Macrocolony of NDM-1 Producing Enterobacter hormaechei subsp. oharae Generates Subpopulations with Different Features Regarding the Response of Antimicrobial Agents and Biofilm Formation
Abstract
1. Introduction
2. Results
2.1. Bacterial Macrocolonies Generate Subpopulations with Distinct Susceptibility to Meropenem and Ability to Form Biofilm
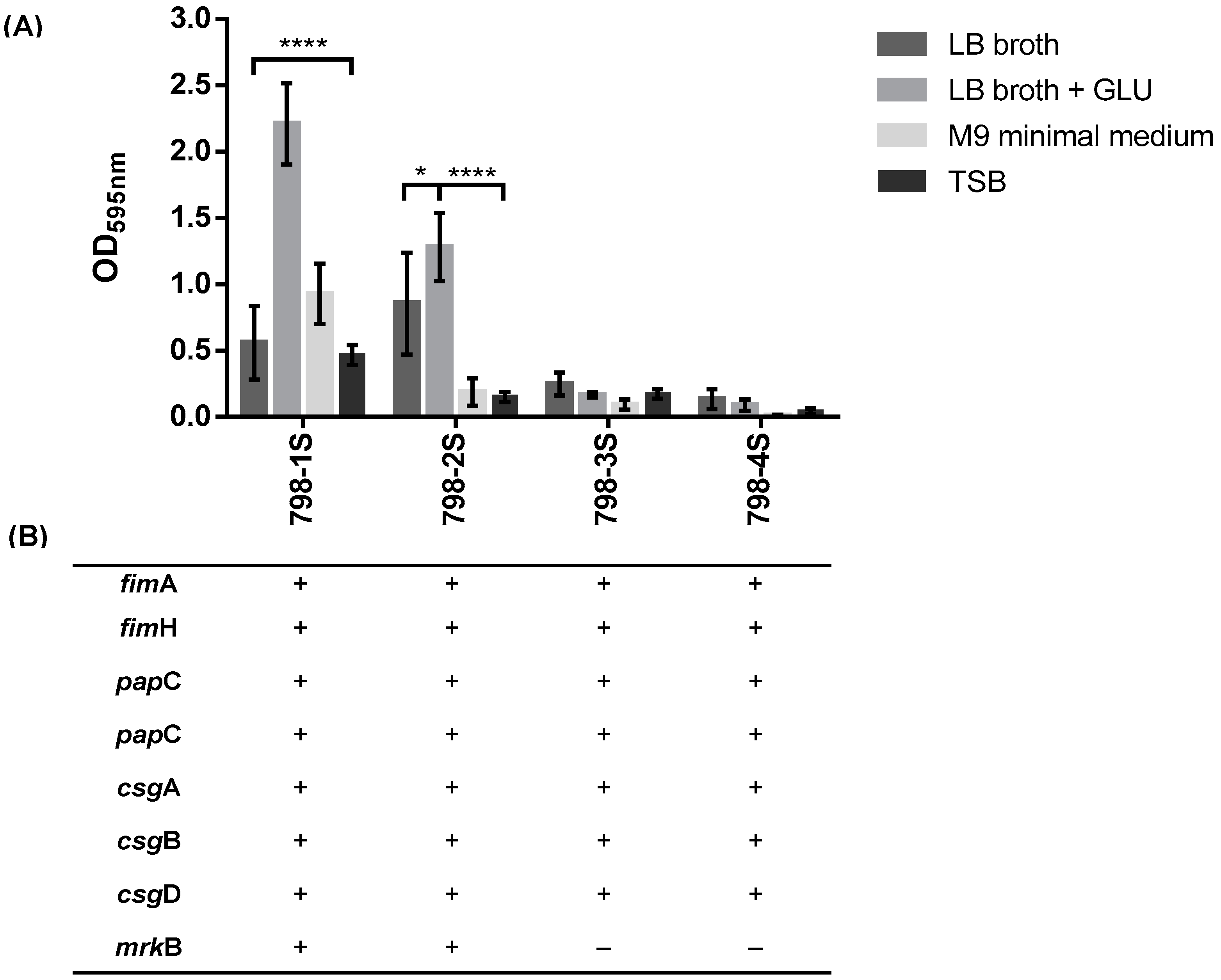
2.2. Subpopulations of 798 Macrocolony
2.2.1. Type 3 Fimbriae are Important for Biofilm Formation in E. hormachei subsp. oharae
2.2.2. Checkerboard Assay: Triple Combination (meropenem–rifampicin–polymyxin B) is Effective against All Subpopulations
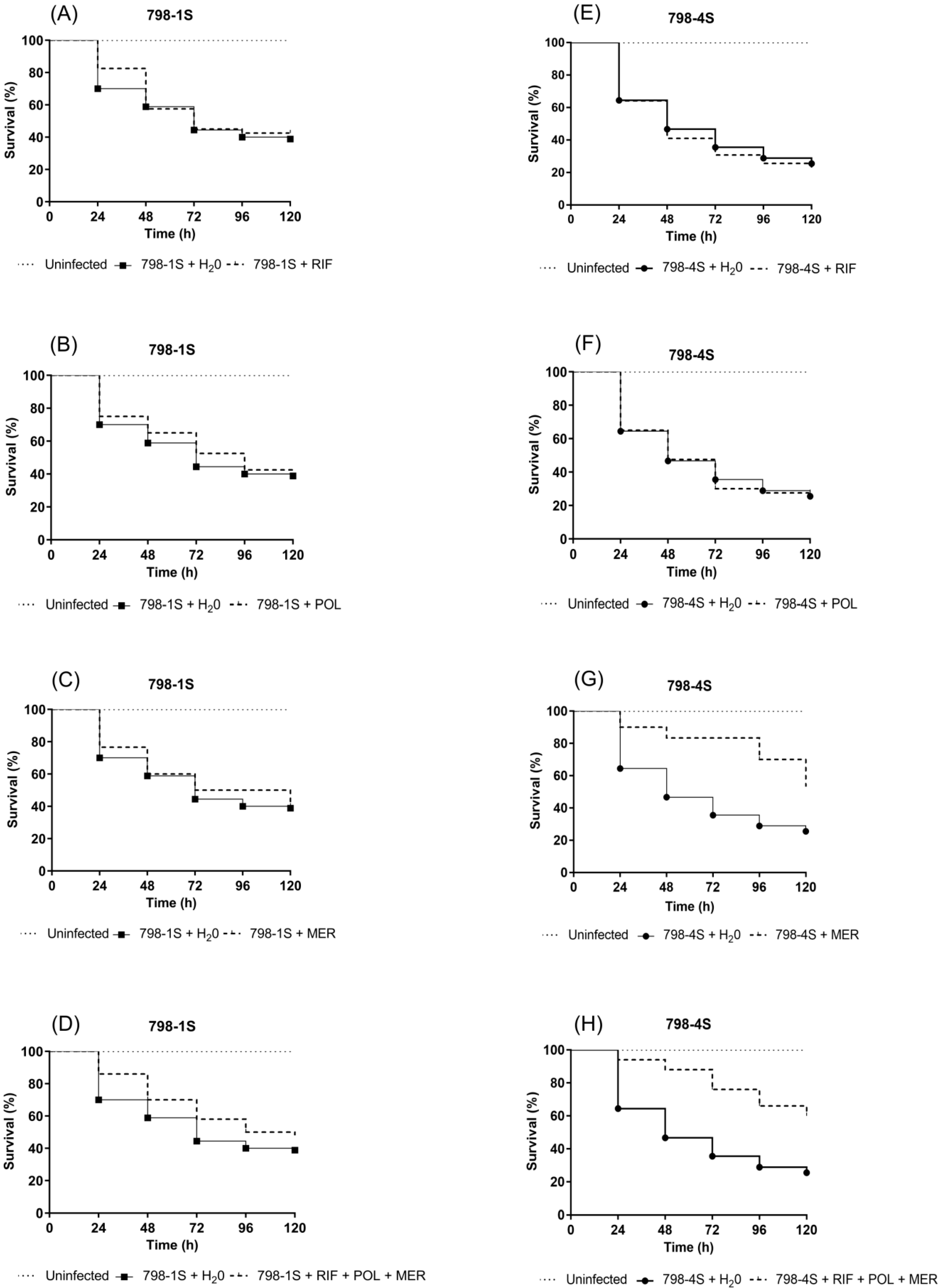
2.2.3. Galleria mellonella Infection Model: Differences in Response to Antimicrobial Treatment between 798-1S and 798-4S Subpopulations
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth of Macrocolonies
4.2. Biofilm Formation: Microtiter Plates Assay
4.3. Polymerase Chain Reaction (PCR):Fimbrial Genes Detection
4.4. Minimum Inhibitory Concentration (MIC): Agar Dilution Method
4.5. Checkerboard Assay
4.6. Galleria mellonella Model Studies
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef]
- Morgan, D.J.; Lomotan, L.L.; Agnes, K.; McGrail, L.; Roghmann, M.C. Characteristics of healthcare-associated infections contributing to unexpected in-hospital deaths. Infect. Control Hosp. Epidemiol. 2010, 31, 864–866. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef]
- Chavda, K.D.; Chen, L.; Fouts, D.E.; Sutton, G.; Brinkac, L.; Jenkins, S.G.; Bonomo, R.A.; Adams, M.D.; Kreiswirth, B.N. Comprehensive Genome Analysis of Carbapenemase-Producing Enterobacter spp.: New Insights into Phylogeny, Population Structure, and Resistance Mechanisms. MBio 2016, 7. [Google Scholar] [CrossRef]
- Moradigaravand, D.; Reuter, S.; Martin, V.; Peacock, S.J.; Parkhill, J. The dissemination of multidrug-resistant Enterobacter cloacae throughout the UK and Ireland. Nat. Microbiol. 2016, 1, 16173. [Google Scholar] [CrossRef] [PubMed]
- Lazarovitch, T.; Amity, K.; Coyle, J.R.; Ackerman, B.; Tal-Jasper, R.; Ofer-Friedman, H.; Hayakawa, K.; Bogan, C.; Lephart, P.R.; Kaplansky, T.; et al. The Complex Epidemiology of Carbapenem-Resistant Enterobacter Infections: A Multicenter Descriptive Analysis. Infect. Control Hosp. Epidemiol. 2015, 36, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.S.; Hsueh, P.R.; Group, S.A.