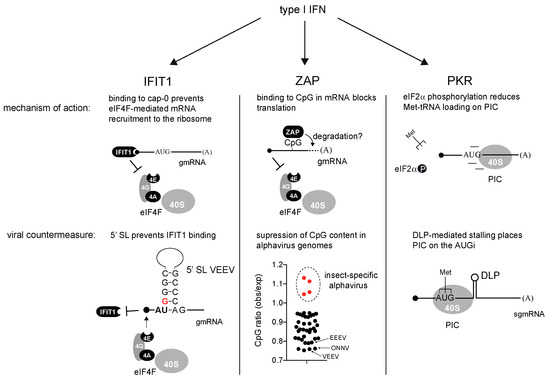
Abstract
Alphaviruses can replicate in arthropods and in many vertebrate species including humankind, but only in vertebrate cells do infections with these viruses result in a strong inhibition of host translation and transcription. Translation shutoff by alphaviruses is a multifactorial process that involves both host- and virus-induced mechanisms, and some of them are not completely understood. Alphavirus genomes contain cis-acting elements (RNA structures and dinucleotide composition) and encode protein activities that promote the translational and transcriptional resistance to type I IFN-induced antiviral effectors. Among them, IFIT1, ZAP and PKR have played a relevant role in alphavirus evolution, since they have promoted the emergence of multiple viral evasion mechanisms at the translational level. In this review, we will discuss how the adaptations of alphaviruses to vertebrate hosts likely involved the acquisition of new features in viral mRNAs and proteins to overcome the effect of type I IFN.
1. Alphavirus Replication, Tropism and Interference with Host Gene Expression
The Alphavirus genus includes over 30 viral species that are classified in seven complexes based on their antigenic characteristics (ICTV Taxonomy 2022 release) (Figure 1A). Most alphaviruses are transmitted between vertebrate hosts (e.g., nonhuman primates, birds, rodents and marsupials) by hematophagous arthropods, mainly mosquitoes [1,2]. In general, the host tropism in alphaviruses is broad, and many zoonotic transmissions to humans have been documented. Thus, the chikungunya virus (CHIKV) has evolved from a sylvatic cycle between primates and forest mosquitoes to an urbanized transmission that involves mosquitoes of the Aedes genus (e.g., tiger mosquito) and humans [3,4,5]. The emergence of Venezuelan equine encephalitis virus (VEEV) strains causing infection in humans is also frequent [6]. Historically, mosquito-borne alphaviruses have been divided into Old World and New World viruses according not only to their geographical distribution, but also to the clinical manifestations they cause in humans [1]. Thus, Old World alphaviruses include the CHIKV, Sindbis virus (SINV), Semliki Forest virus (SFV), Ross River virus (RRV) and o’nyong-nyong virus (ONNV), which were first isolated in Africa and Australia and mainly cause arthritis. New World alphaviruses include the Venezuelan (VEEV), eastern (EEEV) and western (WEEV) equine encephalitis viruses, which are endemic to North, Central and South America and cause diseases in horses and humans with neurological symptoms [7].
Alphaviruses are enveloped, and their genomes consist of a positive-sense single-stranded RNA (ssRNA) of approximately 12 kilobases that encodes four nonstructural proteins (nsP1-4) and five structural proteins (capsid, E3, E2, 6k and E1) in two large open reading frames that are translated as polyproteins (Figure 1B) [8,9]. The nonstructural polyprotein is translated directly from the genomic RNA (gmRNA), whereas the structural polyprotein is translated from a subgenomic mRNA (sgmRNA) that is transcribed by the viral replicase from an internal promotor. Both gmRNA and sgmRNA are 5′ capped (m7Gppp or cap0) and 3′ polyadenylated (Figure 1B) [10]. The nsp1 has both N-7 methyltransferase and guanyltransferase activities that are responsible for the capping of a fraction of both the gmRNA and sgmRNA during RNA transcription [11,12,13]. The nonstructural protein 2 (nsp2) is a multifunctional protein involved in viral RNA synthesis, polyprotein processing, the blockade of host gene expression and viral pathogenesis [9,14]. In addition to its role as the main viral protease that cleaves the nonstructural polyprotein precursor (p1234), nsp2 also contains a NTPase/RNA helicase domain involved in viral RNA synthesis and a SAM (S-adenosyl-methionine)-dependent methyltransferase-like domain that cooperates with the RNA helicase domain [9]. The role of nsp3 in alphavirus replication involves the recruitment of multiple host factors towards the viral replication complex (vRCs), including members of the Ras–GAP SH3 domain-binding proteins family (G3BP) in mammals and the corresponding ortholog in mosquito (Rasputin) [15,16]. Interestingly, since G3BP is involved in stress granule formation upon stress-induced eIF2α phosphorylation, G3BPs hijacking by nsp3 has been proposed to induce the rapid disassembly of stress granules that are formed early in response to an infection, although very recent reports have questioned this idea [17,18,19]. Nsp4 is the viral RNA-dependent RNA polymerase involved in negative- and positive-stranded RNA synthesis to generate gmRNA and sgmRNA [20]. The regulation of these activities involves the sequential processing of the nsP1-4 precursor by the nsp2 protease [21]. In the early stages of infection, the partially processed nsPs (P123 + nsp4) preferentially synthesize a negative RNA strand to form double-stranded RNA intermediates (dsRNA). Later, the fully processed nsPs produce gmRNA and sgmRNA [21,22]. Recent data suggest that viral RNA synthesis initiates in the discrete foci associated with the plasma membrane to further translocate into the cytoplasm to form bigger membrane-bound spherules (or cytopathic vacuoles) containing the mature vRCs [23,24,25]. Although dsRNA replicative forms tend to accumulate into the spherules to serve as templates for the synthesis of new gmRNA molecules, the fact that antiviral sensors such as PKR, RIG-I and MDA5 become activated after alphavirus infection suggests that some viral dsRNA molecules are exposed to these antiviral sensors (Figure 1B) [26].
