google Scholar] [CrossRef]
- Clavarino, G.; Cláudio, N.; Couderc, T.; Dalet, A.; Judith, D.; Camosseto, V.; Schmidt, E.K.; Wenger, T.; Lecuit, M.; Gatti, E.; et al. Induction of GADD34 Is Necessary for dsRNA-Dependent Interferon-β Production and Participates in the Control of Chikungunya Virus Infection. PLoS Pathog. 2012, 8, e1002708. [Google Scholar] [CrossRef]
- Minami, K.; Tambe, Y.; Watanabe, R.; Isono, T.; Haneda, M.; Isobe, K.-I.; Kobayashi, T.; Hino, O.; Okabe, H.; Chano, T.; et al. Suppression of Viral Replication by Stress-Inducible GADD34 Protein via the Mammalian Serine/Threonine Protein Kinase mTOR Pathway. J. Virol. 2007, 81, 11106–11115. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Liao, Y.; Yap, P.L.; Png, K.J.; Tam, J.P.; Liu, D.X. Inhibition of Protein Kinase R Activation and Upregulation of GADD34 Expression Play a Synergistic Role in Facilitating Coronavirus Replication by Maintaining De Novo Protein Synthesis in Virus-Infected Cells. J. Virol. 2009, 83, 12462–12472. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, M.; Marshall, H.; Natarajan, V. GADD34 Attenuates HIV-1 Replication by Viral 5’-UTR TAR RNA-Mediated Translational Inhibition. Virology 2020, 540, 119–131. [Google Scholar] [CrossRef]
- Brush, M.H.; Shenolikar, S. Control of Cellular GADD34 Levels by the 26S Proteasome. Mol. Cell. Biol. 2008, 28, 6989–7000. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, W.; Zhang, S.; Qiu, M.; Li, Z.; Lin, Y.; Tan, J.; Qiao, W. Enterovirus 71 Activates GADD34 via Precursor 3CD to Promote IRES-Mediated Viral Translation. Microbiol. Spectr. 2022, 10, e01388-21. [Google Scholar] [CrossRef] [PubMed]
- Venkataramanan, S.; Gadek, M.; Calviello, L.; Wilkins, K.; Floor, S.N. DDX3X and DDX3Y Are Redundant in Protein Synthesis. RNA 2021, 27, 1577–1588. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M. Human DEAD-Box Protein 3 Has Multiple Functions in Gene Regulation and Cell Cycle Control and Is a Prime Target for Viral Manipulation. Biochem. Pharmacol. 2010, 79, 297–306. [Google Scholar] [CrossRef]