-P. Distribution of ESBLs, AmpC beta-lactamases and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008-14: Results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 2017, 72, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Kontopidou, F.; Giamarellou, H.; Katerelos, P.; Maragos, A.; Kioumis, I.; Trikka-Graphakos, E.; Valakis, C.; Maltezou, H.C.; Group for the Study of KPC-producing Klebsiella pneumoniae infections in intensive care units. Infections caused by carbapenem-resistant Klebsiella pneumoniae among patients in intensive care units in Greece: A multi-centre study on clinical outcome and therapeutic options. Clin. Microbiol. Infect. 2014, 20, O117–O123. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, S.M.; McKinnell, J.A.; Mueller, L.E.; Miller, L.G.; Gohil, S.K.; Huang, S.S.; Lee, B.Y. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin. Microbiol. Infect. 2017, 23, 48.e9–48.e16. [Google Scholar] [CrossRef] [PubMed]
- Thabit, A.K.; Crandon, J.L.; Nicolau, D.P. Antimicrobial resistance: Impact on clinical and economic outcomes and the need for new antimicrobials. Expert Opin. Pharmacother. 2015, 16, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Poirel, L.; Nordmann, P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed. Res. Int. 2014, 2014, 249856. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Morrill, H.J.; Pogue, J.M.; Kaye, K.S.; LaPlante, K.L. Treatment Options for Carbapenem-Resistant Enterobacteriaceae Infections. Open Forum Infect. Dis. 2015, 2, ofv050. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Lourida, P.; Poulikakos, P.; Rafailidis, P.I.; Tansarli, G.S. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: Systematic evaluation of the available evidence. Antimicrob. Agents Chemother. 2014, 58, 654–663. [Google Scholar] [CrossRef]
- Trecarichi, E.M.; Tumbarello, M. Therapeutic options for carbapenem-resistant Enterobacteriaceae infections. Virulence 2017, 8, 470–484. [Google Scholar] [CrossRef]
- Chung, P.Y. The emerging problems of Klebsiella pneumoniae infections: Carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Perez, F.; El Chakhtoura, N.G.; Papp-Wallace, K.M.; Wilson, B.M.; Bonomo, R.A. Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: Can we apply “precision medicine” to antimicrobial chemotherapy? Expert Opin. Pharmacother. 2016, 17, 761–781. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Coote, P.J. Utility of Greater Wax Moth Larva (Galleria mellonella) for Evaluating the Toxicity and Efficacy of New Antimicrobial Agents. Adv. Appl. Microbiol. 2012, 78, 25–53. [Google Scholar] [CrossRef] [PubMed]