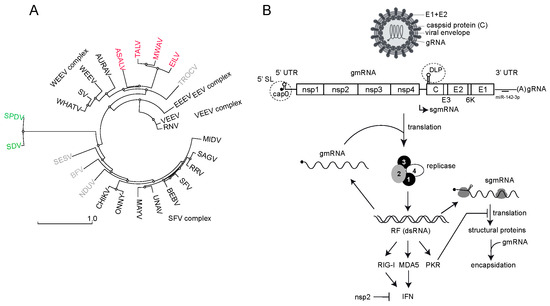
Figure 1.
(A) Phylogenetic tree of some members of the Alphavirus genus. Structural polyprotein sequences (C-E3-E2-6K-E1) were aligned using Muscle, and the resulting distance-based phylogenetic tree was built using the tools available in the NGPhylogeny.fr suite. Some of the main complexes of the genus are shown. Insect-specific viruses close to the WEEV complex are in red. Aquatic alphaviruses infecting fish are in green. Members of other complexes are in light gray. (B) Schematic representation of alphavirus replication cycle including virion composition and genomic organization. The nonstructural (nsP1-3) and structural coding sequences (C-E3-E2-6K-E1) are indicated, as well as some of the cis-acting structures located in the 5′UTR of gmRNA and sgmRNA that are relevant to this review (dashed circles). Viral replicase is involved in the synthesis of (−) ssRNA, (+) gmRNA and sgmRNA from dsRNA intermediaries (RF). gmRNA and sgmRNA are translated independently to produce nonstructural and structural proteins, respectively. Capsid protein recruits and assembles gmRNA to produce new virus particles. The accumulation of dsRNA molecules can trigger the activation of antiviral sensors (PKR, RIG-I and MDA5) that results in an antiviral response and the synthesis of type I IFN. As a countermeasure, viral nsp2 blocks transcription of the IFN gene in mammalian cells. Binding of a specific microRNA (miR-142-3p) to the 3′ NC region of EEEV that restricts the replication of this virus in hematopoietic cells is also shown [27].
Although alphavirus replication has been studied mainly in mammalian cells, comparative analyses also found similar vRCs associated with the membrane-bound spherules in insect cells infected with SINV and other alphaviruses, suggesting that the basic aspects of alphavirus replication are conserved among these hosts [28,29]. However, the outcome of virus replication in mammalian and insect cells is different, suggesting the existence of underlying differences in the way virus replication impacts the host cell physiology. Alphavirus replication in mammalian cells is highly productive, and it is generally associated with a strong cytopathic effect that precedes cell lysis. In mosquito cells, however, alphaviruses easily establish nonlytic, persistent infections, where an active viral replication occurs without signs of a cytopathic effect (Figure 2) [28,30,31]. This fact nicely reflects how alphaviruses and other arboviruses use insects both as reservoirs to persist in nature and as vectors to ensure transmission to vertebrate hosts. At a molecular level, the replication of alphaviruses in mammalian cells is associated with a profound inhibition of both host transcription and translation that ultimately contributes to the cytopathogenesis. Thus, mammalian cells infected with representative Old World and New World members almost exclusively synthesize viral RNAs and proteins at later times of infection. In insect cells, on the contrary, the synthesis of large amounts of viral RNAs and proteins is well tolerated and occurs without significant interference with host transcription and translation [32,33]. This suggests that instead of relying solely on the replication process, alphaviruses employ specific mechanisms to halt gene expression in mammalian cells.
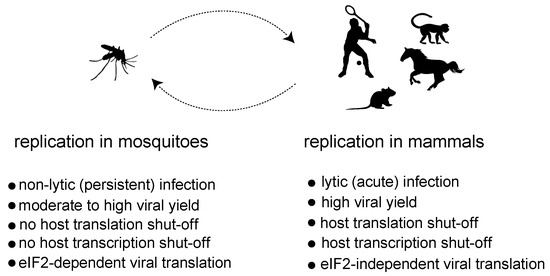
Figure 2.
Comparison of the main characteristics associated with replication of alphaviruses in mosquitoes and mammalian hosts.
