Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation to Elucidate the Molecular Targets and Potential Mechanism of Phoenix dactylifera (Ajwa Dates) against Candidiasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identifying the Potential Targets of Compounds and Diseases
2.2. Finding and Acquiring Potential Targets
2.3. Construction and Analysis of Protein–Protein Interaction Network
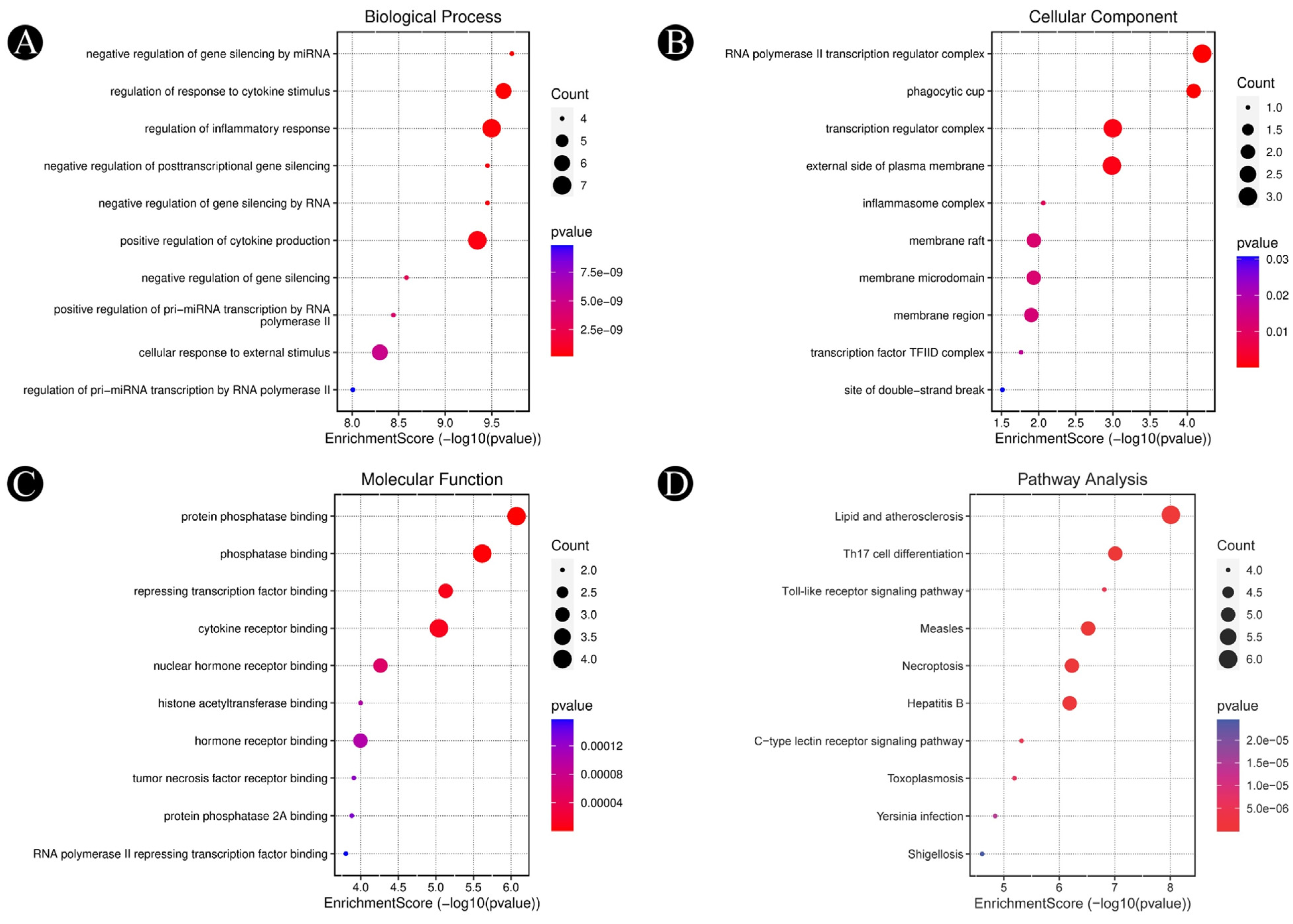
2.4. Findings of Hub-Genes and GO-KEGG Pathway Enrichment Analysis
2.5. Molecular Docking Analysis
2.6. Molecular Dynamics Simulation
2.7. Binding Free Energy Calculations
Where, ∆GMM = ∆GColoumb (electrostatic interaction) + ∆Gvdw and
∆Gsol = ∆Gpolar + ∆Gnonpolar
3. Results
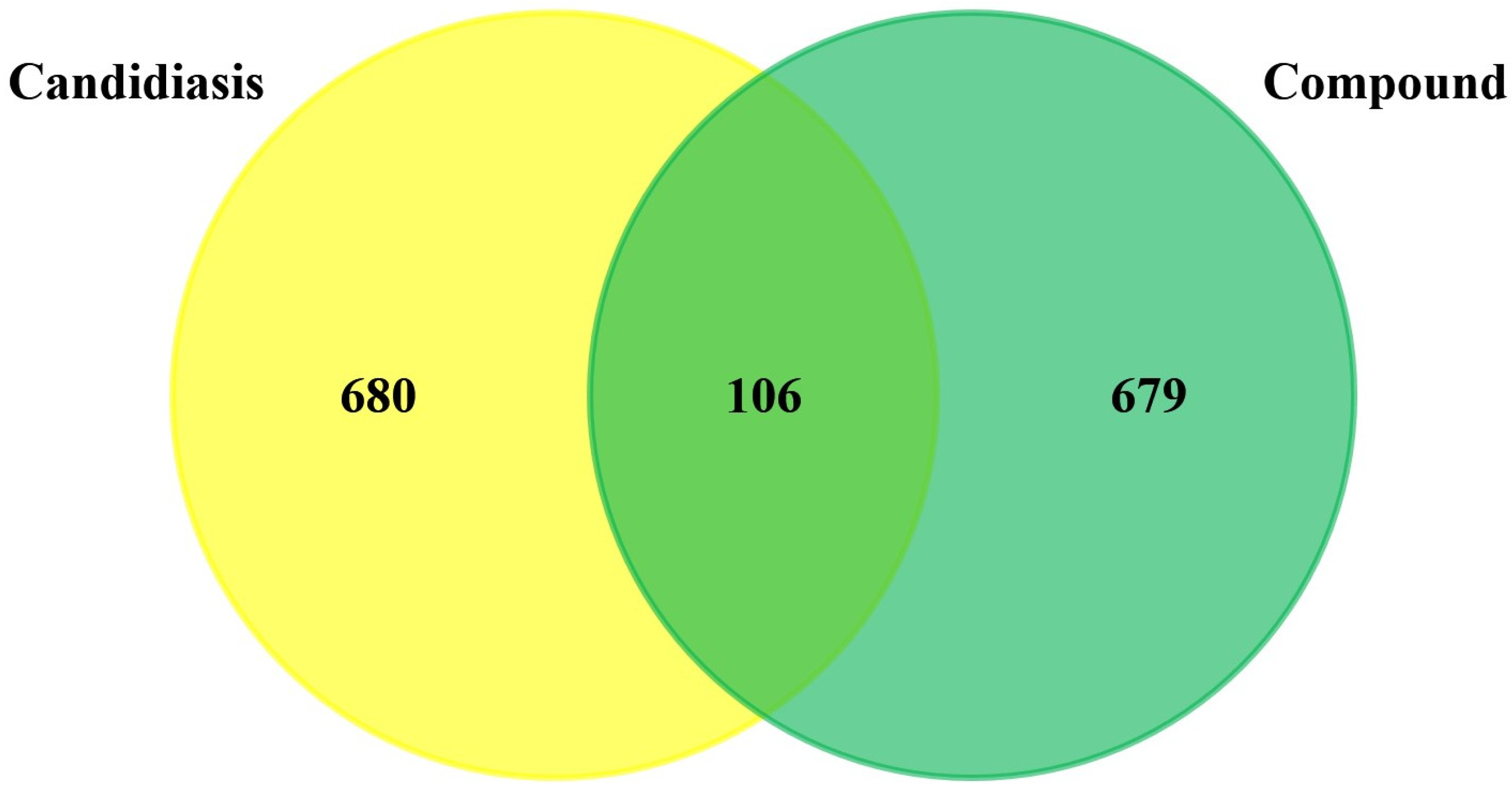
3.1. Prediction and Screening of Compound-Diseases Targets
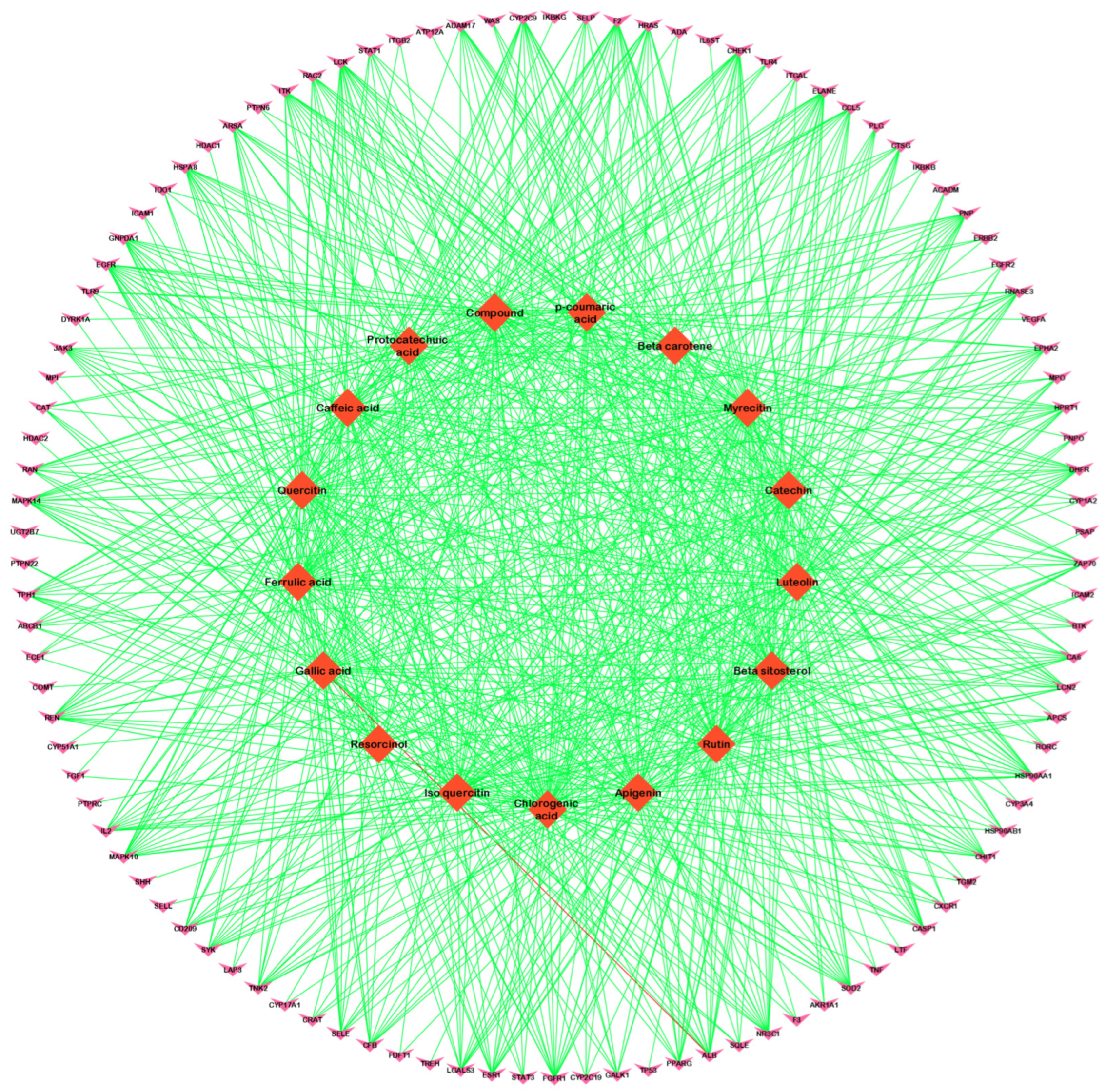
3.2. Compound–Disease Common Target Network Construction and Analysis
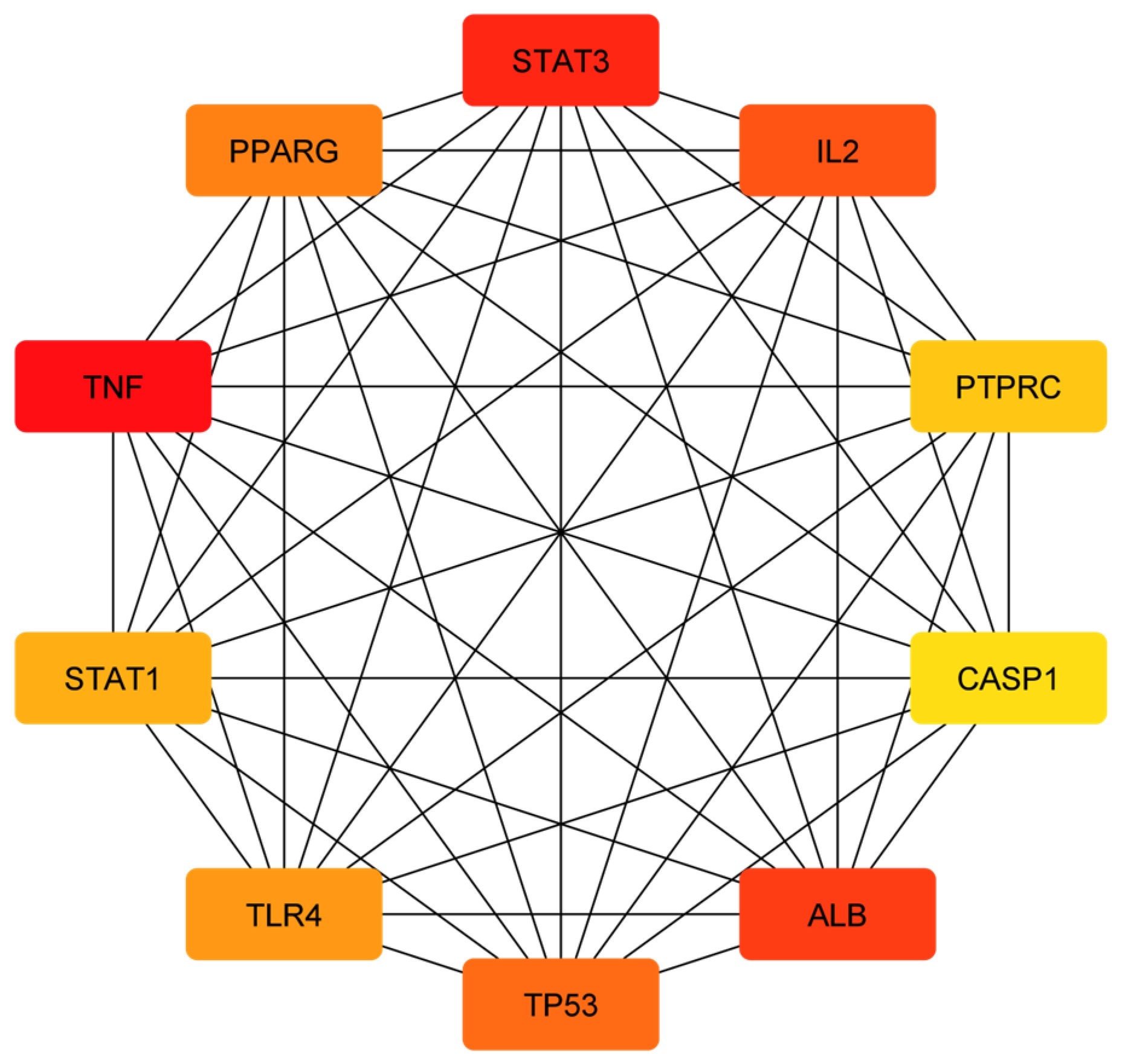
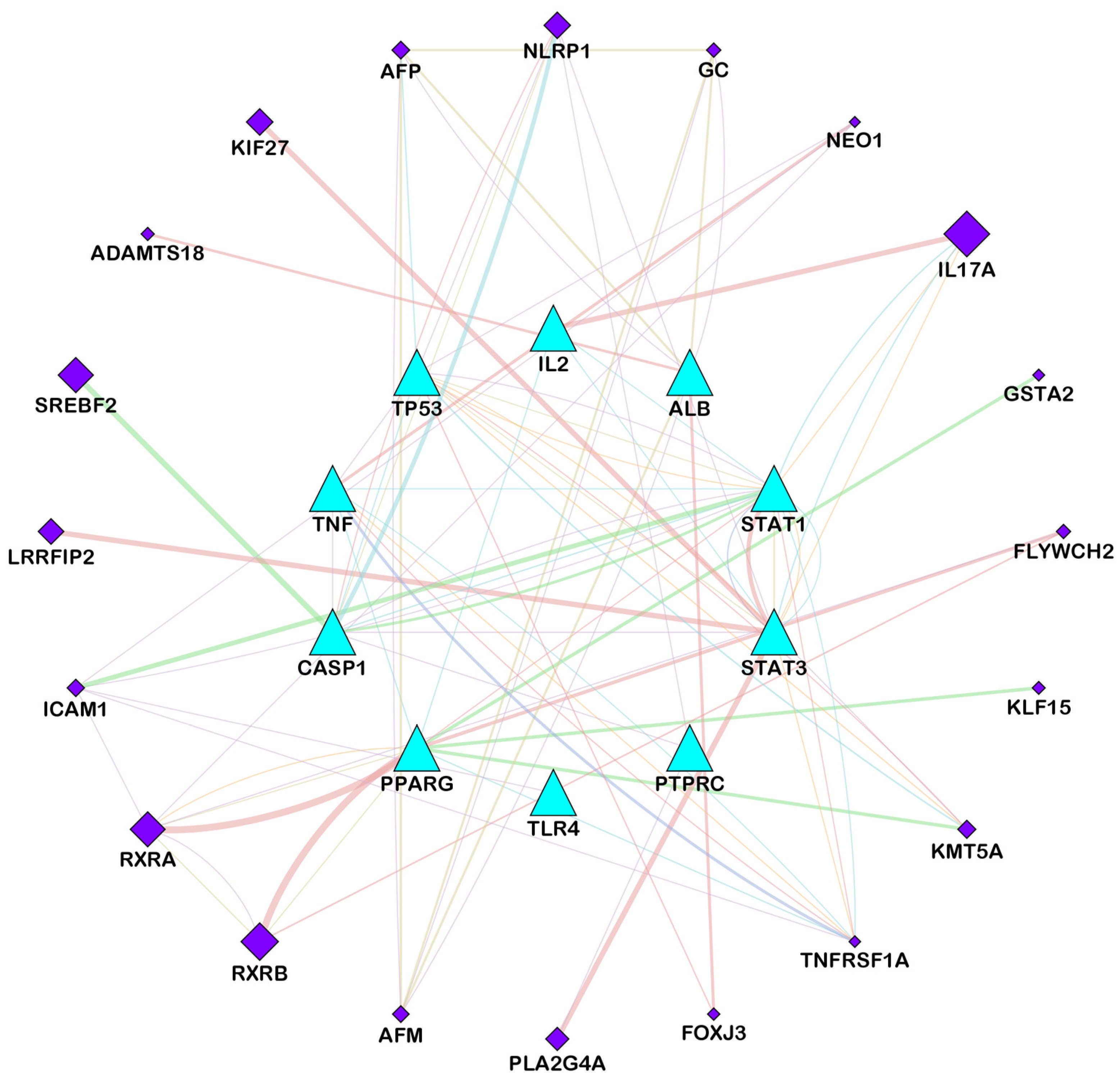
3.3. Analysis of Functional and Pathway Enrichment
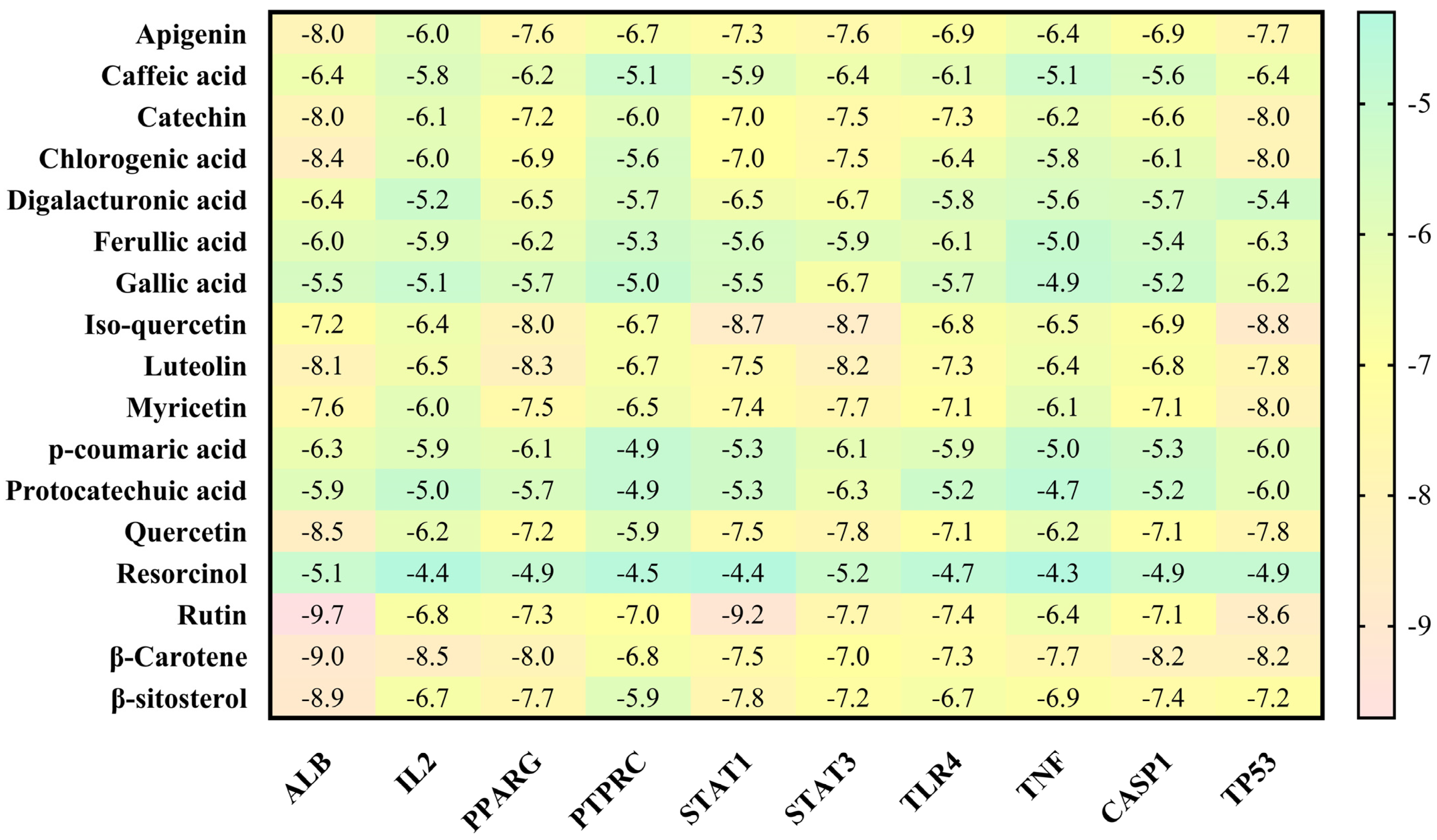
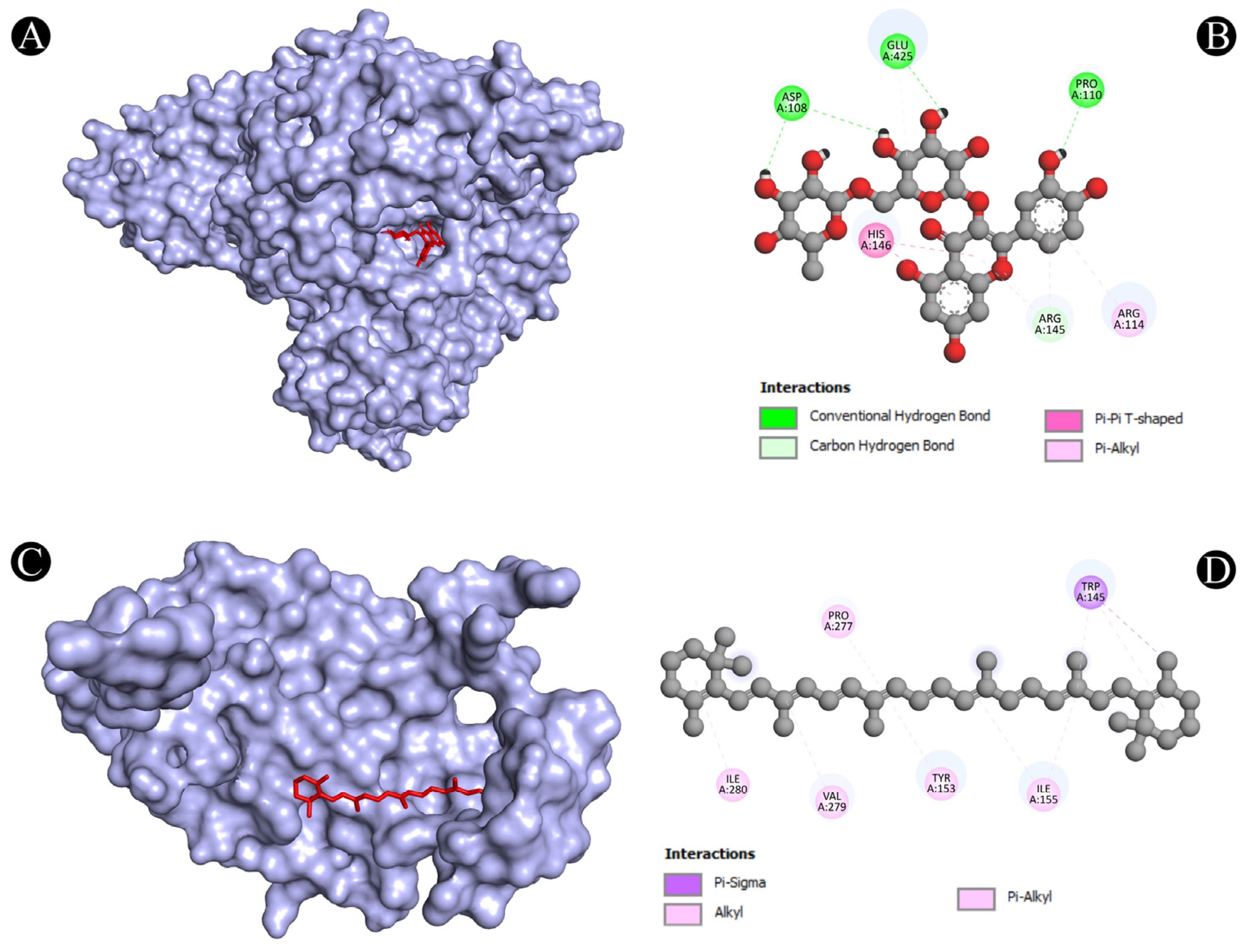
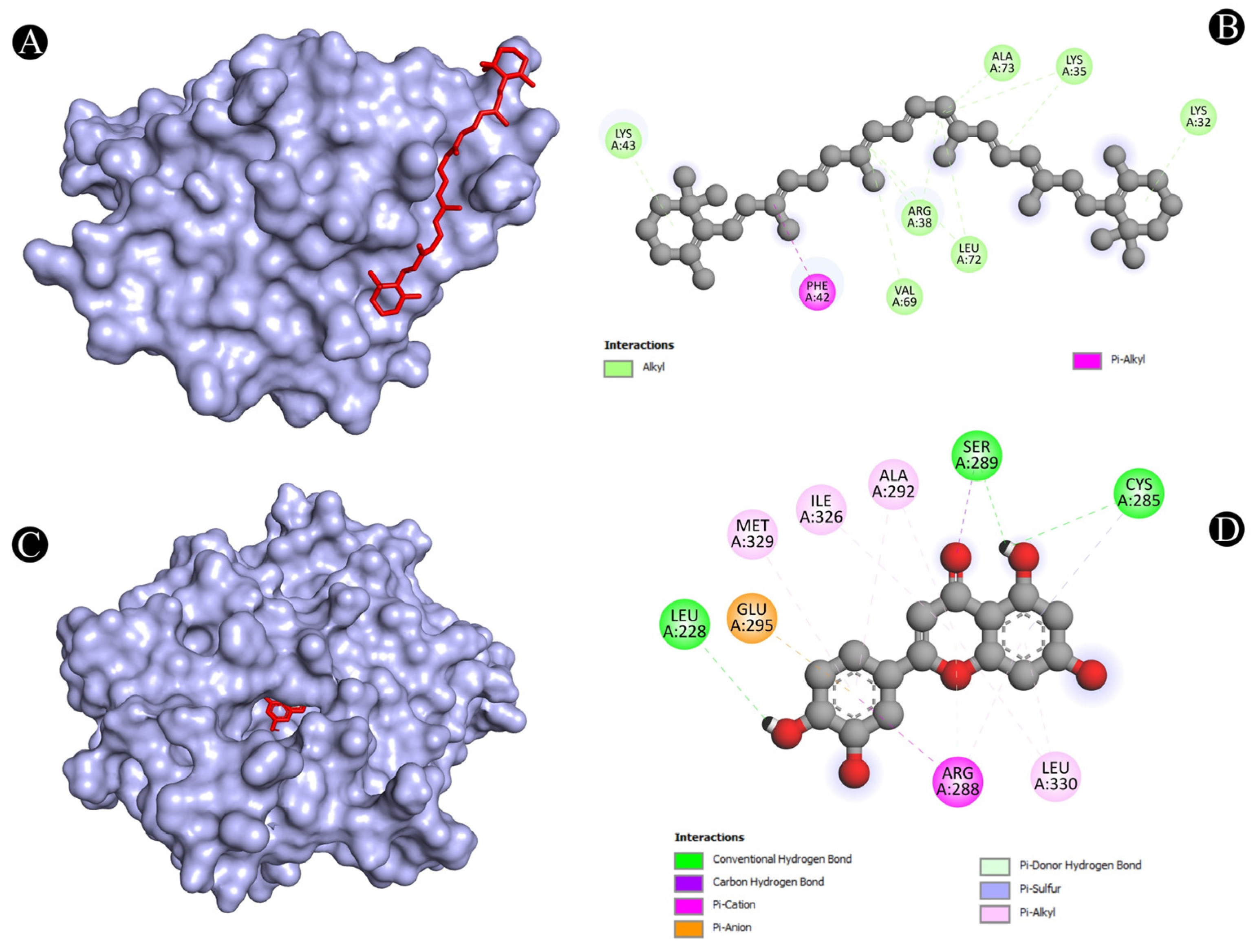
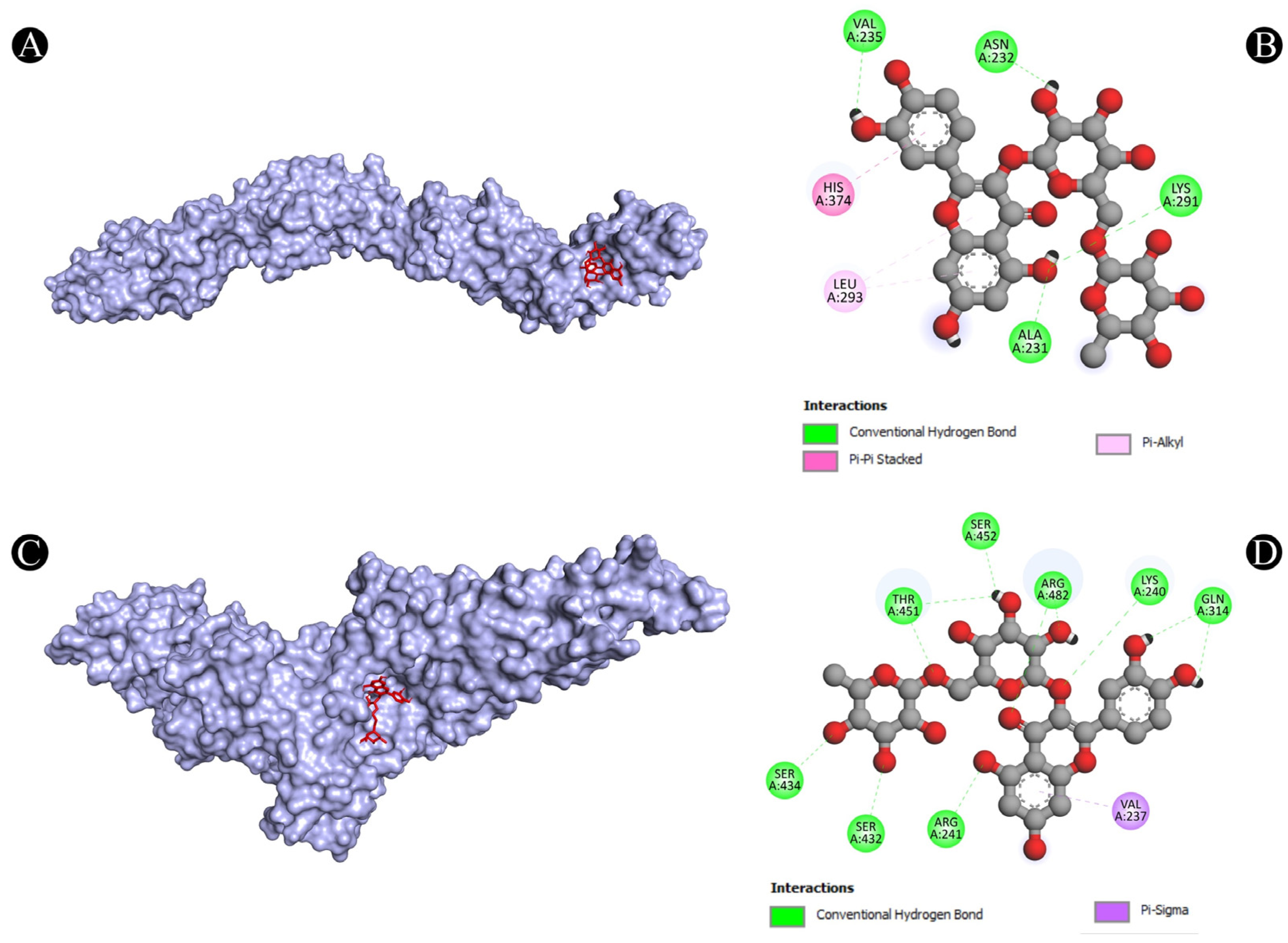
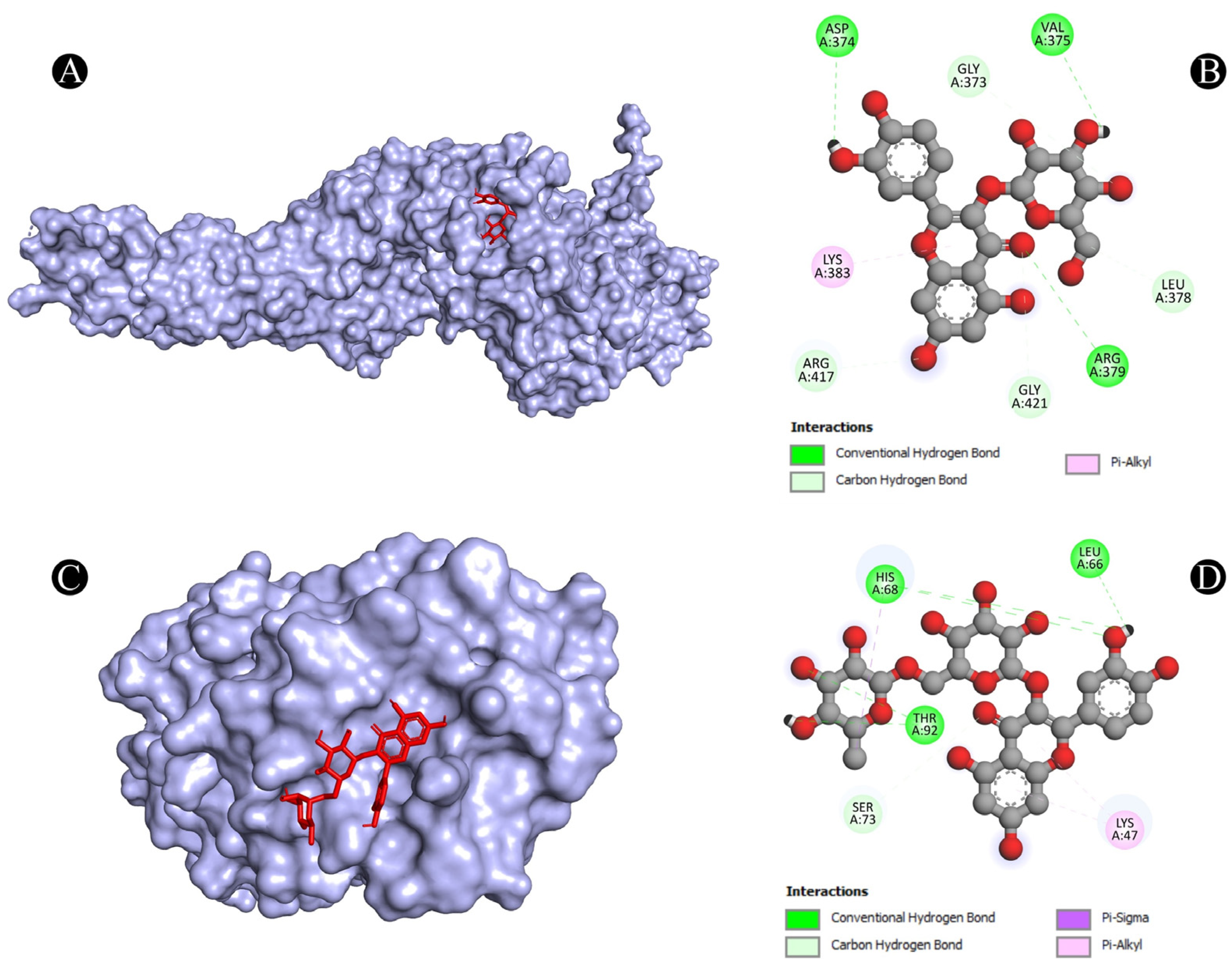
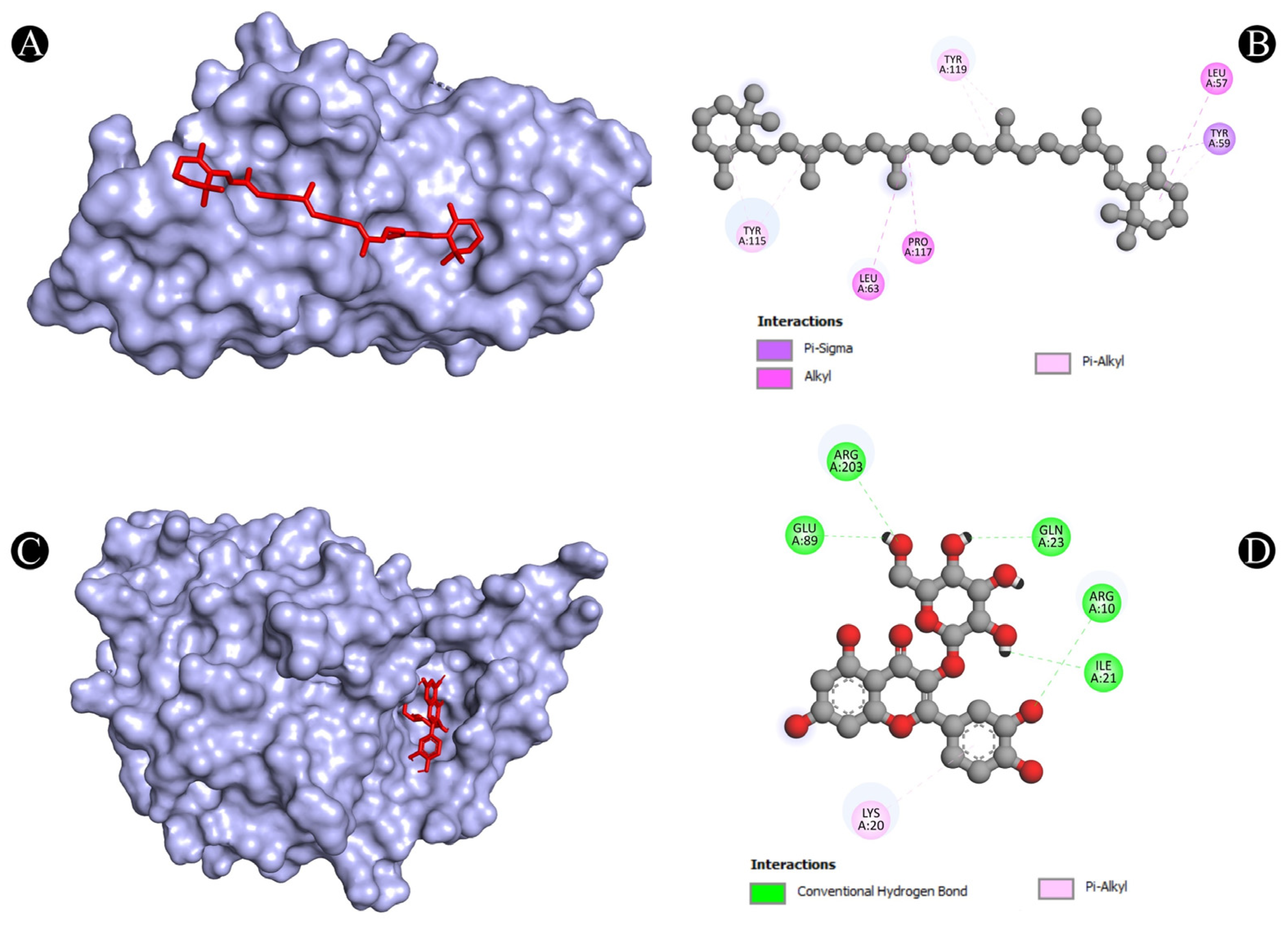
3.4. Molecular Docking Analysis
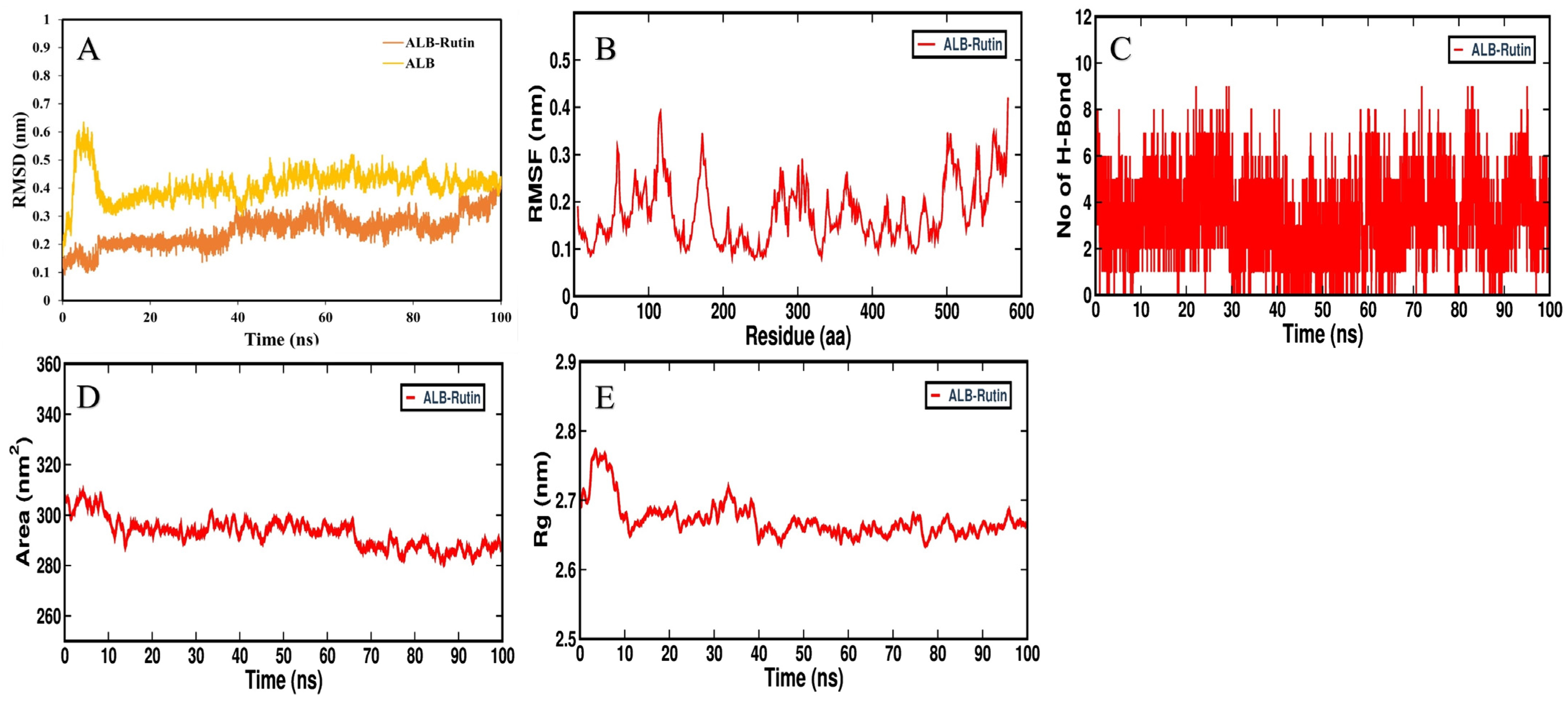
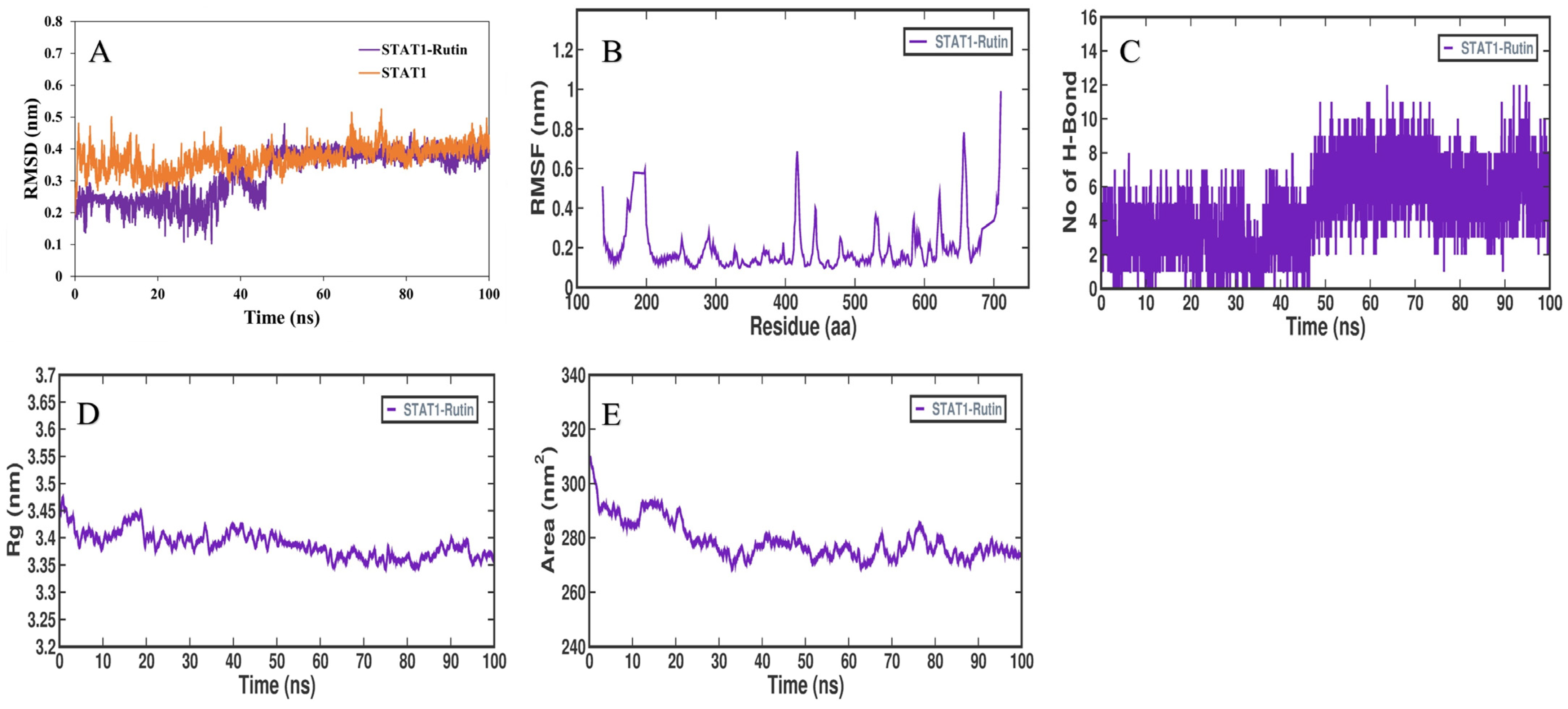
3.5. MD Simulation Analysis
3.6. MMPBSA Binding Free Energy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Friedman, D.Z.; Schwartz, I.S. Emerging fungal infections: New patients, new patterns, and new pathogens. J. Fungi 2019, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Saini, S. Candidiasis: Past, present and future. Int. J. Infect. Trop. Dis. 2015, 2, 12–24. [Google Scholar]
- Sharma, M.; Chakrabarti, A. Candidiasis and Other Emerging Yeasts. Curr. Fungal Infect. Rep. 2023, 17, 15–24. [Google Scholar] [CrossRef]
- Lopes, J.P.; Lionakis, M.S. Pathogenesis and virulence of Candida albicans. Virulence 2022, 13, 89–121. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Sonnenborn, U. Yeasts in the gut: From commensals to infectious agents. Dtsch. Ärzteblatt Int. 2009, 106, 837. [Google Scholar]
- Vila, T.; Sultan, A.S.; Montelongo-Jauregui, D.; Jabra-Rizk, M.A. Oral candidiasis: A disease of opportunity. J. Fungi 2020, 6, 15. [Google Scholar] [CrossRef]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-S.; Lee, S.S.-J.; Chen, W.-C.; Tseng, C.-H.; Lee, N.-Y.; Chen, P.-L.; Li, M.-C.; Syue, L.-S.; Lo, C.-L.; Ko, W.-C. COVID-19-associated candidiasis and the emerging concern of Candida auris infections. J. Microbiol. Immunol. Infect. 2022, 56, 672–679. [Google Scholar] [CrossRef]
- Jain, A.; Jain, S.; Rawat, S. Emerging fungal infections among children: A review on its clinical manifestations, diagnosis, and prevention. J. Pharm. Bioallied Sci. 2010, 2, 314. [Google Scholar] [CrossRef]
- Samaranayake, L.P.; Cheung, L.K.; Samaranayake, Y.H. Candidiasis and other fungal diseases of the mouth. Dermatol. Ther. 2002, 15, 251–269. [Google Scholar] [CrossRef]
- Strollo, S.; Lionakis, M.S.; Adjemian, J.; Steiner, C.A.; Prevots, D.R. Epidemiology of hospitalizations associated with invasive candidiasis, United States, 2002–2012. Emerg. Infect. Dis. 2017, 23, 7. [Google Scholar] [CrossRef]
- Latha, G.S.; Veeresh Babu, D.V.; Thejraj, H.K. Clinical profile of Candidiasis in neonates. Int. J. Contemp. Pediatr. 2017, 4, 1875–1880. [Google Scholar]
- Gaitán-Cepeda, L.A.; Sánchez-Vargas, O.; Castillo, N. Prevalence of oral candidiasis in HIV/AIDS children in highly active antiretroviral therapy era. A literature analysis. Int. J. STD AIDS 2015, 26, 625–632. [Google Scholar] [CrossRef]
- Grossi, P.A. Clinical aspects of invasive candidiasis in solid organ transplant recipients. Drugs 2009, 69, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Ramla, S.; Sharma, V.; Patel, M. Influence of cancer treatment on the Candida albicans isolated from the oral cavities of cancer patients. Support. Care Cancer 2016, 24, 2429–2436. [Google Scholar] [CrossRef]
- Richardson, M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005, 56, i5–i11. [Google Scholar] [CrossRef] [PubMed]
- Kordalewska, M.; Perlin, D.S. Identification of drug resistant Candida auris. Front. Microbiol. 2019, 10, 1918. [Google Scholar] [CrossRef]
- Sarma, S.; Upadhyay, S. Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect. Drug Resist. 2017, 10, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A. Anticandidal Agents; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- von Lilienfeld-Toal, M.; Wagener, J.; Einsele, H.; Cornely, O.A.; Kurzai, O. Invasive fungal infection: New treatments to meet new challenges. Dtsch. Ärzteblatt Int. 2019, 116, 271. [Google Scholar]