- Gruber, T.M.; Gottig, S.; Mark, L.; Christ, S.; Kempf, V.A.; Wichelhaus, T.A.; Hamprecht, A. Pathogenicity of pan-drug-resistant Serratia marcescens harbouring blaNDM-1. J. Antimicrob. Chemother. 2015, 70, 1026–1030. [Google Scholar] [CrossRef]
- Hoffmann, H.; Roggenkamp, A. Population genetics of the nomenspecies Enterobacter cloacae. Appl. Environ. Microbiol. 2003, 69, 5306–5318. [Google Scholar] [CrossRef] [PubMed]
- Ohad, S.; Block, C.; Kravitz, V.; Farber, A.; Pilo, S.; Breuer, R.; Rorman, E. Rapid identification of Enterobacter hormaechei and Enterobacter cloacae genetic cluster III. J. Appl. Microbiol. 2014, 116, 1315–1321. [Google Scholar] [CrossRef]
- Morand, P.C.; Billoet, A.; Rottman, M.; Sivadon-Tardy, V.; Eyrolle, L.; Jeanne, L.; Tazi, A.; Anract, P.; Courpied, J.P.; Poyart, C.; et al. Specific distribution within the Enterobacter cloacae complex of strains isolated from infected orthopedic implants. J. Clin. Microbiol. 2009, 47, 2489–2495. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Pages, J.M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 2015, 6, 392. [Google Scholar] [CrossRef]
- Beyrouthy, R.; Barets, M.; Marion, E.; Dananche, C.; Dauwalder, O.; Robin, F.; Gauthier, L.; Jousset, A.; Dortet, L.; Guerin, F.; et al. Novel Enterobacter Lineage as Leading Cause of Nosocomial Outbreak Involving Carbapenemase-Producing Strains. Emerg. Infect. Dis. 2018, 24, 1505–1515. [Google Scholar] [CrossRef]
- Paauw, A.; Caspers, M.P.; Leverstein-van Hall, M.A.; Schuren, F.H.; Montijn, R.C.; Verhoef, J.; Fluit, A.C. Identification of resistance and virulence factors in an epidemic Enterobacter hormaechei outbreak strain. Microbiology 2009, 155, 1478–1488. [Google Scholar] [CrossRef][Green Version]
- Monahan, L.G.; DeMaere, M.Z.; Cummins, M.L.; Djordjevic, S.P.; Roy Chowdhury, P.; Darling, A.E. High contiguity genome sequence of a multidrug-resistant hospital isolate of Enterobacter hormaechei. Gut Pathog. 2019, 11, 3. [Google Scholar] [CrossRef]
- Mezzatesta, M.L.; Gona, F.; Stefani, S. Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiol. 2012, 7, 887–902. [Google Scholar] [CrossRef]
- Haussler, S.; Fuqua, C. Biofilms 2012: New discoveries and significant wrinkles in a dynamic field. J. Bacteriol. 2013, 195, 2947–2958. [Google Scholar] [CrossRef]