2. Mechanisms of Virus-Induced Host Translation Shutoff
Like many cytolytic viruses that infect mammals, an alphavirus infection often results in a strong blockade of host gene expression at both the transcriptional and translational levels [34]. The causative mechanisms of this interference or “shut-off” have been studied in both Old World (SINV and CHIKV) and New World (VEEV) members of the genus, using highly susceptible cell lines of human and murine origins [14,35,36,37]. Thus, the infection of BHK21 or murine embryonic fibroblasts (MEFs) with SINV and SFV at a high multiplicity of infection resulted in a complete shutoff of host translation at 3–4 h postinfection, concomitant with an almost exclusive translation of viral mRNAs, which can represent up to 30% of the total translation activity of uninfected cells. In other alphaviruses such as the VEEV and CHIKV, this effect is less dramatic in cell culture models [37,38,39,40]. Both the ongoing and de novo translation initiations of host mRNAs are abrogated in SINV-infected cells, suggesting that both the initiation and reinitiation (by ribosomal recycling) of mRNAs are blocked [35].
Different causative mechanisms have been proposed to explain the shutoff induced by alphaviruses [14,41,42]. Regarding mechanisms that impact the activities of translation factors, the strong phosphorylation of eIF2α by dsRNA-activated kinase (PKR) activation observed in cells infected with the SINV, SFV and VEEV could explain the translational block of the vast majority of mRNAs in infected cells [35,36,43]. eIF2α phosphorylation and host translation shutoff was also confirmed in mouse brain and organotypic cultures infected with the SINV [44]. However, SINV and CHIKV infections of PKR-knockout MEFs or PKR-knockdown human fibroblasts (HF), respectively, still induced a shutoff comparable to the control cells, suggesting the existence of an underlying mechanism(s) to halt host translation [35,36]. As discussed below, eIF2α phosphorylation in response to infection must be interpreted as an attempt of the host cell to prevent the translation of viral mRNAs in the context of an IFN response. More recently, a significant phosphorylation of elongation factor 2 (eEF2) has been detected in cells infected with many alphaviruses, including the CHIKV, SINV, SFV and VEEV [45]. eEF2 phosphorylation at the T56 residue reduces the activity of this factor, thus decreasing the rate of translation elongation [46]. The helicase activity associated with the NTPase domains of the CHIKV and VEEV nsp2s was sufficient to induce eEF2 phosphorylation and translation inhibition, although the contribution of this modification to the host shutoff induced by the CHIKV and other alphaviruses remains to be determined [45]. The activity of the nsp2s of Old World alphaviruses has also been linked to translation shutoff, since the SINV and CHIKV with point mutations in nsp2 that abrogated the host transcriptional inhibitory activity also failed to induce a complete inhibition of host translation in infected cells [14,36]. However, it is not clear if this defect in the translation shutoff could be an indirect consequence of a lesser impact of the mutated nsp2 on RNAPII activity that could make more mRNA available in the cytoplasm for translation.
Other indirect causes of shutoff have been proposed, although the precise mechanisms involved and their contributions to the shutoff phenomenon are still an open question. Thus, the large quantities of subgenomic mRNAs (sgmRNAs) accumulated in cells infected with the SINV and SFV could sequester translation components, such as ribosomes and translation factors, to redirect translation towards sgmRNAs [35,47]. Although sgmRNA is efficiently translated in infected cells by acting as a super competitor mRNA, the fact that replicons of the SINV and VEEV, which lack the entire sgmRNA, still repressed host translation, which seriously limits the contribution of sgmRNA accumulation to the translation shutoff by these viruses [48,49]. Related to this fact, the viral RNA synthesis directed by alphaviral replicases has been correlated to host translation inhibition in SINV-infected cells. Thus, the addition of RNA synthesis inhibitors such as 6-aza-uridine or ribavirin reduced the accumulation of both genomic RNA (gmRNA) and sgmRNA in SINV-infected cells and partially prevented or delayed the inhibition of host translation [47]. Some authors also reported the release of nuclear proteins to the cytoplasm in SINV-infected cells that could be interfering with translation by sequestering host mRNAs [47,50]. However, a direct causative correlation between these observations and the shutoff phenomenon is still lacking, so their contribution remains to be measured. Early reports also described an inhibitory effect of the capsid protein of the SFV on host translation [51]. Finally, alterations in the plasma membrane permeability observed in cells infected with alphaviruses and other animal viruses have also been proposed as a cause of translation shutoff some decades ago, but currently, these lines of investigation have been discontinued.
5. Conclusions and Future Directions
The interference of alphaviruses with host translation seems to be a multifactorial process that results from the intricate balance between host-induced measures and virus-induced countermeasures. The interplay between alphavirus-induced transcription and translation shutoff deserves to be explored in the future, as well as the impact that the cytoplasmic accumulation of many RNA binding proteins observed in infected cells could have on host translation. To what extent IFN evasion mechanisms have shaped the genome of alphavirus also deserves further investigation to obtain clues on some aspects of the origin and evolutionary history of alphavirus that still remain controversial. This will require a better understanding of ZAP specificity that will allow us to predict the existence of ZAP binding sites in viral genomes.