- Soto-Rifo, R.; Rubilar, P.S.; Limousin, T.; de Breyne, S.; Décimo, D.; Ohlmann, T. DEAD-Box Protein DDX3 Associates with eIF4F to Promote Translation of Selected mRNAs. EMBO J. 2012, 31, 3745–3756. [Google Scholar] [CrossRef]
- Su, Y.-S.; Tsai, A.-H.; Ho, Y.-F.; Huang, S.-Y.; Liu, Y.-C.; Hwang, L.-H. Stimulation of the Internal Ribosome Entry Site (IRES)-Dependent Translation of Enterovirus 71 by DDX3X RNA Helicase and Viral 2A and 3C Proteases. Front. Microbiol. 2018, 9, 1324. [Google Scholar] [CrossRef]
- Gao, J.; Choudhry, H.; Cao, W. Apolipoprotein B mRNA Editing Enzyme Catalytic Polypeptide-like Family Genes Activation and Regulation during Tumorigenesis. Cancer Sci. 2018, 109, 2375–2382. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, M.; Li, Y.; Li, K.; Wu, S.; Guo, T.; Cen, S.; Jiang, J.; Li, Z.; Li, Y. APOBEC3G Is a Restriction Factor of EV71 and Mediator of IMB-Z Antiviral Activity. Antivir. Res. 2019, 165, 23–33. [Google Scholar] [CrossRef]
- Fehrholz, M.; Kendl, S.; Prifert, C.; Weissbrich, B.; Lemon, K.; Rennick, L.; Duprex, P.W.; Rima, B.K.; Koning, F.A.; Holmes, R.K.; et al. The Innate Antiviral Factor APOBEC3G Targets Replication of Measles, Mumps and Respiratory Syncytial Viruses. J. Gen. Virol. 2012, 93, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.; Hu, J. Reverse Transcriptase- and RNA Packaging Signal-Dependent Incorporation of APOBEC3G into Hepatitis B Virus Nucleocapsids. J. Virol. 2008, 82, 6852–6861. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Iwatani, Y. APOBEC3G-Mediated G-to-A Hypermutation of the HIV-1 Genome: The Missing Link in Antiviral Molecular Mechanisms. Front. Microbiol. 2016, 7, 2027. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ning, S.; Su, X.; Liu, X.; Wang, H.; Liu, Y.; Zheng, W.; Zheng, B.; Yu, X.-F.; Zhang, W. Enterovirus 71 Antagonizes the Inhibition of the Host Intrinsic Antiviral Factor A3G. Nucleic Acids Res. 2018, 46, 11514–11527. [Google Scholar] [CrossRef] [PubMed]