- Serra, D.O.; Hengge, R. Stress responses go three dimensional—The spatial order of physiological differentiation in bacterial macrocolony biofilms. Environ. Microbiol. 2014, 16, 1455–1471. [Google Scholar] [CrossRef]
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2016, 2. [Google Scholar] [CrossRef]
- Ferreira, M.L.; Araujo, B.F.; Goncalves, I.R.; Royer, S.; Campos, P.A.; Machado, L.G.; Batistao, D.W.F.; Brito, C.S.; Gontijo-Filho, P.P.; Ribas, R.M. Association of Colistin-Resistant KPC Clonal Strains with Subsequent Infections and Colonization and Biofilm Production. Microb. Drug Resist. 2018. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 7.1; European Committee on Antimicrobial Susceptibility Testing: Basel, Switzerland, 2017. [Google Scholar]
- Allen, B.L.; Gerlach, G.F.; Clegg, S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 1991, 173, 916–920. [Google Scholar] [CrossRef][Green Version]
- Duguid, J.P. Fimbriae and adhesive properties in Klebsiella strains. J. Gen. Microbiol. 1959, 21, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Adegbola, R.A.; Old, D.C. Fimbrial haemagglutinins in Enterobacter species. J. Gen. Microbiol. 1983, 129, 2175–2180. [Google Scholar] [CrossRef]
- Hornick, D.B.; Allen, B.L.; Horn, M.A.; Clegg, S. Fimbrial types among respiratory isolates belonging to the family Enterobacteriaceae. J. Clin. Microbiol. 1991, 29, 1795–1800. [Google Scholar] [PubMed]
- Burmolle, M.; Bahl, M.I.; Jensen, L.B.; Sorensen, S.J.; Hansen, L.H. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 2008, 154, 187–195. [Google Scholar] [CrossRef]
- Hornick, D.B.; Thommandru, J.; Smits, W.; Clegg, S. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect. Immun. 1995, 63, 2026–2032. [Google Scholar]
- Boll, E.J.; Marti, R.; Hasman, H.; Overballe-Petersen, S.; Stegger, M.; Ng, K.; Knochel, S.; Krogfelt, K.A.; Hummerjohann, J.; Struve, C. Turn Up the Heat-Food and Clinical Escherichia coli Isolates Feature Two Transferrable Loci of Heat Resistance. Front. Microbiol. 2017, 8, 579. [Google Scholar] [CrossRef]
- Jagnow, J.; Clegg, S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 2003, 149, 2397–2405. [Google Scholar] [CrossRef]
- Di Martino, P.; Cafferini, N.; Joly, B.; Darfeuille-Michaud, A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 2003, 154, 9–16. [Google Scholar] [CrossRef]