Author Contributions
I.V. conceptualized this work. I.V., J.J.B., R.T. and I.D.-L. wrote this review together. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Spanish Ministry of Science and Innovation (PID2021-125844OB-I00).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No specific data were generated to support reported results.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Powers, A.M.; Brault, A.C.; Shirako, Y.; Strauss, E.G.; Kang, W.; Strauss, J.H.; Weaver, S.C. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 2001, 75, 10118–10131. [Google Scholar] [CrossRef]
- Weaver, S.C.; Barrett, A.D.T. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2004, 2, 789–801. [Google Scholar] [CrossRef]
- Weaver, S.C.; Winegar, R.; Manger, I.D.; Forrester, N.L. Alphaviruses: Population genetics and determinants of emergence. Antivir. Res. 2012, 94, 242–257. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Chen, R.; Sherman, M.B.; Weaver, S.C. Chikungunya virus: Evolution and genetic determinants of emergence. Curr. Opin. Virol. 2011, 1, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Chen, R.; Weaver, S.C. Interspecies transmission and chikungunya virus emergence. Curr. Opin. Virol. 2016, 16, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Zacks, M.A.; Paessler, S. Encephalitic alphaviruses. Vet. Microbiol. 2010, 140, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Kafai, N.M.; Diamond, M.S.; Fox, J.M. Distinct Cellular Tropism and Immune Responses to Alphavirus Infection. Annu. Rev. Immunol. 2022, 40, 615–649. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Ahola, T.; McInerney, G.; Merits, A. Alphavirus RNA replication in vertebrate cells. Adv. Virus Res. 2021, 111, 111–156. [Google Scholar] [CrossRef]
- Pettersson, R.F.; Soderlund, H.; Kaariainen, L. The nucleotide sequences of the 5′-terminal T1 oligonucleotides of Semliki-Forest-virus 42-S and 26-S RNAs are different. Eur. J. Biochem. 1980, 105, 435–443. [Google Scholar] [CrossRef]
- Ahola, T.; Kaariainen, L. Reaction in alphavirus mRNA capping: Formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 1995, 92, 507–511. [Google Scholar] [CrossRef]
- Ahola, T.; Laakkonen, P.; Vihinen, H.; Kaariainen, L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 1997, 71, 392–397. [Google Scholar] [CrossRef]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011, 10, 51–65. [Google Scholar] [CrossRef]
- Gorchakov, R.; Frolova, E.; Frolov, I. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 2005, 79, 9397–9409. [Google Scholar] [CrossRef]
- Gorchakov, R.; Garmashova, N.; Frolova, E.; Frolov, I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 2008, 82, 10088–10101. [Google Scholar] [CrossRef]
- Fros, J.J.; Geertsema, C.; Zouache, K.; Baggen, J.; Domeradzka, N.; van Leeuwen, D.M.; Flipse, J.; Vlak, J.M.; Failloux, A.; Pijlman, G.P. Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus. Parasit. Vectors 2015, 8, 464. [Google Scholar] [CrossRef]
- Panas, M.D.; Varjak, M.; Lulla, A.; Eng, K.E.; Merits, A.; Karlsson Hedestam, G.B.; McInerney, G.M. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell 2012, 23, 4701–4712. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, A.K.; Adivarahan, S.; Koppula, A.; Abraham, R.; Batish, M.; Zenklusen, D.; Griffin, D.E.; Leung, A.K.L. Stress granule formation, disassembly, and composition are regulated by alphavirus ADP-ribosylhydrolase activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2021719118. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.I.; Palchevska, O.; Dominguez, F.; Frolov, I. Alphavirus-induced transcriptional and translational shutoffs play major roles in blocking the formation of stress granules. J. Virol. 2023, 97, e0097923-23. [Google Scholar] [CrossRef] [PubMed]
- Pietila, M.K.; Hellstrom, K.; Ahola, T. Alphavirus polymerase and RNA replication. Virus Res. 2017, 234, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Lemm, J.A.; Rumenapf, T.; Strauss, E.G.; Strauss, J.H.; Rice, C.M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994, 13, 2925–2934. [Google Scholar] [CrossRef]
- Lulla, V.; Karo-Astover, L.; Rausalu, K.; Saul, S.; Merits, A.; Lulla, A. Timeliness of Proteolytic Events Is Prerequisite for Efficient Functioning of the Alphaviral Replicase. J. Virol. 2018, 92, e00151-18. [Google Scholar] [CrossRef]
- Frolova, E.I.; Gorchakov, R.; Pereboeva, L.; Atasheva, S.; Frolov, I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010, 84, 11679–11695. [Google Scholar] [CrossRef]
- Froshauer, S.; Kartenbeck, J.; Helenius, A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988, 107, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Laurent, T.; Kumar, P.; Liese, S.; Zare, F.; Jonasson, M.; Carlson, A.; Carlson, L. Architecture of the chikungunya virus replication organelle. eLife 2022, 11, e83042. [Google Scholar] [CrossRef] [PubMed]