- Awadelkareem, A.M.; Siddiqui, A.J.; Noumi, E.; Ashraf, S.A.; Hadi, S.; Snoussi, M.; Badraoui, R.; Bardakci, F.; Ashraf, M.S.; Danciu, C. Biosynthesized Silver Nanoparticles Derived from Probiotic Lactobacillus rhamnosus (AgNPs-LR) Targeting Biofilm Formation and Quorum Sensing-Mediated Virulence Factors. Antibiotics 2023, 12, 986. [Google Scholar] [CrossRef]
- Bazaid, A.S.; Alsolami, A.; Patel, M.; Khateb, A.M.; Aldarhami, A.; Snoussi, M.; Almusheet, S.M.; Qanash, H. Antibiofilm, Antimicrobial, Anti-Quorum Sensing, and Antioxidant Activities of Saudi Sidr Honey: In Vitro and Molecular Docking Studies. Pharmaceutics 2023, 15, 2177. [Google Scholar] [CrossRef] [PubMed]
- Ranjutha, V.; Chen, Y.; Al-Keridis, L.A.; Patel, M.; Alshammari, N.; Adnan, M.; Sahreen, S.; Gopinath, S.C.; Sasidharan, S. Synergistic Antimicrobial Activity of Ceftriaxone and Polyalthia longifolia Methanol (MEPL) Leaf Extract against Methicillin-Resistant Staphylococcus aureus and Modulation of mecA Gene Presence. Antibiotics 2023, 12, 477. [Google Scholar] [CrossRef]
- Al-Alawi, R.A.; Al-Mashiqri, J.H.; Al-Nadabi, J.S.; Al-Shihi, B.I.; Baqi, Y. Date palm tree (Phoenix dactylifera L.): Natural products and therapeutic options. Front. Plant Sci. 2017, 8, 845. [Google Scholar] [CrossRef]
- Younas, A.; Naqvi, S.A.; Khan, M.R.; Shabbir, M.A.; Jatoi, M.A.; Anwar, F.; Inam-Ur-Raheem, M.; Saari, N.; Aadil, R.M. Functional food and nutra-pharmaceutical perspectives of date (Phoenix dactylifera L.) fruit. J. Food Biochem. 2020, 44, e13332. [Google Scholar] [CrossRef]
- Abdullah, N.; Ishak, N.M.; Shahida, W.W. In-vitro antibacterial activities of Ajwa dates fruit (Phoenix dactylifera L.) extract against selected gram-negative bacteria causing gastroenteritis. Int. J. Pharm. Sci. Res. 2019, 10, 2951–2955. [Google Scholar]
- Al-Shwyeh, H.A. Date palm (Phoenix dactylifera L.) fruit as potential antioxidant and antimicrobial agents. J. Pharm. Bioallied Sci. 2019, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Al-Tamimi, A.; Alfarhan, A.; Rajagopal, R. Antimicrobial and anti-biofilm activities of polyphenols extracted from different Saudi Arabian date cultivars against human pathogens. J. Infect. Public Health 2021, 14, 1783–1787. [Google Scholar] [CrossRef]
- Bokhari, N.A.; Perveen, K. In vitro inhibition potential of Phoenix dactylifera L. extracts on the growth of pathogenic fungi. J. Med. Plants Res. 2012, 6, 1083–1088. [Google Scholar]
- Boulenouar, N.; Marouf, A.; Cheriti, A. Antifungal activity and phytochemical screening of extracts from Phoenix dactylifera L. cultivars. Nat. Prod. Res. 2011, 25, 1999–2002. [Google Scholar] [CrossRef]
- Elasbali, A.M.; Al-Soud, W.A.; Mousa Elayyan, A.E.; Al-Oanzi, Z.H.; Alhassan, H.H.; Mohamed, B.M.; Alanazi, H.H.; Ashraf, M.S.; Moiz, S.; Patel, M. Integrating network pharmacology approaches for the investigation of multi-target pharmacological mechanism of 6-shogaol against cervical cancer. J. Biomol. Struct. Dyn. 2023, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, B.; Chen, S.; Lin, M.; Chen, Y.; Jin, S.; Chen, W.; Zhang, Y. Applications of network pharmacology in traditional Chinese medicine research. Evid.-Based Complement. Altern. Med. 2020, 2020, 1646905. [Google Scholar] [CrossRef] [PubMed]
- Baothman, O.A.; Altayb, H.N.; Zeyadi, M.A.; Hosawi, S.B.; Abo-Golayel, M.K. Phytochemical analysis and nephroprotective potential of Ajwa dates in doxorubicin-induced nephrotoxicity rats: Biochemical and molecular docking approaches. Food Sci. Nutr. 2023, 11, 1584–1598. [Google Scholar] [CrossRef]
- Khalid, S.; Khalid, N.; Khan, R.S.; Ahmed, H.; Ahmad, A. A review on chemistry and pharmacology of Ajwa dates fruit and pit. Trends Food Sci. Technol. 2017, 63, 60–69. [Google Scholar] [CrossRef]
- Qadir, A.; Singh, S.P.; Akhtar, J.; Ali, A.; Arif, M. Phytochemical and GC-MS analysis of Saudi Arabian Ajwa variety of date seed oil and extracts obtained by the slow pyrolysis method. Orient. Pharm. Exp. Med. 2017, 17, 81–87. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, gkw943. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using autodock for ligand-receptor docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar] [CrossRef]
- Studio, D. Discovery studio. Accelrys [2.1]. 2008. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 15 February 2023).
- Adnan, M.; Anwar, S.; DasGupta, D.; Patel, M.; Elasbali, A.M.; Alhassan, H.H.; Shafie, A.; Siddiqui, A.J.; Bardakci, F.; Snoussi, M. Targeting inhibition of microtubule affinity regulating kinase 4 by Harmaline: Strategy to combat Alzheimer’s disease. Int. J. Biol. Macromol. 2023, 224, 188–195. [Google Scholar] [CrossRef]
- Adnan, M.; Shamsi, A.; Elasbali, A.M.; Siddiqui, A.J.; Patel, M.; Alshammari, N.; Alharethi, S.H.; Alhassan, H.H.; Bardakci, F.; Hassan, M.I. Structure-guided approach to discover tuberosin as a potent activator of pyruvate kinase M2, targeting cancer therapy. Int. J. Mol. Sci. 2022, 23, 13172. [Google Scholar] [CrossRef]