- Azevedo, P.A.A.; Furlan, J.P.R.; Oliveira-Silva, M.; Nakamura-Silva, R.; Gomes, C.N.; Costa, K.R.C.; Stehling, E.G.; Pitondo-Silva, A. Detection of virulence and beta-lactamase encoding genes in Enterobacter aerogenes and Enterobacter cloacae clinical isolates from Brazil. Braz. J. Microbiol. 2018, 49 (Suppl. 1), 224–228. [Google Scholar] [CrossRef]
- Serra, D.O.; Richter, A.M.; Klauck, G.; Mika, F.; Hengge, R. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio 2013, 4, e00103-13. [Google Scholar] [CrossRef]
- Schaufler, K.; Semmler, T.; Pickard, D.J.; de Toro, M.; de la Cruz, F.; Wieler, L.H.; Ewers, C.; Guenther, S. Carriage of Extended-Spectrum Beta-Lactamase-Plasmids Does Not Reduce Fitness but Enhances Virulence in Some Strains of Pandemic E. coli Lineages. Front. Microbiol 2016, 7, 336. [Google Scholar] [CrossRef] [PubMed]
- Bokranz, W.; Wang, X.; Tschape, H.; Romling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Cabeen, M.T.; Leiman, S.A.; Losick, R. Colony-morphology screening uncovers a role for the Pseudomonas aeruginosa nitrogen-related phosphotransferase system in biofilm formation. Mol. Microbiol. 2016, 99, 557–570. [Google Scholar] [CrossRef]
- Zogaj, X.; Bokranz, W.; Nimtz, M.; Romling, U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 2003, 71, 4151–4158. [Google Scholar] [CrossRef]
- Richter, A.M.; Povolotsky, T.L.; Wieler, L.H.; Hengge, R. Cyclic-di-GMP signalling and biofilm-related properties of the Shiga toxin-producing 2011 German outbreak Escherichia coli O104:H4. EMBO Mol. Med. 2014, 6, 1622–1637. [Google Scholar] [CrossRef]
- Freese, P.D.; Korolev, K.S.; Jimenez, J.I.; Chen, I.A. Genetic drift suppresses bacterial conjugation in spatially structured populations. Biophys. J. 2014, 106, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Krone, S.M.; Lu, R.; Fox, R.; Suzuki, H.; Top, E.M. Modelling the spatial dynamics of plasmid transfer and persistence. Microbiology 2007, 153, 2803–2816. [Google Scholar] [CrossRef][Green Version]
- Tangden, T.; Hickman, R.A.; Forsberg, P.; Lagerback, P.; Giske, C.G.; Cars, O. Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time-kill experiments. Antimicrob. Agents Chemother. 2014, 58, 1757–1762. [Google Scholar] [CrossRef]
- Urban, C.; Mariano, N.; Rahal, J.J. In vitro double and triple bactericidal activities of doripenem, polymyxin B, and rifampin against multidrug-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli. Antimicrob. Agents Chemother. 2010, 54, 2732–2734. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Doi, Y. Therapy of Infections due to Carbapenem-Resistant Gram-Negative Pathogens. Infect. Chemother. 2014, 46, 149–164. [Google Scholar] [CrossRef]