- Akhrymuk, I.; Frolov, I.; Frolova, E.I. Both RIG-I and MDA5 detect alphavirus replication in concentration-dependent mode. Virology 2016, 487, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Trobaugh, D.W.; Gardner, C.L.; Sun, C.; Haddow, A.D.; Wang, E.; Chapnik, E.; Mildner, A.; Weaver, S.C.; Ryman, K.D.; Klimstra, W.B. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 2014, 506, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Taylor, A.B.; Kuhn, R.J. Spatial and Temporal Analysis of Alphavirus Replication and Assembly in Mammalian and Mosquito Cells. mBio 2017, 8, e02294-16. [Google Scholar] [CrossRef] [PubMed]
- Gliedman, J.B.; Smith, J.F.; Brown, D.T. Morphogenesis of Sindbis virus in cultured Aedes albopictus cells. J. Virol. 1975, 16, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Dalgarno, L. Semliki Forest virus replication in cultured Aedes albopictus cells: Studies on the establishment of persistence. J. Gen. Virol. 1974, 24, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Rumenapf, T.; Strauss, E.G.; Strauss, J.H. Regulation of Semliki Forest virus RNA replication: A model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 2004, 323, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Ventoso, I. Adaptive changes in alphavirus mRNA translation allowed colonization of vertebrate hosts. J. Virol. 2012, 86, 9484–9494. [Google Scholar] [CrossRef] [PubMed]
- Akhrymuk, I.; Kulemzin, S.V.; Frolova, E.I. Evasion of the innate immune response: The Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J. Virol. 2012, 86, 7180–7191. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Thompson, S.R.; Mathews, M.B.; Mohr, I. Translational Control in Virus-Infected Cells. Cold Spring Harb. Perspect. Biol. 2019, 11, a033001. [Google Scholar] [CrossRef] [PubMed]
- Ventoso, I.; Sanz, M.A.; Molina, S.; Berlanga, J.J.; Carrasco, L.; Esteban, M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006, 20, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Gorchakov, R.; Frolova, E.; Williams, B.R.G.; Rice, C.M.; Frolov, I. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 2004, 78, 8455–8467. [Google Scholar] [CrossRef] [PubMed]
- Meshram, C.D.; Lukash, T.; Phillips, A.T.; Akhrymuk, I.; Frolova, E.I.; Frolov, I. Lack of nsP2-specific nuclear functions attenuates chikungunya virus replication both in vitro and in vivo. Virology 2019, 534, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.; Hauer, D.; McPherson, R.L.; Utt, A.; Kirby, I.T.; Cohen, M.S.; Merits, A.; Leung, A.K.L.; Griffin, D.E. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. USA 2018, 115, E10457–E10466. [Google Scholar] [CrossRef]
- Dominguez, F.; Palchevska, O.; Frolova, E.I.; Frolov, I. Alphavirus-based replicons demonstrate different interactions with host cells and can be optimized to increase protein expression. J. Virol. 2023, 97, e0122523. [Google Scholar] [CrossRef]
- Bhalla, N.; Sun, C.; Metthew Lam, L.K.; Gardner, C.L.; Ryman, K.D.; Klimstra, W.B. Host translation shutoff mediated by non-structural protein 2 is a critical factor in the antiviral state resistance of Venezuelan equine encephalitis virus. Virology 2016, 496, 147–165. [Google Scholar] [CrossRef]
- Carrasco, L.; Sanz, M.A.; Gonzalez-Almela, E. The Regulation of Translation in Alphavirus-Infected Cells. Viruses 2018, 10, 70. [Google Scholar] [CrossRef]
- Fros, J.J.; Pijlman, G.P. Alphavirus Infection: Host Cell Shut-Off and Inhibition of Antiviral Responses. Viruses 2016, 8, 166. [Google Scholar] [CrossRef]
- McInerney, G.M.; Kedersha, N.L.; Kaufman, R.J.; Anderson, P.; Liljestrom, P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 2005, 16, 3753–3763. [Google Scholar] [CrossRef] [PubMed]
- Toribio, R.; Ventoso, I. Inhibition of host translation by virus infection in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 9837–9842. [Google Scholar] [CrossRef] [PubMed]
- Treffers, E.E.; Tas, A.; Scholte, F.E.M.; de Ru, A.H.; Snijder, E.J.; van Veelen, P.A.; van Hemert, M.J. The alphavirus nonstructural protein 2 NTPase induces a host translational shut-off through phosphorylation of eEF2 via cAMP-PKA-eEF2K signaling. PLoS Pathog. 2023, 19, e1011179. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, U.; Nilsson, A.; Nygard, O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990, 191, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.A.; Garcia-Moreno, M.; Carrasco, L. Inhibition of host protein synthesis by Sindbis virus: Correlation with viral RNA replication and release of nuclear proteins to the cytoplasm. Cell. Microbiol. 2015, 17, 520–541. [Google Scholar] [CrossRef] [PubMed]