- Wickham, L.; Duchaîne, T.; Luo, M.; Nabi, I.R.; DesGroseillers, L. Mammalian Staufen Is a Double-Stranded-RNA- and Tubulin-Binding Protein Which Localizes to the Rough Endoplasmic Reticulum. Mol. Cell. Biol. 1999, 19, 2220–2230. [Google Scholar] [CrossRef]
- Kim, Y.K.; Furic, L.; Desgroseillers, L.; Maquat, L.E. Mammalian Staufen1 Recruits Upf1 to Specific mRNA 3’UTRs so as to Elicit mRNA Decay. Cell 2005, 120, 195–208. [Google Scholar] [CrossRef]
- Furic, L.; Maher-Laporte, M.; DesGroseillers, L. A Genome-Wide Approach Identifies Distinct but Overlapping Subsets of Cellular mRNAs Associated with Staufen1- and Staufen2-Containing Ribonucleoprotein Complexes. RNA 2008, 14, 324–335. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Ou, B.-T.; Chen, C.-Y.; Chan, H.-H.; Chen, C.-J.; Wang, R.Y. Staufen1 Protein Participates Positively in the Viral RNA Replication of Enterovirus 71. Viruses 2019, 11, 142. [Google Scholar] [CrossRef]
- Blackham, S.L.; McGarvey, M.J. A Host Cell RNA-Binding Protein, Staufen1, Has a Role in Hepatitis C Virus Replication before Virus Assembly. J. Gen. Virol. 2013, 94 Pt 11, 2429–2436. [Google Scholar] [CrossRef]
- Abrahamyan, L.G.; Chatel-Chaix, L.; Ajamian, L.; Milev, M.P.; Monette, A.; Clément, J.-F.; Song, R.; Lehmann, M.; DesGroseillers, L.; Laughrea, M.; et al. Novel Staufen1 Ribonucleoproteins Prevent Formation of Stress Granules but Favour Encapsidation of HIV-1 Genomic RNA. J. Cell Sci. 2010, 123 Pt 3, 369–383. [Google Scholar] [CrossRef]