- Bahaman, A.H.; Wahab, R.A.; Abdul Hamid, A.A.; Abd Halim, K.B.; Kaya, Y. Molecular docking and molecular dynamics simulations studies on β-glucosidase and xylanase Trichoderma asperellum to predict degradation order of cellulosic components in oil palm leaves for nanocellulose preparation. J. Biomol. Struct. Dyn. 2021, 39, 2628–2641. [Google Scholar] [CrossRef] [PubMed]
- Bouali, N.; Ahmad, I.; Patel, H.; Alhejaili, E.B.; Hamadou, W.S.; Badraoui, R.; Hadj Lajimi, R.; Alreshidi, M.; Siddiqui, A.J.; Adnan, M. GC–MS screening of the phytochemical composition of Ziziphus honey: ADME properties and in vitro/in silico study of its antimicrobial activity. J. Biomol. Struct. Dyn. 2023, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Páll, S.; Abraham, M.J.; Kutzner, C.; Hess, B.; Lindahl, E. Tackling exascale software challenges in molecular dynamics simulations with GROMACS. In Proceedings of the Solving Software Challenges for Exascale: International Conference on Exascale Applications and Software, EASC 2014, Stockholm, Sweden, 2–3 April 2014; pp. 3–27. [Google Scholar]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Altaf, M.M.; Khan, M.S.A.; Ahmad, I. Diversity of bioactive compounds and their therapeutic potential. In New Look to Phytomedicine; Elsevier: Amsterdam, The Netherlands, 2019; pp. 15–34. [Google Scholar]
- Cordier, C.; Morton, D.; Murrison, S.; Nelson, A.; O’Leary-Steele, C. Natural products as an inspiration in the diversity-oriented synthesis of bioactive compound libraries. Nat. Prod. Rep. 2008, 25, 719–737. [Google Scholar] [CrossRef]
- Bu, Q.-R.; Bao, M.-Y.; Yang, Y.; Wang, T.-M.; Wang, C.-Z. Targeting Virulence Factors of Candida albicans with Natural Products. Foods 2022, 11, 2951. [Google Scholar] [CrossRef]
- Coleman, J.J.; Okoli, I.; Tegos, G.P.; Holson, E.B.; Wagner, F.F.; Hamblin, M.R.; Mylonakis, E. Characterization of plant-derived saponin natural products against Candida albicans. ACS Chem. Biol. 2010, 5, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Zida, A.; Bamba, S.; Yacouba, A.; Ouedraogo-Traore, R.; Guiguemdé, R. Anti-Candida albicans natural products, sources of new antifungal drugs: A review. J. Mycol. Medicale 2017, 27, 1–19. [Google Scholar]
- Gharibpour, F.; Shirban, F.; Bagherniya, M.; Nosouhian, M.; Sathyapalan, T.; Sahebkar, A. The effects of nutraceuticals and herbal medicine on candida albicans in oral candidiasis: A comprehensive review. Adv. Exp. Med. Biol. 2021, 1308, 225–248. [Google Scholar] [PubMed]
- Soliman, S.; Alnajdy, D.; El-Keblawy, A.A.; Mosa, K.A.; Khoder, G.; Noreddin, A.M. Plants’ natural products as alternative promising anti-Candida drugs. Pharmacogn. Rev. 2017, 11, 104. [Google Scholar]
- Helmstädter, A.; Staiger, C. Traditional use of medicinal agents: A valid source of evidence. Drug Discov. Today 2014, 19, 4–7. [Google Scholar] [CrossRef]
- Augostine, C.R.; Avery, S.V. Discovery of natural products with antifungal potential through combinatorial synergy. Front. Microbiol. 2022, 13, 1261. [Google Scholar]
- Heard, S.C.; Wu, G.; Winter, J.M. Antifungal natural products. Curr. Opin. Biotechnol. 2021, 69, 232–241. [Google Scholar] [CrossRef]
- Jacob, M.R.; Walker, L.A. Natural products and antifungal drug discovery. Methods Mol. Med. 2005, 118, 83–109. [Google Scholar] [PubMed]
- Hussain, M.I.; Semreen, M.H.; Shanableh, A.; Khattak, M.N.K.; Saadoun, I.; Ahmady, I.M.; Mousa, M.; Darwish, N.; Radeef, W.; Soliman, S.S. Phenolic composition and antimicrobial activity of different Emirati Date (Phoenix dactylifera L.) pits: A comparative study. Plants 2019, 8, 497. [Google Scholar]
- Selim, S.; Abdel-Mawgoud, M.; Al-Sharary, T.; Almuhayawi, M.S.; Alruhaili, M.H.; Al Jaouni, S.K.; Warrad, M.; Mohamed, H.S.; Akhtar, N.; AbdElgawad, H. Pits of date palm: Bioactive composition, antibacterial activity and antimutagenicity potentials. Agronomy 2021, 12, 54. [Google Scholar]
- Anwar, S.; Raut, R.; Alsahli, M.A.; Almatroudi, A.; Alfheeaid, H.; Alzahrani, F.M.; Khan, A.A.; Allemailem, K.S.; Almatroodi, S.A.; Rahmani, A.H. Role of Ajwa dates fruit pulp and seed in the management of diseases through in vitro and in silico analysis. Biology 2022, 11, 78. [Google Scholar]
- Smeekens, S.P.; Ng, A.; Kumar, V.; Johnson, M.D.; Plantinga, T.S.; Van Diemen, C.; Arts, P.; Verwiel, E.T.; Gresnigt, M.S.; Fransen, K. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat. Commun. 2013, 4, 1342. [Google Scholar]
- Steen, H.C.; Gamero, A.M. STAT2 phosphorylation and signaling. Jak-Stat 2013, 2, e25790. [Google Scholar] [PubMed]
- Djeu, J.; Liu, J.H.; Wei, S.; Rui, H.; Pearson, C.; Leonard, W.; Blanchard, D. Function associated with IL-2 receptor-beta on human neutrophils. Mechanism of activation of antifungal activity against Candida albicans by IL-2. J. Immunol. (Baltim. Md. 1950) 1993, 150, 960–970. [Google Scholar]
- Li, M.; Yu, Y. Innate immune receptor clustering and its role in immune regulation. J. Cell Sci. 2021, 134, jcs249318. [Google Scholar] [PubMed]