- Frolov, I.; Schlesinger, S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 1994, 68, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.A.; Castello, A.; Carrasco, L. Viral translation is coupled to transcription in Sindbis virus-infected cells. J. Virol. 2007, 81, 7061–7068. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.D.; Moon, S.L.; Emch, A.W.; Wilusz, C.J.; Wilusz, J. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 2013, 5, 909–917. [Google Scholar] [CrossRef]
- van Steeg, H.; Kasperaitis, M.; Voorma, H.O.; Benne, R. Infection of neuroblastoma cells by Semliki Forest virus. The interference of viral capsid protein with the binding of host messenger RNAs into initiation complexes is the cause of the shut-off of host protein synthesis. Eur. J. Biochem. 1984, 138, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Frolov, I.; Akhrymuk, M.; Akhrymuk, I.; Atasheva, S.; Frolova, E.I. Early events in alphavirus replication determine the outcome of infection. J. Virol. 2012, 86, 5055–5066. [Google Scholar] [CrossRef] [PubMed]
- Ryman, K.D.; Klimstra, W.B. Host responses to alphavirus infection. Immunol. Rev. 2008, 225, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Schilte, C.; Couderc, T.; Chretien, F.; Sourisseau, M.; Gangneux, N.; Guivel-Benhassine, F.; Kraxner, A.; Tschopp, J.; Higgs, S.; Michault, A.; et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 2010, 207, 429–442. [Google Scholar] [CrossRef]
- Grieder, F.B.; Vogel, S.N. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology 1999, 257, 106–118. [Google Scholar] [CrossRef]
- Ryman, K.D.; Klimstra, W.B.; Nguyen, K.B.; Biron, C.A.; Johnston, R.E. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 2000, 74, 3366–3378. [Google Scholar] [CrossRef]
- Friedman, R.M.; Fantes, K.H.; Levy, H.B.; Carter, W.B. Interferon action on parental Semliki forest virus ribonucleic acid. J. Virol. 1967, 1, 1168–1173. [Google Scholar] [CrossRef]
- Friedman, R.M. Inhibition of arbovirus protein synthesis by interferon. J. Virol. 1968, 2, 1081–1085. [Google Scholar] [CrossRef]
- Toribio, R.; Diaz-Lopez, I.; Berlanga, J.J.; Molina-Jimenez, F.; Majano, P.; Ventoso, I. Naturally Occurring and Engineered Alphaviruses Sensitive to Double-Stranded-RNA-Activated Protein Kinase Show Restricted Translation in Mammalian Cells, Increased Sensitivity to Interferon, and Marked Oncotropism. J. Virol. 2020, 94, e01630-19. [Google Scholar] [CrossRef]
- Frolova, E.I.; Fayzulin, R.Z.; Cook, S.H.; Griffin, D.E.; Rice, C.M.; Frolov, I. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 2002, 76, 11254–11264. [Google Scholar] [CrossRef]
- Atasheva, S.; Kim, D.Y.; Frolova, E.I.; Frolov, I. Venezuelan equine encephalitis virus variants lacking transcription inhibitory functions demonstrate highly attenuated phenotype. J. Virol. 2015, 89, 71–82. [Google Scholar] [CrossRef]
- Her, Z.; Malleret, B.; Chan, M.; Ong, E.K.S.; Wong, S.; Kwek, D.J.C.; Tolou, H.; Lin, R.T.P.; Tambyah, P.A.; Renia, L.; et al. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J. Immunol. 2010, 184, 5903–5913. [Google Scholar] [CrossRef]
- Sadler, A.J.; Williams, B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef]
- Zhang, Y.; Burke, C.W.; Ryman, K.D.; Klimstra, W.B. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 2007, 81, 11246–11255. [Google Scholar] [CrossRef]
- McDougal, M.B.; De Maria, A.M.; Ohlson, M.B.; Kumar, A.; Xing, C.; Schoggins, J.W. Interferon inhibits a model RNA virus via a limited set of inducible effector genes. EMBO Rep. 2023, 24, e56901. [Google Scholar] [CrossRef] [PubMed]
- Tesfay, M.Z.; Yin, J.; Gardner, C.L.; Khoretonenko, M.V.; Korneeva, N.L.; Rhoads, R.E.; Ryman, K.D.; Klimstra, W.B. Alpha/beta interferon inhibits cap-dependent translation of viral but not cellular mRNA by a PKR-independent mechanism. J. Virol. 2008, 82, 2620–2630. [Google Scholar] [CrossRef] [PubMed]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Hyde, J.L.; Gardner, C.L.; Kimura, T.; White, J.P.; Liu, G.; Trobaugh, D.W.; Huang, C.; Tonelli, M.; Paessler, S.; Takeda, K.; et al. A viral RNA structural element alters host recognition of nonself RNA. Science 2014, 343, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Lassnig, C.; Eberle, C.; Gorna, M.W.; Baumann, C.L.; Burkard, T.R.; Burckstummer, T.; Stefanovic, A.; Krieger, S.; Bennett, K.L.; et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011, 12, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.M.; Pichlmair, A.; Gorna, M.W.; Superti-Furga, G.; Nagar, B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 2013, 494, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sweeney, T.R.; Skabkin, M.A.; Skabkina, O.V.; Hellen, C.U.T.; Pestova, T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp-mRNAs. Nucleic Acids Res. 2014, 42, 3228–3245. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; VanBlargan, L.A.; Xu, W.; White, J.P.; Shan, C.; Shi, P.; Zhang, R.; Adhikari, J.; Gross, M.L.; Leung, D.W.; et al. Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability. Immunity 2018, 48, 487–499.e5. [Google Scholar] [CrossRef] [PubMed]