- de Lucas, S.; Peredo, J.; Marión, R.M.; Sánchez, C.; Ortín, J. Human Staufen1 Protein Interacts with Influenza Virus Ribonucleoproteins and Is Required for Efficient Virus Multiplication. J. Virol. 2010, 84, 7603–7612. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Lin, C.; Garcia-Blanco, M.A. hnRNP H and hnRNP F Complex with Fox2 To Silence Fibroblast Growth Factor Receptor 2 Exon IIIc. Mol. Cell. Biol. 2008, 28, 5403–5419. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xie, N.; Su, Y.; Sun, Z.; Liang, Y.; Zhang, N.; Liu, D.; Jia, S.; Xing, X.; Han, L.; et al. HnRNP F/H Associate with hTERC and Telomerase Holoenzyme to Modulate Telomerase Function and Promote Cell Proliferation. Cell Death Differ. 2020, 27, 1998–2013. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Cibelli, G. Regulation of Life and Death by the Zinc Finger Transcription Factor Egr-1. J. Cell. Physiol. 2002, 193, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.-C.; Yu, S.-L.; Chen, J.J.W.; Chang, S.-Y.; Yan, B.-S.; Hong, Q.-S.; Singh, S.; Kao, C.-L.; Chen, H.-Y.; Su, K.-Y.; et al. Enterovirus-Induced miR-141 Contributes to Shutoff of Host Protein Translation by Targeting the Translation Initiation Factor eIF4E. Cell Host Microbe 2011, 9, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Cheng, X.; Yang, X.; Zhao, R.; Wang, P.; Han, Y.; Luo, Z.; Cao, Y.; Zhu, C.; Xiong, Y.; et al. Early Growth Response-1 Facilitates Enterovirus 71 Replication by Direct Binding to the Viral Genome RNA. Int. J. Biochem. Cell Biol. 2015, 62, 36–46. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-Binding Proteins TIA-1 and TIAR Link the Phosphorylation of eIF-2 Alpha to the Assembly of Mammalian Stress Granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef]
- Gottschald, O.R.; Malec, V.; Krasteva, G.; Hasan, D.; Kamlah, F.; Herold, S.; Rose, F.; Seeger, W.; Hänze, J. TIAR and TIA-1 mRNA-Binding Proteins Co-Aggregate under Conditions of Rapid Oxygen Decline and Extreme Hypoxia and Suppress the HIF-1α Pathway. J. Mol. Cell Biol. 2010, 2, 345–356. [Google Scholar] [CrossRef]