- Miryala, S.K.; Anbarasu, A.; Ramaiah, S. Organ-specific host differential gene expression analysis in systemic candidiasis: A systems biology approach. Microb. Pathog. 2022, 169, 105677. [Google Scholar] [PubMed]
- Okada, S.; Asano, T.; Moriya, K.; Boisson-Dupuis, S.; Kobayashi, M.; Casanova, J.-L.; Puel, A. Human STAT1 gain-of-function heterozygous mutations: Chronic mucocutaneous candidiasis and type I interferonopathy. J. Clin. Immunol. 2020, 40, 1065–1081. [Google Scholar] [PubMed]
- Man, S.M.; Karki, R.; Kanneganti, T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017, 277, 61–75. [Google Scholar] [PubMed]
- Wilde, B.; Katsounas, A. Immune dysfunction and albumin-related immunity in liver cirrhosis. Mediat. Inflamm. 2019, 2019, 7537649. [Google Scholar] [CrossRef]
- Levine, A.J. P53 and the immune response: 40 years of exploration—A plan for the future. Int. J. Mol. Sci. 2020, 21, 541. [Google Scholar] [PubMed]
- Blasi, E.; Mucci, A.; Neglia, R.; Pezzini, F.; Colombari, B.; Radzioch, D.; Cossarizza, A.; Lugli, E.; Volpini, G.; Giudice, G.D. Biological importance of the two Toll-like receptors, TLR2 and TLR4, in macrophage response to infection with Candida albicans. FEMS Immunol. Med. Microbiol. 2005, 44, 69–79. [Google Scholar] [PubMed]
- Ohta, H.; Tanimoto, T.; Taniai, M.; Taniguchi, M.; Ariyasu, T.; Arai, S.; Ohta, T.; Fukuda, S. Regulation of Candida albicans morphogenesis by tumor necrosis factor-alpha and potential for treatment of oral candidiasis. In Vivo 2007, 21, 25–32. [Google Scholar] [PubMed]
- Galès, A.; Conduché, A.; Bernad, J.; Lefevre, L.; Olagnier, D.; Béraud, M.; Martin-Blondel, G.; Linas, M.-D.; Auwerx, J.; Coste, A. PPARγ controls Dectin-1 expression required for host antifungal defense against Candida albicans. PLoS Pathog. 2010, 6, e1000714. [Google Scholar] [CrossRef]
- Hernández-Santos, N.; Gaffen, S.L. Th17 cells in immunity to Candida albicans. Cell Host Microbe 2012, 11, 425–435. [Google Scholar]
- Tasaki, S.; Cho, T.; Nagao, J.-i.; Ikezaki, S.; Narita, Y.; Arita-Morioka, K.-i.; Yasumatsu, K.; Toyoda, K.; Kojima, H.; Tanaka, Y. Th17 cells differentiated with mycelial membranes of Candida albicans prevent oral candidiasis. FEMS Yeast Res. 2018, 18, foy018. [Google Scholar]
- Gil, M.L.; Gozalbo, D. Role of Toll-like receptors in systemic Candida albicans infections. Front. Biosci. 2009, 14, 570–582. [Google Scholar] [CrossRef]
- Calich, V.L.; Pina, A.; Felonato, M.; Bernardino, S.; Costa, T.A.; Loures, F.V. Toll-like receptors and fungal infections: The role of TLR2, TLR4 and MyD88 in paracoccidioidomycosis. FEMS Immunol. Med. Microbiol. 2008, 53, 1–7. [Google Scholar]
- Domínguez-Andrés, J.; Arts, R.J.; Ter Horst, R.; Gresnigt, M.S.; Smeekens, S.P.; Ratter, J.M.; Lachmandas, E.; Boutens, L.; van de Veerdonk, F.L.; Joosten, L.A. Rewiring monocyte glucose metabolism via C-type lectin signaling protects against disseminated candidiasis. PLoS Pathog. 2017, 13, e1006632. [Google Scholar]
- Hardison, S.E.; Brown, G.D. C-type lectin receptors orchestrate antifungal immunity. Nat. Immunol. 2012, 13, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Lin, G.; Langdon, W.Y.; Tao, L.; Zhang, J. Regulation of C-type lectin receptor-mediated antifungal immunity. Front. Immunol. 2018, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Lei, L.; Wang, S.; Hu, J.; Zhang, G. Necroptosis and its role in infectious diseases. Apoptosis 2020, 25, 169–178. [Google Scholar] [CrossRef]
- Chen, Y.; Ren, W.; Wang, Q.; He, Y.; Ma, D.; Cai, Z. The regulation of necroptosis by ubiquitylation. Apoptosis 2022, 27, 668–684. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Wu, Z.; Lou, Q.; Lu, W.; Zhang, J.; Li, Q.; Zhang, Y.; Yao, Y.; Zhao, Q.; Li, M. Dectin-1-induced RIPK1 and RIPK3 activation protects host against Candida albicans infection. Cell Death Differ. 2019, 26, 2622–2636. [Google Scholar] [CrossRef]
- Xia, T.; Zhang, M.; Lei, W.; Yang, R.; Fu, S.; Fan, Z.; Yang, Y.; Zhang, T. Advances in the role of STAT3 in macrophage polarization. Front. Immunol. 2023, 14, 1160719. [Google Scholar] [CrossRef]
- Tsai, M.H.; Pai, L.M.; Lee, C.K. Fine-tuning of type I interferon response by STAT3. Front. Immunol. 2019, 10, 1448. [Google Scholar] [CrossRef]
- Piaszyk-Borychowska, A.; Széles, L.; Csermely, A.; Chiang, H.C.; Wesoły, J.; Lee, C.K.; Nagy, L.; Bluyssen, H.A. Signal integration of IFN-I and IFN-II with TLR4 involves sequential recruitment of STAT1-complexes and NFκB to enhance pro-inflammatory transcription. Front. Immunol. 2019, 10, 1253. [Google Scholar] [CrossRef]
- Wilmes, S.; Jeffrey, P.A.; Martinez-Fabregas, J.; Hafer, M.; Fyfe, P.K.; Pohler, E.; Gaggero, S.; López-García, M.; Lythe, G.; Taylor, C.; et al. Competitive binding of STATs to receptor phospho-Tyr motifs accounts for altered cytokine responses. eLife 2021, 10, e66014. [Google Scholar] [CrossRef]

| Sr. No. | Name | PubChem ID | MF | MW | Canonical SMILES | Structure |