- Fleith, R.C.; Mears, H.V.; Leong, X.Y.; Sanford, T.J.; Emmott, E.; Graham, S.C.; Mansur, D.S.; Sweeney, T.R. IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Res. 2018, 46, 5269–5285. [Google Scholar] [CrossRef] [PubMed]
- Reynaud, J.M.; Kim, D.Y.; Atasheva, S.; Rasalouskaya, A.; White, J.P.; Diamond, M.S.; Weaver, S.C.; Frolova, E.I.; Frolov, I. IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon. PLoS Pathog. 2015, 11, e1004863. [Google Scholar] [CrossRef] [PubMed]
- Bick, M.J.; Carroll, J.N.; Gao, G.; Goff, S.P.; Rice, C.M.; MacDonald, M.R. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 2003, 77, 11555–11562. [Google Scholar] [CrossRef]
- Ficarelli, M.; Neil, S.J.D.; Swanson, C.M. Targeted Restriction of Viral Gene Expression and Replication by the ZAP Antiviral System. Annu. Rev. Virol. 2021, 8, 265–283. [Google Scholar] [CrossRef]
- Goncalves-Carneiro, D.; Takata, M.A.; Ong, H.; Shilton, A.; Bieniasz, P.D. Origin and evolution of the zinc finger antiviral protein. PLoS Pathog. 2021, 17, e1009545. [Google Scholar] [CrossRef]
- Gao, G.; Guo, X.; Goff, S.P. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 2002, 297, 1703–1706. [Google Scholar] [CrossRef]
- Guo, X.; Carroll, J.N.; Macdonald, M.R.; Goff, S.P.; Gao, G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 2004, 78, 12781–12787. [Google Scholar] [CrossRef]
- Schwerk, J.; Soveg, F.W.; Ryan, A.P.; Thomas, K.R.; Hatfield, L.D.; Ozarkar, S.; Forero, A.; Kell, A.M.; Roby, J.A.; So, L.; et al. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat. Immunol. 2019, 20, 1610–1620. [Google Scholar] [CrossRef]
- Li, M.M.H.; Aguilar, E.G.; Michailidis, E.; Pabon, J.; Park, P.; Wu, X.; de Jong, Y.P.; Schneider, W.M.; Molina, H.; Rice, C.M.; et al. Characterization of Novel Splice Variants of Zinc Finger Antiviral Protein (ZAP). J. Virol. 2019, 93, e00715-19. [Google Scholar] [CrossRef]
- Kerns, J.A.; Emerman, M.; Malik, H.S. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008, 4, e21. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, X.; Goff, S.P.; Gao, G. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J. 2012, 31, 4236–4246. [Google Scholar] [CrossRef]
- Takata, M.A.; Goncalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 2017, 550, 124–127. [Google Scholar] [CrossRef]
- Luo, X.; Wang, X.; Gao, Y.; Zhu, J.; Liu, S.; Gao, G.; Gao, P. Molecular Mechanism of RNA Recognition by Zinc-Finger Antiviral Protein. Cell Rep. 2020, 30, 46–52.e4. [Google Scholar] [CrossRef]
- Meagher, J.L.; Takata, M.; Goncalves-Carneiro, D.; Keane, S.C.; Rebendenne, A.; Ong, H.; Orr, V.K.; MacDonald, M.R.; Stuckey, J.A.; Bieniasz, P.D.; et al. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc. Natl. Acad. Sci. USA 2019, 116, 24303–24309. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Xia, W.; Baillie, J.K.; McKinnon, K. Modelling mutational and selection pressures on dinucleotides in eukaryotic phyla—Selection against CpG and UpA in cytoplasmically expressed RNA and in RNA viruses. BMC Genom. 2013, 14, 610. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.P.; Aldana, K.S.; Yang, E.; Yao, Z.; Li, M.M.H. Alphavirus Evasion of Zinc Finger Antiviral Protein (ZAP) Correlates with CpG Suppression in a Specific Viral nsP2 Gene Sequence. Viruses 2023, 15, 830. [Google Scholar] [CrossRef] [PubMed]
- Goonawardane, N.; Nguyen, D.; Simmonds, P. Association of Zinc Finger Antiviral Protein Binding to Viral Genomic RNA with Attenuation of Replication of Echovirus 7. mSphere 2021, 6, e01138-20. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Nguyen, L.P.; Wisherop, C.A.; Kan, R.L.; Li, M.M.H. The Role of ZAP and TRIM25 RNA Binding in Restricting Viral Translation. Front. Cell. Infect. Microbiol. 2022, 12, 886929. [Google Scholar] [CrossRef]
- Kozaki, T.; Takahama, M.; Misawa, T.; Matsuura, Y.; Akira, S.; Saitoh, T. Role of zinc-finger anti-viral protein in host defense against Sindbis virus. Int. Immunol. 2015, 27, 357–364. [Google Scholar] [CrossRef]
- Li, M.M.H.; Lau, Z.; Cheung, P.; Aguilar, E.G.; Schneider, W.M.; Bozzacco, L.; Molina, H.; Buehler, E.; Takaoka, A.; Rice, C.M.; et al. TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP). PLoS Pathog. 2017, 13, e1006145. [Google Scholar] [CrossRef]
- Roberts, W.K.; Hovanessian, A.; Brown, R.E.; Clemens, M.J.; Kerr, I.M. Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature 1976, 264, 477–480. [Google Scholar] [CrossRef]