- White, J.P.; Lloyd, R.E. Poliovirus Unlinks TIA1 Aggregation and mRNA Stress Granule Formation. J. Virol. 2011, 85, 12442–12454. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Lin, L.; Si, X.; Wang, T.; Zhong, X.; Tong, L.; Luan, Y.; Chen, Y.; Li, X.; et al. Protease 2A Induces Stress Granule Formation during Coxsackievirus B3 and Enterovirus 71 Infections. Virol. J. 2014, 11, 192. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Li, Y.; Jin, Y.; Chu, Y.; Su, A.; Wu, Z. TIA-1 and TIAR Interact with 5′-UTR of Enterovirus 71 Genome and Facilitate Viral Replication. Biochem. Biophys. Res. Commun. 2015, 466, 254–259. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Kung, Y.-A.; Shih, S.-R. Antivirals and Vaccines for Enterovirus A71. J. Biomed. Sci. 2019, 26, 65. [Google Scholar] [CrossRef]
- de Breyne, S.; Yu, Y.; Unbehaun, A.; Pestova, T.V.; Hellen, C.U.T. Direct Functional Interaction of Initiation Factor eIF4G with Type 1 Internal Ribosomal Entry Sites. Proc. Natl. Acad. Sci. USA 2009, 106, 9197–9202. [Google Scholar] [CrossRef] [PubMed]
- Beckham, S.A.; Matak, M.Y.; Belousoff, M.J.; Venugopal, H.; Shah, N.; Vankadari, N.; Elmlund, H.; Nguyen, J.H.C.; Semler, B.L.; Wilce, M.C.J.; et al. Structure of the PCBP2/Stem-Loop IV Complex Underlying Translation Initiation Mediated by the Poliovirus Type I IRES. Nucleic Acids Res. 2020, 48, 8006–8021. [Google Scholar] [CrossRef] [PubMed]
- Walter, B.L.; Parsley, T.B.; Ehrenfeld, E.; Semler, B.L. Distinct Poly(rC) Binding Protein KH Domain Determinants for Poliovirus Translation Initiation and Viral RNA Replication. J. Virol. 2002, 76, 12008–12022. [Google Scholar] [CrossRef] [PubMed]
- Blyn, L.B.; Swiderek, K.M.; Richards, O.; Stahl, D.C.; Semler, B.L.; Ehrenfeld, E. Poly(rC) Binding Protein 2 Binds to Stem-Loop IV of the Poliovirus RNA 5′ Noncoding Region: Identification by Automated Liquid Chromatography-Tandem Mass Spectrometry. Proc. Natl. Acad. Sci. USA 1996, 93, 11115–11120. [Google Scholar] [CrossRef]