- Tascini, C.; Tagliaferri, E.; Giani, T.; Leonildi, A.; Flammini, S.; Casini, B.; Lewis, R.; Ferranti, S.; Rossolini, G.M.; Menichetti, F. Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2013, 57, 3990–3993. [Google Scholar] [CrossRef]
- Elemam, A.; Rahimian, J.; Doymaz, M. In vitro evaluation of antibiotic synergy for polymyxin B-resistant carbapenemase-producing Klebsiella pneumoniae. J. Clin. Microbiol 2010, 48, 3558–3562. [Google Scholar] [CrossRef]
- Yang, H.; Lv, N.; Hu, L.; Liu, Y.; Cheng, J.; Ye, Y.; Li, J. In vivo activity of vancomycin combined with colistin against multidrug-resistant strains of Acinetobacter baumannii in a Galleria mellonella model. Infect. Dis. (Lond.) 2016, 48, 189–194. [Google Scholar] [CrossRef]
- Yang, H.; Chen, G.; Hu, L.; Liu, Y.; Cheng, J.; Ye, Y.; Li, J. Enhanced efficacy of imipenem-colistin combination therapy against multiple-drug-resistant Enterobacter cloacae: In vitro activity and a Galleria mellonella model. J. Microbiol. Immunol. Infect. 2018, 51, 70–75. [Google Scholar] [CrossRef]
- Benthall, G.; Touzel, R.E.; Hind, C.K.; Titball, R.W.; Sutton, J.M.; Thomas, R.J.; Wand, M.E. Evaluation of antibiotic efficacy against infections caused by planktonic or biofilm cultures of Pseudomonas aeruginosa and Klebsiella pneumoniae in Galleria mellonella. Int. J. Antimicrob. Agents 2015, 46, 538–545. [Google Scholar] [CrossRef]
- Rozales, F.P.; Ribeiro, V.B.; Magagnin, C.M.; Pagano, M.; Lutz, L.; Falci, D.R.; Machado, A.; Barth, A.L.; Zavascki, A.P. Emergence of NDM-1-producing Enterobacteriaceae in Porto Alegre, Brazil. Int. J. Infect. Dis. 2014, 25, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Trentin, D.S.; Giordani, R.B.; Zimmer, K.R.; da Silva, A.G.; da Silva, M.V.; Correia, M.T.; Baumvol, I.J.; Macedo, A.J. Potential of medicinal plants from the Brazilian semi-arid region (Caatinga) against Staphylococcus epidermidis planktonic and biofilm lifestyles. J. Ethnopharmacol. 2011, 137, 327–335. [Google Scholar] [CrossRef]
- Wang, Y.; Bao, W.; Guo, N.; Chen, H.; Cheng, W.; Jin, K.; Shen, F.; Xu, J.; Zhang, Q.; Wang, C.; et al. Antimicrobial activity of the imipenem/rifampicin combination against clinical isolates of Acinetobacter baumannii grown in planktonic and biofilm cultures. World J. Microbiol. Biotechnol. 2014, 30, 3015–3025. [Google Scholar] [CrossRef]