|---|---|---|---|---|---|---|
| 1 | Apigenin | 5280443 | C15H10O5 | 270.24 | C1=CC(=CC=C1C2=CC(=O)C3=C(C=C(C=C3O2)O)O)O | 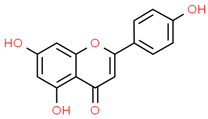 |
| 2 | Caffeic acid | 689043 | C9H8O4 | 180.16 | C1=CC(=C(C=C1C=CC(=O)O)O)O | 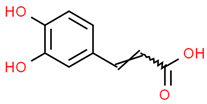 |
| 3 | Catechin | 9064 | C15H14O6 | 290.27 | C1C(C(OC2=CC(=CC(=C21)O)O)C3=CC(=C(C=C3)O)O)O |  |
| 4 | Chlorogenic acid | 1794427 | C16H18O9 | 354.31 | C1C(C(C(CC1(C(=O)O)O)OC(=O)C=CC2=CC(=C(C=C2)O)O)O)O |  |
| 5 | Digalacturonic acid | 439694 | C12H18O13 | 370.26 | C1(C(C(OC(C1O)OC2C(C(C(OC2C(=O)O)O)O)O)C(=O)O)O)O | 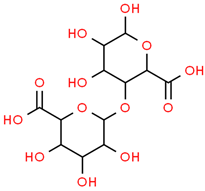 |
| 6 | Ferullic acid | 445858 | C10H10O4 | 194.18 | COC1=C(C=CC(=C1)C=CC(=O)O)O |  |
| 7 | Gallic acid | 370 | C7H6O5 | 170.12 | C1=C(C=C(C(=C1O)O)O)C(=O)O | 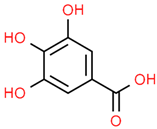 |
| 8 | Iso-quercetin | 10813969 | C21H20O12 | 464.4 | C1=CC(=C(C=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)OC4C(C(C(C(O4)CO)O)O)O)O)O | 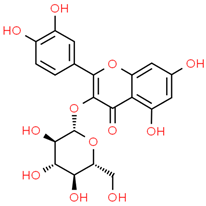 |
| 9 | Luteolin | 5280445 | C15H10O6 | 286.24 | C1=CC(=C(C=C1C2=CC(=O)C3=C(C=C(C=C3O2)O)O)O)O |  |
| 10 | Myricetin | 5281672 | C15H10O8 | 318.23 | C1=C(C=C(C(=C1O)O)O)C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O | 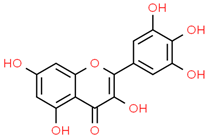 |
| 11 | p-coumaric acid | 637542 | C9H8O3 | 164.16 | C1=CC(=CC=C1C=CC(=O)O)O |  |
| 12 | Protocatechuic acid | 72 | C7H6O4 | 154.12 | C1=CC(=C(C=C1C(=O)O)O)O | 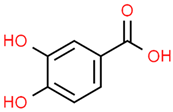 |
| 13 | Quercetin | 5280343 | C15H10O7 | 302.23 | C1=CC(=C(C=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O)O)O | 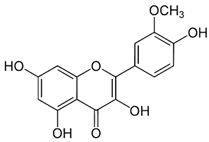 |
| 14 | Resorcinol | 5054 | C6H6O2 | 110.11 | C1=CC(=CC(=C1)O)O | 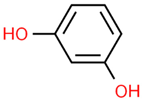 |
| 15 | Rutin | 5280805 | C27H30O16 | 610.5 | CC1C(C(C(C(O1)OCC2C(C(C(C(O2)OC3=C(OC4=CC(=CC(=C4C3=O)O)O)C5=CC(=C(C=C5)O)O)O)O)O)O)O)O | 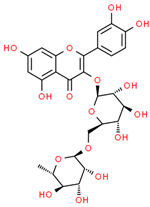 |
| 16 | β-Carotene | 5280489 | C40H56 | 536.9 | CC1=C(C(CCC1)(C)C)C=CC(=CC=CC(=CC=CC=C(C)C=CC=C(C)C=CC2=C(CCCC2(C)C)C)C)C | 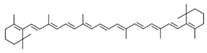 |
| 17 |  | 222284 | C29H50O | 414.7 | CCC(CCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C)C)C(C)C | 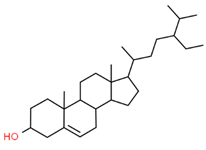 |
| Sr. No. | Genes | Degree | Betweenness | Closeness |
|---|---|---|---|---|
| 1 | TNF | 65 | 988.208 | 0.15648286 |
| 2 | ALB | 64 | 1225.0367 | 0.15695067 |
| 3 | STAT3 | 54 | 470.05515 | 0.15418503 |
| 4 | EGFR | 53 | 668.1908 | 0.15441176 |
| 5 | VEGFA | 50 | 370.19366 | 0.15328467 |
| 6 | TP53 | 50 | 373.23392 | 0.15328467 |
| 7 | TLR4 | 47 | 222.14207 | 0.15239477 |
| 8 | PTPRC | 45 | 248.6846 | 0.15086207 |
| 9 | IL2 | 45 | 268.14774 | 0.1517341 |
| 10 | STAT1 | 42 | 167.48207 | 0.15086207 |
| 11 | HRAS | 41 | 266.0002 | 0.15129682 |
| 12 | ICAM1 | 38 | 276.51984 | 0.1502146 |
| 13 | HSP90AA1 | 37 | 441.82166 | 0.15064563 |
| 14 | PPARG | 35 | 175.21269 | 0.14957266 |
| 15 | SYK | 33 | 68.06617 | 0.14767933 |
| 16 | CCL5 | 33 | 62.759113 | 0.14767933 |
| 17 | ERBB2 | 33 | 128.60736 | 0.14872521 |
| 18 | ESR1 | 32 | 262.75586 | 0.14957266 |
| 19 | MAPK14 | 32 | 76.563156 | 0.14872521 |
| 20 | MPO | 32 | 104.47263 | 0.1480959 |
| 21 | CAT | 30 | 590.64325 | 0.14872521 |
| 22 | SELL | 29 | 68.562836 | 0.14623955 |
| 23 | LCK | 28 | 69.942894 | 0.1474719 |
| 24 | CASP1 | 27 | 38.12457 | 0.1474719 |
| 25 | NR3C1 | 26 | 135.64508 | 0.1474719 |
| 26 | ITGB2 | 25 | 38.64269 | 0.1446281 |
| 27 | SELE | 24 | 31.730501 | 0.14623955 |
| 28 | SELP | 24 | 28.364697 | 0.14563107 |
| 29 | BTK | 24 | 38.74685 | 0.14502762 |
| 30 | TLR9 | 23 | 26.082947 | 0.14522822 |
| 31 | IKBKB | 23 | 16.278957 | 0.14644352 |
| 32 | HDAC1 | 23 | 75.05072 | 0.14644352 |
| 33 | PLG | 23 | 93.70508 | 0.14664805 |
| 34 | HSP90AB1 | 23 | 156.0722 | 0.1474719 |
| 35 | REN | 22 | 101.38719 | 0.14583333 |
| 36 | ZAP70 | 22 | 22.437016 | 0.14482759 |
| 37 | ABCB1 | 21 | 110.38468 | 0.14644352 |
| 38 | RAC2 | 21 | 30.51389 | 0.14383562 |
| 39 | ELANE | 20 | 15.914155 | 0.14344262 |
| 40 | LGALS3 | 20 | 203.58853 | 0.1446281 |
| 41 | CXCR1 | 20 | 11.40124 | 0.14403293 |
| 42 | ITGAL | 20 | 17.016296 | 0.14363885 |
| 43 | CYP3A4 | 19 | 90.02866 | 0.14603616 |
| 44 | F2 | 19 | 33.436974 | 0.14403293 |
| 45 | F3 | 18 | 12.540235 | 0.14403293 |
| 46 | JAK3 | 16 | 12.340351 | 0.14363885 |
| 47 | HSPA8 | 16 | 51.118656 | 0.1446281 |
| 48 | PTPN6 | 15 | 3.2728631 | 0.1420839 |
| 49 | CTSG | 15 | 10.223254 | 0.14227642 |
| 50 | LCN2 | 15 | 11.1281185 | 0.14383562 |
| 51 | SOD2 | 14 | 26.54373 | 0.1446281 |
| 52 | SHH | 14 | 19.630491 | 0.14403293 |
| 53 | ADAM17 | 13 | 3.4592445 | 0.14324693 |
| 54 | PTPN22 | 13 | 3.366081 | 0.1418919 |
| 55 | FGFR1 | 13 | 4.2498856 | 0.14383562 |
| 56 | CYP1A2 | 13 | 239.40514 | 0.14246947 |
| 57 | HDAC2 | 13 | 5.7672634 | 0.14383562 |
| 58 | CD209 | 13 | 4.428888 | 0.14266305 |