- Andreev, D.E.; Hirnet, J.; Terenin, I.M.; Dmitriev, S.E.; Niepmann, M.; Shatsky, I.N. Glycyl-tRNA Synthetase Specifically Binds to the Poliovirus IRES to Activate Translation Initiation. Nucleic Acids Res. 2012, 40, 5602–5614. [Google Scholar] [CrossRef]
- Meerovitch, K.; Svitkin, Y.V.; Lee, H.S.; Lejbkowicz, F.; Kenan, D.J.; Chan, E.K.; Agol, V.I.; Keene, J.D.; Sonenberg, N. La Autoantigen Enhances and Corrects Aberrant Translation of Poliovirus RNA in Reticulocyte Lysate. J. Virol. 1993, 67, 3798–3807. [Google Scholar] [CrossRef]
- Bedard, K.M.; Daijogo, S.; Semler, B.L. A Nucleo-Cytoplasmic SR Protein Functions in Viral IRES-Mediated Translation Initiation. EMBO J. 2007, 26, 459–467. [Google Scholar] [CrossRef]
- Hunt, S.L.; Hsuan, J.J.; Totty, N.; Jackson, R.J. Unr, a Cellular Cytoplasmic RNA-Binding Protein with Five Cold-Shock Domains, Is Required for Internal Initiation of Translation of Human Rhinovirus RNA. Genes Dev. 1999, 13, 437–448. [Google Scholar] [CrossRef]
- Davila-Calderon, J.; Penumutchu, S.; Tolbert, B. A Regulatory Interplay: hnRNP A1 and AUF1 Compete for the Same IRES Domain to Regulate Viral Translation in EV71. FASEB J. 2019, 33, 625.5. [Google Scholar] [CrossRef]
- Courtney, D.G. Post-Transcriptional Regulation of Viral RNA through Epitranscriptional Modification. Cells 2021, 10, 1129. [Google Scholar] [CrossRef] [PubMed]
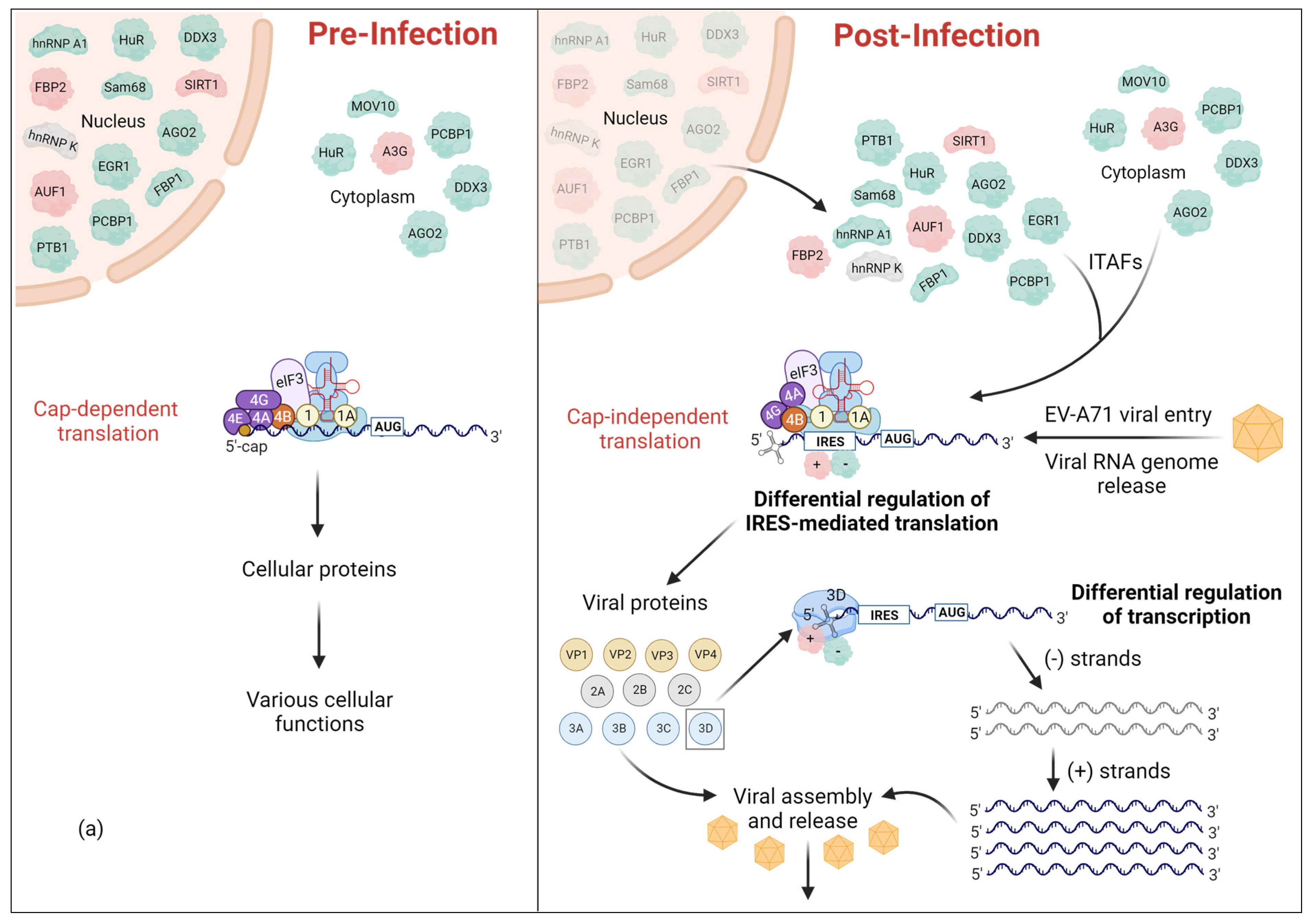
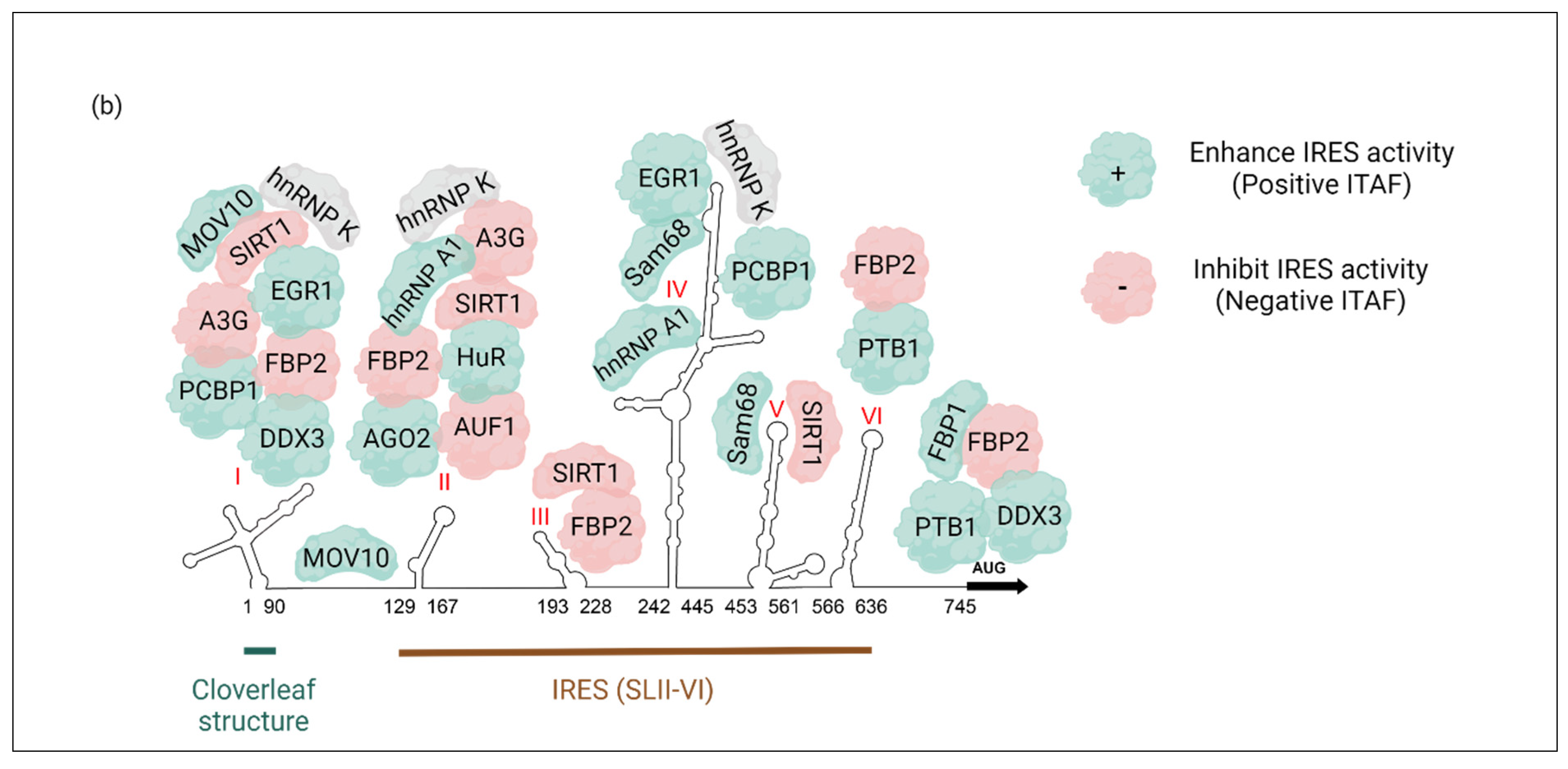
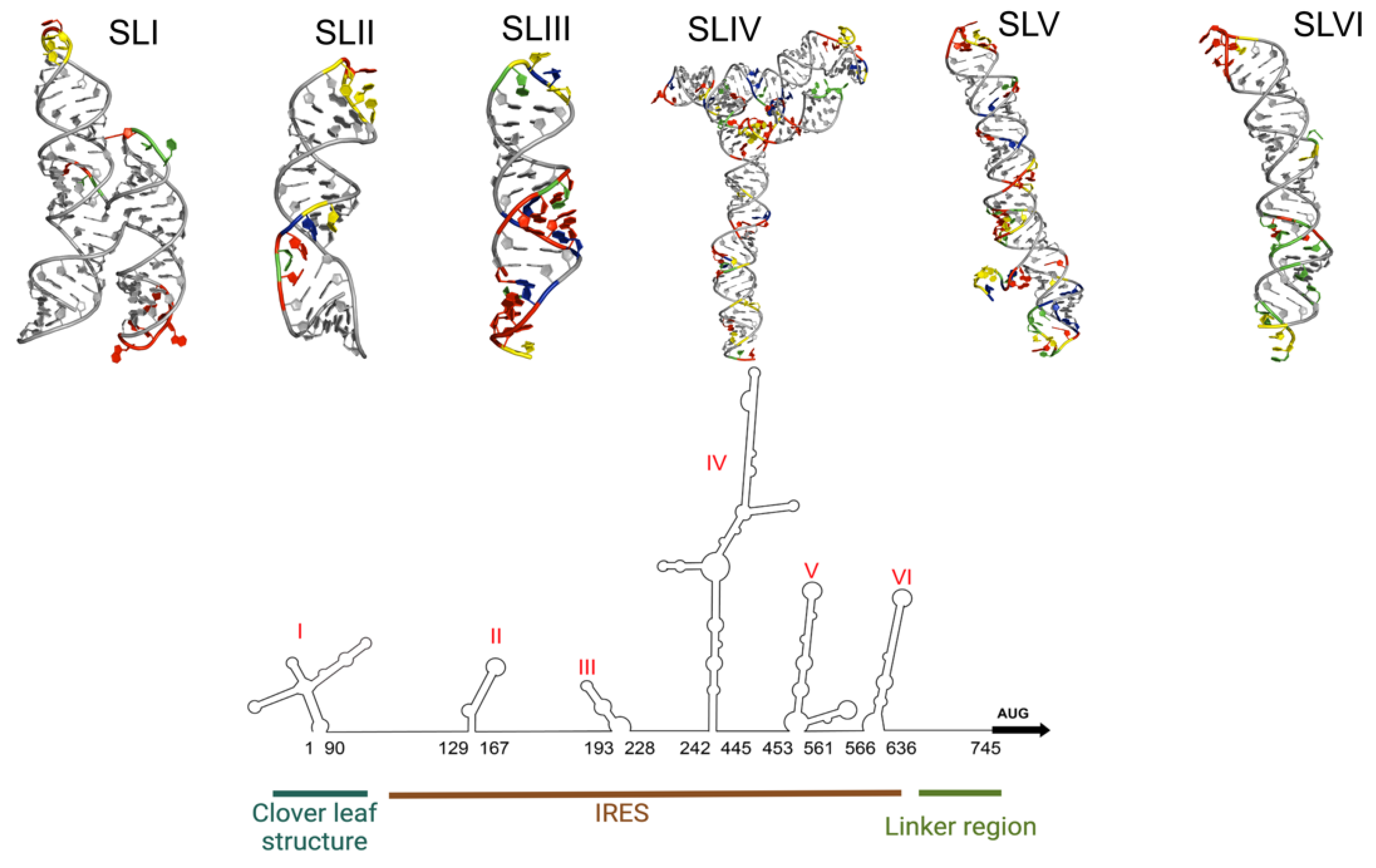
| ITAF | Other Names | Cellular Distribution | Regulatory Site | IRES Activity | PDB ID(s) | |
|---|---|---|---|---|---|---|
| Pre-Infection | Post-Infection | |||||
| Heterogeneous nuclear ribonucleoprotein A1 | hnRNP A1 | Nucleus | Cytoplasm | SLII and IV | Enhancement | 4YOE: 1.92 Å 5MPL: NMR 5MPG: NMR |