- White, R.L.; Burgess, D.S.; Manduru, M.; Bosso, J.A. Comparison of three different in vitro methods of detecting synergy: Time-kill, checkerboard, and E test. Antimicrob. Agents Chemother. 1996, 40, 1914–1918. [Google Scholar] [CrossRef] [PubMed]
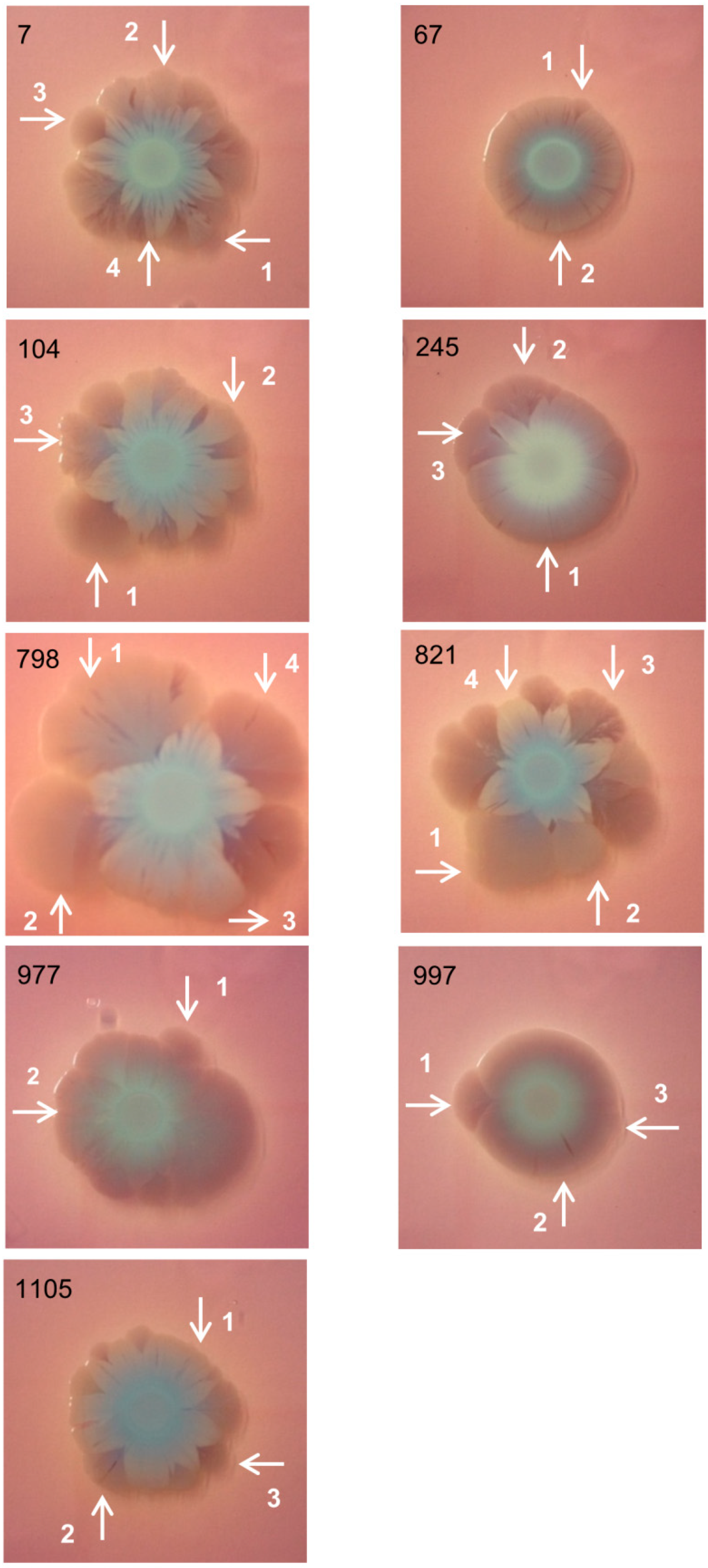
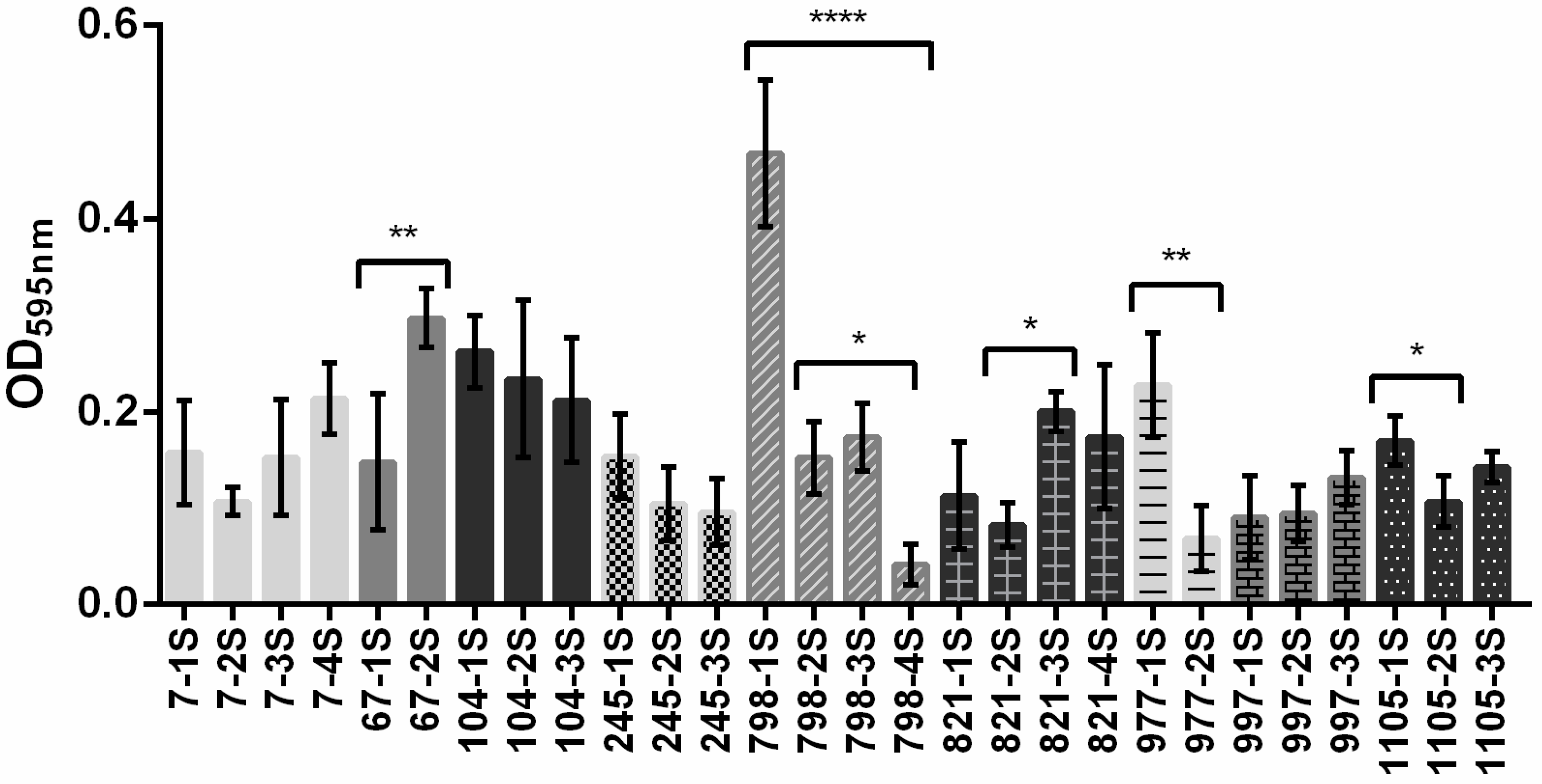

| Subpopulations | CAZ | CIP | GEN | MER |
|---|---|---|---|---|
| 7-1 S | >256 | 8 | ≤2 | 32 |
| 7-2 S | >256 | 8 | ≤2 | 64 |
| 7-3 S | 32 | 8 | ≤2 | ≤2 |
| 7-4 S | >256 | 8 | ≤2 | 32 |
| 67-1 S | >256 | >256 | 256 | 64 |
| 67-2 S | >256 | >256 | 256 | 256 |
| 104-1 S | 32 | 128 | 32 | ≤2 |
| 104-2 S | 32 | 128 | 16 | ≤2 |
| 104-3 S | 32 | 128 | 16 | ≤2 |
| 245-1 S | 128 | 64 | >256 | ≤2 |
| 245-2 S | 64 | 64 | >256 | ≤2 |
| 245-3 S | 64 | 64 | >256 | ≤2 |
| 798-1 S | >256 | 16 | 256 | 32 |
| 798-2 S | >256 | 32 | >256 | 32 |
| 798-3 S | >256 | 32 | 256 | 16 |
| 798-4 S | >256 | 8 | 128 | 16 |
| 821-1 S | 64 | >256 | >256 | ≤2 |
| 821-2 S | >256 | 256 | >256 | 32 |
| 821-3 S | >256 | 128 | 32 | 64 |
| 821-4 S | >256 | 256 | >256 | 32 |
| 977-1 S | 256 | 8 | 256 | ≤2 |
| 977-2 S | >256 | 8 | 256 | ≤2 |
| 997-1 S | >256 | 8 | ≤2 | ≤2 |
| 997-2 S | >256 | 8 | ≤2 | ≤2 |
| 1105-1 S | >256 | 64 | >256 | 32 |
| 1105-2 S | >256 | 64 | 256 | 64 |
| 1105-3 S | 128 | 64 | 256 | ≤2 |
| Subpopulation | MIC (µg/mL) | Checkerboard (interpretation) | |||||
|---|---|---|---|---|---|---|---|
| MER | POL | RIF * | MER/POL | MER/RIF | POL/RIF | MER/POL/RIF | |
| 798-1S | 32 (R) | 2 (S) | 128 | 4/0.5 (SE) | 2/8 (SE) | 1/64 (NI) | 2/0.25/4(SE) |
| 798-2S | 32 (R) | 1 (S) | 128 | 8/0.5 (NI) | 0.5/64 (NI) | 1/1 (NI) | 1/0.25/4 (SE) |
| 798-3S | 16 (R) | 1 (S) | 128 | 4/0.5 (NI) | 1/64 (NI) | 0.5/64 (NI) | 1/0.25/4 (SE) |
| 798-4S | 16 (R) | 1 (S) | 128 | 1/1 (NI) | 2/64 (NI) | 0.5/64 (NI) | 1/0.25/4 (SE) |
| E. hormaechei subsp. oharae Strains | Description |
|---|---|
| 1 (245) | Sink isolate |
| 2 (7) | Rectal swab isolate |
| 3 (67) | Rectal swab isolate |
| 4 (104) | Rectal swab isolate |
| 5 (798) | Urine isolate |
| 6 (821) | Cerebrospinal fluid isolate |
| 9 (977) | Rectal swab isolate |
| 10 (997) | Rectal swab isolate |
| 11 (1105) | Rectal swab isolate |
| Gene | Encoding Protein | Primer Sequence (5’ to 3’) | Size (bp) |
|---|---|---|---|