| 59 | FGF1 | 13 | 7.1389694 | 0.14423077 |
| 60 | IKBKG | 13 | 2.9129455 | 0.14285715 |
| 61 | HPRT1 | 13 | 41.86046 | 0.14403293 |
| 62 | FGFR2 | 13 | 62.313843 | 0.14285715 |
| 63 | IDO1 | 12 | 114.35072 | 0.14363885 |
| 64 | WAS | 12 | 9.96579 | 0.13962767 |
| 65 | CYP2C9 | 12 | 28.406013 | 0.14131898 |
| 66 | EPHA2 | 12 | 110.74736 | 0.1420839 |
| 67 | CHEK1 | 11 | 3.897872 | 0.14344262 |
| 68 | COMT | 11 | 44.608868 | 0.13636364 |
| 69 | ATP12A | 11 | 20.04787 | 0.14266305 |
| 70 | ITK | 11 | 2.3593173 | 0.13962767 |
| 71 | DHFR | 10 | 143.46443 | 0.14324693 |
| 72 | CYP2C19 | 10 | 51.41783 | 0.14056225 |
| 73 | ICAM2 | 10 | 1.6470731 | 0.13981359 |
| 74 | CYP17A1 | 9 | 132.31921 | 0.136897 |
| 75 | RNASE3 | 8 | 0.6450318 | 0.14075068 |
| 76 | MAPK10 | 7 | 1.7650008 | 0.13833992 |
| 77 | UGT2B7 | 7 | 2.0575106 | 0.13944224 |
| 78 | LTF | 7 | 0.71694976 | 0.14018692 |
| 79 | CYP51A1 | 6 | 265.89664 | 0.13530928 |
| 80 | RORC | 6 | 0 | 0.13981359 |
| 81 | TGM2 | 6 | 8.930667 | 0.14112903 |
| 82 | ADA | 6 | 15.780002 | 0.14112903 |
| 83 | TNK2 | 6 | 3.9851854 | 0.1392573 |
| 84 | IL6ST | 5 | 0 | 0.13779527 |
| 85 | LAP3 | 5 | 98.4974 | 0.13707572 |
| 86 | MPI | 5 | 223.96904 | 0.13027295 |
| 87 | PNP | 4 | 83.11256 | 0.136897 |
| 88 | ACADM | 4 | 6.846439 | 0.13358779 |
| 89 | CFB | 4 | 0.9110306 | 0.13981359 |
| 90 | DYRK1A | 4 | 1.15 | 0.13636364 |
| 91 | APCS | 3 | 0 | 0.13907285 |
| 92 | GALK1 | 3 | 19.146414 | 0.12962963 |
| 93 | TPH1 | 3 | 5.359037 | 0.13092269 |
| 94 | SQLE | 3 | 14.4429655 | 0.12382075 |
| 95 | RAN | 3 | 0 | 0.13307984 |
| 96 | FDFT1 | 2 | 0 | 0.12041284 |
| 97 | CRAT | 2 | 0 | 0.13059701 |
| 98 | TREH | 1 | 0 | 0.13043478 |
| 99 | CHIT1 | 1 | 0 | 0.13636364 |
| 100 | ARSA | 1 | 0 | 0.00952381 |
| 101 | ECE1 | 1 | 0 | 0.12727273 |
| 102 | PSAP | 1 | 0 | 0.00952381 |
| 103 | GNPDA1 | 1 | 0 | 0.1160221 |
| 104 | CA6 | 0 | 0 | 0.009433962 |
| 105 | PNPO | 0 | 0 | 0.009433962 |
| 106 | AKR1A1 | 0 | 0 | 0.009433962 |
| Sr. No. | Compounds | Degree | Betweenness | Closeness |
|---|---|---|---|---|
| 1 | Myrecitin | 52 | 551.76263 | 0.49593496 |
| 2 | Quercitin | 51 | 523.5583 | 0.4919355 |
| 3 | Luteolin | 52 | 652.4729 | 0.49593496 |
| 4 | Rutin | 57 | 1270.8419 | 0.5169492 |
| 5 | Isoquercitin | 54 | 855.9138 | 0.5041322 |
| 6 | Chlorogenic acid | 58 | 1955.7657 | 0.52136755 |
| 7 | Catechin | 47 | 422.1807 | 0.4765625 |
| 8 | Digalacturonic acid | 56 | 1521.1764 | 0.5126051 |
| 9 | Apigenin | 46 | 393.6386 | 0.4728682 |
| 10 | Ferrulic acid | 51 | 1359.5 | 0.4919355 |
| 11 | Caffeic acid | 47 | 1025.8472 | 0.4765625 |
| 12 | Gallic acid | 44 | 779.46545 | 0.46564886 |
| 13 | Protocatechuic acid | 33 | 176.04279 | 0.42957747 |
| 14 | Beta-sitosterol | 43 | 1956.6543 | 0.46212122 |
| 15 | p-coumaric acid | 38 | 838.4169 | 0.4452555 |
| 16 | Beta-carotene | 35 | 881.8214 | 0.43571427 |
| 17 | Resorcinol | 20 | 932.9409 | 0.3935484 |
| Sr. No. | Protein | Receptor-Ligand | Interaction Type | Distance |
|---|---|---|---|---|
| 1 | ALB | N:UNK1:H-A:PRO110:O | Conventional Hydrogen Bond | 2.41382 |
| N:UNK1:H-A:ASP108:O | Conventional Hydrogen Bond | 1.97207 | ||
| N:UNK1:H-A:GLU425:OE2 | Conventional Hydrogen Bond | 2.20851 | ||
| N:UNK1:H-A:ASP108:O | Conventional Hydrogen Bond | 2.60039 | ||
| A:ARG145:CD-N:UNK1:O | Carbon Hydrogen Bond | 3.09589 | ||
| N:UNK1:C-A:GLU425:OE2 | Carbon Hydrogen Bond | 3.09465 | ||
| A:HIS146-N:UNK1 | Pi-Pi T-Shaped | 5.79161 | ||
| A:HIS146-N:UNK1 | Pi-Pi T-Shaped | 5.91519 | ||
| N:UNK1-A:ARG145 | Pi-Alkyl | 5.23581 | ||
| N:UNK1-A:ARG114 | Pi-Alkyl | 5.19604 | ||
| N:UNK1-A:ARG145 | Pi-Alkyl | 5.49631 | ||
| 2 | IL2 | A:LYS32-N:UNK1 | Alkyl | 4.38514 |
| A:LYS35-N:UNK1 | Alkyl | 4.59242 | ||
| A:LYS35-N:UNK1 | Alkyl | 3.78304 | ||
| A:ARG38-N:UNK1 | Alkyl | 4.70114 | ||
| A:ARG38-N:UNK1 | Alkyl | 4.64384 | ||
| A:LYS43-N:UNK1 | Alkyl | 5.14946 | ||
| A:VAL69-N:UNK1 | Alkyl | 5.48053 | ||
| A:LEU72-N:UNK1 | Alkyl | 4.86731 | ||
| A:ALA73-N:UNK1 | Alkyl | 4.04445 | ||
| N:UNK1-A:LEU72 | Alkyl | 4.75444 | ||
| A:PHE42-N:UNK1 | Pi-Alkyl | 4.28876 | ||
| 3 | PPARG | N:UNK1:H-A:LEU228:O | Conventional Hydrogen Bond | 3.05297 |
| N:UNK1:H-A:CYS285:O | Conventional Hydrogen Bond | 2.84417 | ||
| N:UNK1:H-A:SER289:OG | Conventional Hydrogen Bond | 2.46012 | ||
| A:SER289:CA-N:UNK1:O | Carbon Hydrogen Bond | 3.46788 | ||
| A:ARG288:NH2-N:UNK1 | Pi-Cation | 3.74404 | ||
| A:GLU295:OE1-N:UNK1 | Pi-Anion | 4.02798 | ||
| A:ARG288:NE-N:UNK1 | Pi-Donor Hydrogen Bond | 3.91198 | ||
| A:CYS285:SG-N:UNK1 | Pi-Sulfur | 5.52645 | ||
| N:UNK1-A:ARG288 | Pi-Alkyl | 4.10762 | ||
| N:UNK1-A:ALA292 | Pi-Alkyl | 4.4279 | ||
| N:UNK1-A:ILE326 | Pi-Alkyl | 5.37149 | ||
| N:UNK1-A:LEU330 | Pi-Alkyl | 5.03093 | ||
| N:UNK1-A:ARG288 | Pi-Alkyl | 3.59494 | ||
| N:UNK1-A:LEU330 | Pi-Alkyl | 4.98253 | ||
| N:UNK1-A:ALA292 | Pi-Alkyl | 5.35981 | ||
| N:UNK1-A:MET329 | Pi-Alkyl | 4.6949 | ||