| AU-rich element RNA-binding protein 1 | AUF1, hnRNP D | Nucleus | Cytoplasm | SLII | Inhibition | 1WTB: NMR 1 × 0F: NMR |
| Heterogeneous nuclear ribonucleoprotein K | hnRNP K | Nucleus | Cytoplasm | SLI, II, and IV | - | 1J5K: NMR 1ZZI: 1.80Å 1ZZJ: 2.30 Å 7CRE: 3.00 Å |
| Poly(c)-binding protein 1 | PCBP1, hnRNP E1 | Nucleus, Cytoplasm | Cytoplasm | SLI and IV | Enhancement | 1ZTG: 3 Å 3VKE: 1.77 Å |
| Polypyrimidine tract-binding protein 1 | PTB, PTB1 PTBP1 hnRNP I | Nucleus | Cytoplasm | SLVI + linker (564–742 nt) | Enhancement | 2N3O: NMR 2AD9: NMR 2ADB: NMR 2ADC: NMR |
| Far upstream binding protein 2 | FBP2, FUBP2 KSRP, KHSRP | Nucleus | Cytoplasm | SLI-SLII (1-167) SLII-SLIII (91-228) SLVI + linker (566–745) | Inhibition | 4B8T: NMR |
| C-terminus cleaved FBP2 | FBP21–503 | - | - | 5′-UTR | Enhancement | |
| N-terminus cleaved FBP2 | FBP2190–711 | - | - | 5′-UTR | Inhibition | |
| Far upstream binding protein 1 | FBP1 FUBP1 | Nucleus | Cytoplasm | Linker (686–714 nt) | Enhancement | 1J4W: NMR |
| Cleaved FBP1 | FBP11-371 | - | - | Linker (656–674 nt) | Enhancement | |
| Argonaute 2 | Ago2 | Nucleus, Cytoplasm (P-bodies) | - | SLII | Enhancement | 5KI6: 2.15 Å |
| Human Antigen R | HuR, ELAVL1 | Nucleus, shuttles to Cytoplasm | Cytoplasm | SLII | Enhancement | 4ED5: 2.00 Å 6G2K: 2.00 Å 6GC5: 1.90 Å 6GD2: 1.90 Å |
| Moloney leukemia virus 10 (C-terminus domain) | MOV10 | Cytoplasm | Cytoplasm (P-bodies & aggregates perinuclear) | SLI and IRES (Excluding the linker region) | Enhancement | |
| Silent mating type information regulation 2 homolog 1 | SIRT1 | Nucleus | Cytoplasm | SLI, II, III and V | Inhibition | |
| 68-kDa Src-associated protein in mitosis | Sam68, KHDRBS1 | Nucleus | Cytoplasm | SLIV and V | Enhancement | |
| Far upstream element-binding protein 3 | FUBP3 | Nucleus | Cytoplasm | 5′-UTR | Enhancement | |