| csgA | Major fimbrial subunit | Forward: caacctgatgcacagtcacc | 214 |
| Reverse: tggacagggatctgatgaca | |||
| csgB | Minor subunit | Forward: agccatttgcgactgtctct | 233 |
| Reverse: tgtccgttatttcccaggag | |||
| csgD | Transcriptional regulator of the csgBAC operon | Forward: ccttccttacaagcgacagc | 236 |
| Reverse: tcgcggaaaggatactcatc | |||
| fimA | Major fimbrial subunit | Forward: tgctgtcgaggatctcaatg | 229 |
| Reverse: acggttaatctcggccagta | |||
| fimH | Fimbrial adhesion | Forward: ccccgtccagatagtcgtta | 210 |
| Reverse: acgacctgacggacaaattc | |||
| papC | Fimbrial usher | Forward: ccctgaagaccgatgacaat | 148 |
| Reverse: cggaacggaggtttgataga | |||
| papD | Fimbrial chaperone | Forward: tggatggaagacgagaaagg | 134 |
| Reverse: catccagtacagcgtctcg | |||
| mrkB | Fimbrial chaperone | Forward: ggtggctgaatctgctggaaatt | 514 |
| Reverse: atcacggttttactgttcagggcttt | |||
| Reverse: attggcataagtcgcaatcc |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brust, F.R.; Boff, L.; da Silva Trentin, D.; Pedrotti Rozales, F.; Barth, A.L.; Macedo, A.J. Macrocolony of NDM-1 Producing Enterobacter hormaechei subsp. oharae Generates Subpopulations with Different Features Regarding the Response of Antimicrobial Agents and Biofilm Formation. Pathogens 2019, 8, 49. https://doi.org/10.3390/pathogens8020049
Brust FR, Boff L, da Silva Trentin D, Pedrotti Rozales F, Barth AL, Macedo AJ. Macrocolony of NDM-1 Producing Enterobacter hormaechei subsp. oharae Generates Subpopulations with Different Features Regarding the Response of Antimicrobial Agents and Biofilm Formation. Pathogens. 2019; 8(2):49. https://doi.org/10.3390/pathogens8020049
Chicago/Turabian StyleBrust, Flávia Roberta, Luana Boff, Danielle da Silva Trentin, Franciele Pedrotti Rozales, Afonso Luís Barth, and Alexandre José Macedo. 2019. "Macrocolony of NDM-1 Producing Enterobacter hormaechei subsp. oharae Generates Subpopulations with Different Features Regarding the Response of Antimicrobial Agents and Biofilm Formation" Pathogens 8, no. 2: 49. https://doi.org/10.3390/pathogens8020049
APA StyleBrust, F. R., Boff, L., da Silva Trentin, D., Pedrotti Rozales, F., Barth, A. L., & Macedo, A. J. (2019). Macrocolony of NDM-1 Producing Enterobacter hormaechei subsp. oharae Generates Subpopulations with Different Features Regarding the Response of Antimicrobial Agents and Biofilm Formation. Pathogens, 8(2), 49. https://doi.org/10.3390/pathogens8020049