| 4 | PTPRC | A:LYS291:HZ1-N:UNK1:O | Conventional Hydrogen Bond | 2.49711 |
| N:UNK1:H-A:VAL235:O | Conventional Hydrogen Bond | 2.70571 | ||
| N:UNK1:H-A:ALA231:O | Conventional Hydrogen Bond | 2.54842 | ||
| N:UNK1:H-A:ASN232:O | Conventional Hydrogen Bond | 2.57383 | ||
| A:HIS374-N:UNK1 | Pi-Pi Stacked | 4.36158 | ||
| N:UNK1-A:LEU293 | Pi-Alkyl | 5.26513 | ||
| N:UNK1-A:LEU293 | Pi-Alkyl | 5.26961 | ||
| 5 | STAT1 | A:LYS240:HZ3-N:UNK1:O | Conventional Hydrogen Bond | 2.11392 |
| A:ARG241:HH11-N:UNK1:O | Conventional Hydrogen Bond | 2.78186 | ||
| A:ARG241:HH21-N:UNK1:O | Conventional Hydrogen Bond | 1.92015 | ||
| A:SER432:HG-N:UNK1:O | Conventional Hydrogen Bond | 2.58764 | ||
| A:SER434:HG-N:UNK1:O | Conventional Hydrogen Bond | 2.76816 | ||
| A:THR451:HG1-N:UNK1:O | Conventional Hydrogen Bond | 3.06789 | ||
| A:ARG482:HH11-N:UNK1:O | Conventional Hydrogen Bond | 2.16428 | ||
| A:ARG482:HH11-N:UNK1:O | Conventional Hydrogen Bond | 3.0504 | ||
| A:ARG482:HH21-N:UNK1:O | Conventional Hydrogen Bond | 2.62021 | ||
| N:UNK1:H-A:GLN314:O | Conventional Hydrogen Bond | 2.12922 | ||
| N:UNK1:H-A:GLN314:O | Conventional Hydrogen Bond | 2.11387 | ||
| N:UNK1:H-A:THR451:O | Conventional Hydrogen Bond | 2.07658 | ||
| N:UNK1:H-A:THR451:OG1 | Conventional Hydrogen Bond | 2.24927 | ||
| N:UNK1:H-A:SER452:O | Conventional Hydrogen Bond | 2.74446 | ||
| A:VAL237:CG2-N:UNK1 | Pi-Sigma | 3.65444 | ||
| 6 | STAT3 | A:ARG379:NH1-N:UNK1:O | Conventional Hydrogen Bond | 3.33961 |
| N:UNK1:H-A:VAL375:O | Conventional Hydrogen Bond | 2.64317 | ||
| N:UNK1:H-A:ASP374:OD1 | Conventional Hydrogen Bond | 2.35408 | ||
| A:GLY373:CA-N:UNK1:O | Carbon Hydrogen Bond | 3.45037 | ||
| A:ARG417:CA-N:UNK1:O | Carbon Hydrogen Bond | 3.42956 | ||
| A:GLY421:CA-N:UNK1:O | Carbon Hydrogen Bond | 3.39518 | ||
| N:UNK1:C-A:LEU378:O | Carbon Hydrogen Bond | 3.29566 | ||
| N:UNK1-A:LYS383 | Pi-Alkyl | 5.00243 | ||
| 7 | TLR4 | A:HIS68:ND1-N:UNK1:O | Conventional Hydrogen Bond | 3.03529 |
| A:SER73:CA-N:UNK1:O | Carbon Hydrogen Bond | 2.94848 | ||
| A:THR92:OG1-N:UNK1:O | Conventional Hydrogen Bond | 3.05155 | ||
| N:UNK1-A:LYS47 | Pi-Alkyl | 5.33037 | ||
| N:UNK1-A:LYS47 | Pi-Alkyl | 5.15142 | ||
| N:UNK1:C-A:HIS68 | Pi-Sigma | 3.88961 | ||
| N:UNK1:H-A:HIS68:O | Conventional Hydrogen Bond | 2.20639 | ||
| N:UNK1:H-A:LEU66:O | Conventional Hydrogen Bond | 2.34857 | ||
| N:UNK1:H-A:THR92:OG1 | Conventional Hydrogen Bond | 2.39563 | ||
| 8 | TNF | N:UNK1:C-A:TYR59 | Pi-Sigma | 3.73712 |
| A:LEU57-N:UNK1 | Alkyl | 5.38917 | ||
| A:LEU63-N:UNK1 | Alkyl | 5.26338 | ||
| A:PRO117-N:UNK1 | Alkyl | 4.92982 | ||
| A:TYR59-N:UNK1 | Pi-Alkyl | 4.35802 | ||
| A:TYR115-N:UNK1 | Pi-Alkyl | 4.90592 | ||
| A:TYR115-N:UNK1 | Pi-Alkyl | 4.49189 | ||
| A:TYR119-N:UNK1 | Pi-Alkyl | 4.33167 | ||
| A:TYR119-N:UNK1:C | Pi-Alkyl | 4.13411 | ||
| 9 | TP53 | A:ARG10:NH1-N:UNK1:O | Conventional Hydrogen Bond | 3.15168 |
| A:ARG203:HH2-N:UNK1:O | Conventional Hydrogen Bond | 2.79577 | ||
| N:UNK1:H-A:GLU89:OE2 | Conventional Hydrogen Bond | 2.30967 | ||
| N:UNK1:H-A:GLN23:O | Conventional Hydrogen Bond | 2.28776 | ||
| N:UNK1:H-A:ILE21:O | Conventional Hydrogen Bond | 2.7139 | ||
| N:UNK1-A:LYS20 | Pi-Alkyl | 5.35537 | ||
| 10 | CASP1 | N:UNK1:C-A:TRP145 | Pi-Sigma | 3.79529 |
| A:ILE155-N:UNK1 | Alkyl | 5.20732 | ||
| A:ILE155-N:UNK1 | Alkyl | 4.76342 | ||
| A:PRO277-N:UNK1 | Alkyl | 4.22891 | ||
| A:VAL279-N:UNK1 | Alkyl | 4.21566 | ||
| A:ILE280-N:UNK1 | Alkyl | 5.42589 | ||
| A:TRP145-N:UNK1 | Pi-Alkyl | 5.22269 | ||
| A:TRP145-N:UNK1 | Pi-Alkyl | 4.00488 | ||
| A:TRP145-N:UNK1 | Pi-Alkyl | 4.62996 | ||
| A:TRP145-N:UNK1 | Pi-Alkyl | 5.03064 | ||
| A:TYR153-N:UNK1 | Pi-Alkyl | 5.27671 |
| Biochemical Pathway | Effect on Fungus or Virulence Factor | Explanation | References |
|---|---|---|---|
| STAT3 (signal transducer and activator of transcription 3) | Suppresses inflammation and tissue damage caused by fungal infection | STAT3 is a transcription factor that can modulate the immune response and prevent excessive inflammation and tissue damage. STAT3 can also inhibit the growth and invasion of C. albicans by regulating the expression of anti-fungal genes and enhancing the phagocytosis of fungal cells. Ajwa date extract may stimulate the release of STAT3 and enhance its anti-fungal activity. | [90,91] |
| IL-2 (interleukin-2) | Promotes T cell activation and proliferation against fungal infection | IL-2 is a cytokine that can stimulate the activation and proliferation of T cells, which are immune cells that can recognize and kill infected cells. IL-2 can also enhance the production of other cytokines that have anti-fungal effects, such as IFN-gamma and TNF-alpha. Ajwa date extract may stimulate the release of IL-2 and increase its anti-fungal function. | [69,70] |