| Growth arrest and DNA damage-inducible protein 34 | GADD34, PPP1R15A | ER membrane Mitochondial membrane | - | 5′-UTR | Enhancement | |
| DEAD-box protein 3 | DDX3 | Nucleus Cytoplasm | - | Full 5′-UTR | Enhancement | 6O5F: 2.5 Å |
| Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G | APOBEC3G, A3G | Cytoplasm (mainly) Nucleus P-bodies | Cytoplasm Virions | SLI and II | Inhibition | 5ZVA: 2.30 Å 5ZVB: 2.00 Å 6BUX: 1.86 Å 7UXD: 1.50 Å |
| Staufen homolog 1 | Staufen1 | Rough ER Cytoplasm | - | 5′-UTR | Enhancement | 6HTU: 2.89 Å |
| Heterogeneous nuclear ribonucleoprotein H | HNRNP H | Nucleus | Cytoplasm | 5′-UTR | Enhancement | |
| Heterogeneous nuclear ribonucleoprotein F | HNRNP F | Nucleus | Cytoplasm | 5′-UTR | Enhancement | 2KFY: NMR 2KG0: NMR 2KG1: NMR |
| Early growth response-1 | EGR1 | Nucleus | Cytoplasm | SLI and IV | Enhancement | 4R2A: 1.59 Å |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abedeera, S.M.; Davila-Calderon, J.; Haddad, C.; Henry, B.; King, J.; Penumutchu, S.; Tolbert, B.S. The Repurposing of Cellular Proteins during Enterovirus A71 Infection. Viruses 2024, 16, 75. https://doi.org/10.3390/v16010075
Abedeera SM, Davila-Calderon J, Haddad C, Henry B, King J, Penumutchu S, Tolbert BS. The Repurposing of Cellular Proteins during Enterovirus A71 Infection. Viruses. 2024; 16(1):75. https://doi.org/10.3390/v16010075
Chicago/Turabian StyleAbedeera, Sudeshi M., Jesse Davila-Calderon, Christina Haddad, Barrington Henry, Josephine King, Srinivasa Penumutchu, and Blanton S. Tolbert. 2024. "The Repurposing of Cellular Proteins during Enterovirus A71 Infection" Viruses 16, no. 1: 75. https://doi.org/10.3390/v16010075
APA StyleAbedeera, S. M., Davila-Calderon, J., Haddad, C., Henry, B., King, J., Penumutchu, S., & Tolbert, B. S. (2024). The Repurposing of Cellular Proteins during Enterovirus A71 Infection. Viruses, 16(1), 75. https://doi.org/10.3390/v16010075