- Meurs, E.; Chong, K.; Galabru, J.; Thomas, N.S.; Kerr, I.M.; Williams, B.R.; Hovanessian, A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 1990, 62, 379–390. [Google Scholar] [CrossRef]
- Garcia, M.A.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006, 70, 1032–1060. [Google Scholar] [CrossRef] [PubMed]
- Chaumont, L.; Collet, B.; Boudinot, P. Double-stranded RNA-dependent protein kinase (PKR) in antiviral defence in fish and mammals. Dev. Comp. Immunol. 2023, 145, 104732. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Reis, L.F.; Pavlovic, J.; Aguzzi, A.; Schafer, R.; Kumar, A.; Williams, B.R.; Aguet, M.; Weissmann, C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995, 14, 6095–6106. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, S.; Roberts, P.C.; Brown, L.E.; Truong, H.; Pattnaik, A.K.; Archer, D.R.; Barber, G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 2000, 13, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Gordiyenko, Y.; Llacer, J.L.; Ramakrishnan, V. Structural basis for the inhibition of translation through eIF2alpha phosphorylation. Nat. Commun. 2019, 10, 2640. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, T.; Pavitt, G.D.; Zhang, F.; Dever, T.E.; Hinnebusch, A.G. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 2001, 21, 5018–5030. [Google Scholar] [CrossRef] [PubMed]
- Cesaro, T.; Michiels, T. Inhibition of PKR by Viruses. Front. Microbiol. 2021, 12, 757238. [Google Scholar] [CrossRef]
- Garcia-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, P.V.; Adams, A.P.; Wang, E.; Kang, W.; Carrara, A.; Anishchenko, M.; Frolov, I.; Weaver, S.C. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 2008, 82, 4920–4930. [Google Scholar] [CrossRef] [PubMed]
- Torii, S.; Orba, Y.; Hang’ombe, B.M.; Mweene, A.S.; Wada, Y.; Anindita, P.D.; Phongphaew, W.; Qiu, Y.; Kajihara, M.; Mori-Kajihara, A.; et al. Discovery of Mwinilunga alphavirus: A novel alphavirus in Culex mosquitoes in Zambia. Virus Res. 2018, 250, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, K.; Marklewitz, M.; Zirkel, F.; Overheul, G.J.; Page, R.A.; Loaiza, J.R.; Drosten, C.; van Rij, R.P.; Junglen, S. Agua Salud alphavirus defines a novel lineage of insect-specific alphaviruses discovered in the New World. J. Gen. Virol. 2020, 101, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Nasar, F.; Palacios, G.; Gorchakov, R.V.; Guzman, H.; Da Rosa, A.P.T.; Savji, N.; Popov, V.L.; Sherman, M.B.; Lipkin, W.I.; Tesh, R.B.; et al. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc. Natl. Acad. Sci. USA 2012, 109, 14622–14627. [Google Scholar] [CrossRef]
- Hermanns, K.; Zirkel, F.; Kopp, A.; Marklewitz, M.; Rwego, I.B.; Estrada, A.; Gillespie, T.R.; Drosten, C.; Junglen, S. Discovery of a novel alphavirus related to Eilat virus. J. Gen. Virol. 2017, 98, 43–49. [Google Scholar] [CrossRef]
- Odon, V.; Fiddaman, S.R.; Smith, A.L.; Simmonds, P. Comparison of CpG- and UpA-mediated restriction of RNA virus replication in mammalian and avian cells and investigation of potential ZAP-mediated shaping of host transcriptome compositions. RNA 2022, 28, 1089–1109. [Google Scholar] [CrossRef]
- Toribio, R.; Diaz-Lopez, I.; Boskovic, J.; Ventoso, I. An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res. 2016, 44, 4368–4380. [Google Scholar] [CrossRef]
- Frolov, I.; Schlesinger, S. Translation of Sindbis virus mRNA: Effects of sequences downstream of the initiating codon. J. Virol. 1994, 68, 8111–8117. [Google Scholar] [CrossRef]
- Rathore, A.P.S.; Ng, M.; Vasudevan, S.G. Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2alpha phosphorylation. Virol. J. 2013, 10, 36. [Google Scholar] [CrossRef]
- Garmashova, N.; Gorchakov, R.; Volkova, E.; Paessler, S.; Frolova, E.; Frolov, I. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 2007, 81, 2472–2484. [Google Scholar] [CrossRef]
- Garmashova, N.; Gorchakov, R.; Frolova, E.; Frolov, I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 2006, 80, 5686–5696. [Google Scholar] [CrossRef]
- Atasheva, S.; Garmashova, N.; Frolov, I.; Frolova, E. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in Mammalian but not in mosquito cells. J. Virol. 2008, 82, 4028–4041. [Google Scholar] [CrossRef] [PubMed]
- Akhrymuk, I.; Lukash, T.; Frolov, I.; Frolova, E.I. Novel Mutations in nsP2 Abolish Chikungunya Virus-Induced Transcriptional Shutoff and Make the Virus Less Cytopathic without Affecting Its Replication Rates. J. Virol. 2019, 93, e02062-18. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Fish, A.; Fornerod, M.; Frolova, E.I. Venezuelan equine Encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J. Virol. 2010, 84, 4158–4171. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).