| PTPRC (protein tyrosine phosphatase receptor type C) | Regulates T cell receptor signaling and immune response against fungal infection | PTPRC, also known as CD45, is a protein that can regulate the signaling of T cell receptor (TCR), which is a molecule that recognizes antigens presented by infected cells. PTPRC can modulate the activation and differentiation of T cells and their anti-fungal effector functions. Ajwa date extract may stimulate the release of PTPRC and improve its anti-fungal function. | [71,72] |
| STAT1 (signal transducer and activator of transcription 1) | Activates anti-fungal genes and enhances the phagocytosis of fungal cells | STAT1 is a transcription factor that can activate the expression of genes that are involved in anti-fungal responses, such as IFN-gamma, NOS2 and CXCL10. STAT1 can also enhance the phagocytosis of fungal cells by macrophages, which are immune cells that can engulf and destroy foreign particles. Ajwa date extract may stimulate the release of STAT1 and increase its anti-fungal function. | [90,92] |
| CASP1 (caspase-1) | Induces the pyroptosis (inflammatory cell death) of infected cells and prevents fungal dissemination | CASP1 is a protein that can trigger pyroptosis, which is a process of inflammatory cell death, in the response to fungal infection. Pyroptosis can help eliminate infected cells and prevent the spread of fungal pathogens. Pyroptosis can also release cytokines, such as IL-1beta and IL-18, that have anti-fungal effects. Ajwa date extract may stimulate the release of CASP1 and increase its pyroptotic function. | [74] |
| ALB (albumin) | Binds to fungal toxins and neutralizes their effects | ALB is a protein that can bind to various substances in the blood, including fungal toxins such as gliotoxin and fumagillin. ALB can neutralize the effects of these toxins on immune cells and tissues. Ajwa date extract may stimulate the release of ALB and enhance its anti-toxin activity. | [75] |
| TP53 (tumor protein p53) | Induces the apoptosis (cell death) of infected cells and prevents fungal dissemination | TP53 is a protein that can trigger apoptosis, which is a process of programmed cell death, in response to DNA damage or stress. Apoptosis can help eliminate infected cells and prevent the spread of fungal pathogens. Ajwa date extract may stimulate the release of TP53 and increase its apoptotic function. | [93] |
| TLR4 (Toll-like receptor 4) | Recognizes fungal components and activates the inflammatory response against fungal infection | TLR4 is a protein that can recognize fungal components such as lipopolysaccharide and beta-glucan. TLR4 can activate the inflammatory response against fungal infection by inducing the expression of cytokines such as TNF-alpha, IL-1beta, IL-6, IL-12 and IL-23. Ajwa date extract may stimulate the release of TLR4 and increase its anti-fungal function. | [77] |
| TNF (tumor necrosis factor) | Induces inflammation and cell death against fungal infection | TNF is a cytokine that can induce inflammation and cell death against fungal infection by activating the expression of genes such as NOS2, CXCL10 and ICAM1. TNF can also enhance the phagocytosis of fungal cells by macrophages and neutrophils. Ajwa date extract may stimulate the release of TNF and increase its anti-fungal function. | [78] |
| PPARG (peroxisome proliferator-activated receptor gamma) | Inhibits fungal growth and biofilm formation | PPARG is a protein that can inhibit the growth and biofilm formation of C. albicans by regulating the expression of genes such as EFG1, NRG1 and HWP1. PPARG can also modulate the immune response and inflammation against fungal infection by influencing the production of cytokines such as IL-10, IL-17 and TGF-beta. Ajwa date extract may stimulate the release of PPARG and increase its anti-fungal function. | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnan, M.; Siddiqui, A.J.; Ashraf, S.A.; Bardakci, F.; Alreshidi, M.; Badraoui, R.; Noumi, E.; Tepe, B.; Sachidanandan, M.; Patel, M. Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation to Elucidate the Molecular Targets and Potential Mechanism of Phoenix dactylifera (Ajwa Dates) against Candidiasis. Pathogens 2023, 12, 1369. https://doi.org/10.3390/pathogens12111369
Adnan M, Siddiqui AJ, Ashraf SA, Bardakci F, Alreshidi M, Badraoui R, Noumi E, Tepe B, Sachidanandan M, Patel M. Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation to Elucidate the Molecular Targets and Potential Mechanism of Phoenix dactylifera (Ajwa Dates) against Candidiasis. Pathogens. 2023; 12(11):1369. https://doi.org/10.3390/pathogens12111369
Chicago/Turabian StyleAdnan, Mohd, Arif Jamal Siddiqui, Syed Amir Ashraf, Fevzi Bardakci, Mousa Alreshidi, Riadh Badraoui, Emira Noumi, Bektas Tepe, Manojkumar Sachidanandan, and Mitesh Patel. 2023. "Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation to Elucidate the Molecular Targets and Potential Mechanism of Phoenix dactylifera (Ajwa Dates) against Candidiasis" Pathogens 12, no. 11: 1369. https://doi.org/10.3390/pathogens12111369










