Effect of a 12-Week Polyphenol Rutin Intervention on Markers of Pancreatic β-Cell Function and Gut Microbiota in Adults with Overweight without Diabetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participant Recruitment
2.2. Clinical Intervention Days (CID)
2.3. Anthropometry
2.4. Body Composition
2.5. Biochemistry
2.6. Composition of Rutin Products
2.7. Gut Microbiota
2.7.1. Faecal Sample Collection
2.7.2. DNA Extraction
2.7.3. 16S rRNA Gene-Targeted PCR and Sequencing
2.7.4. Bioinformatics
2.8. Statistical Analyses
2.8.1. Calculation of Power
2.8.2. Demographic, Anthropometric, and Metabolic Data
2.8.3. Gut Microbiota Sequence Data
3. Results
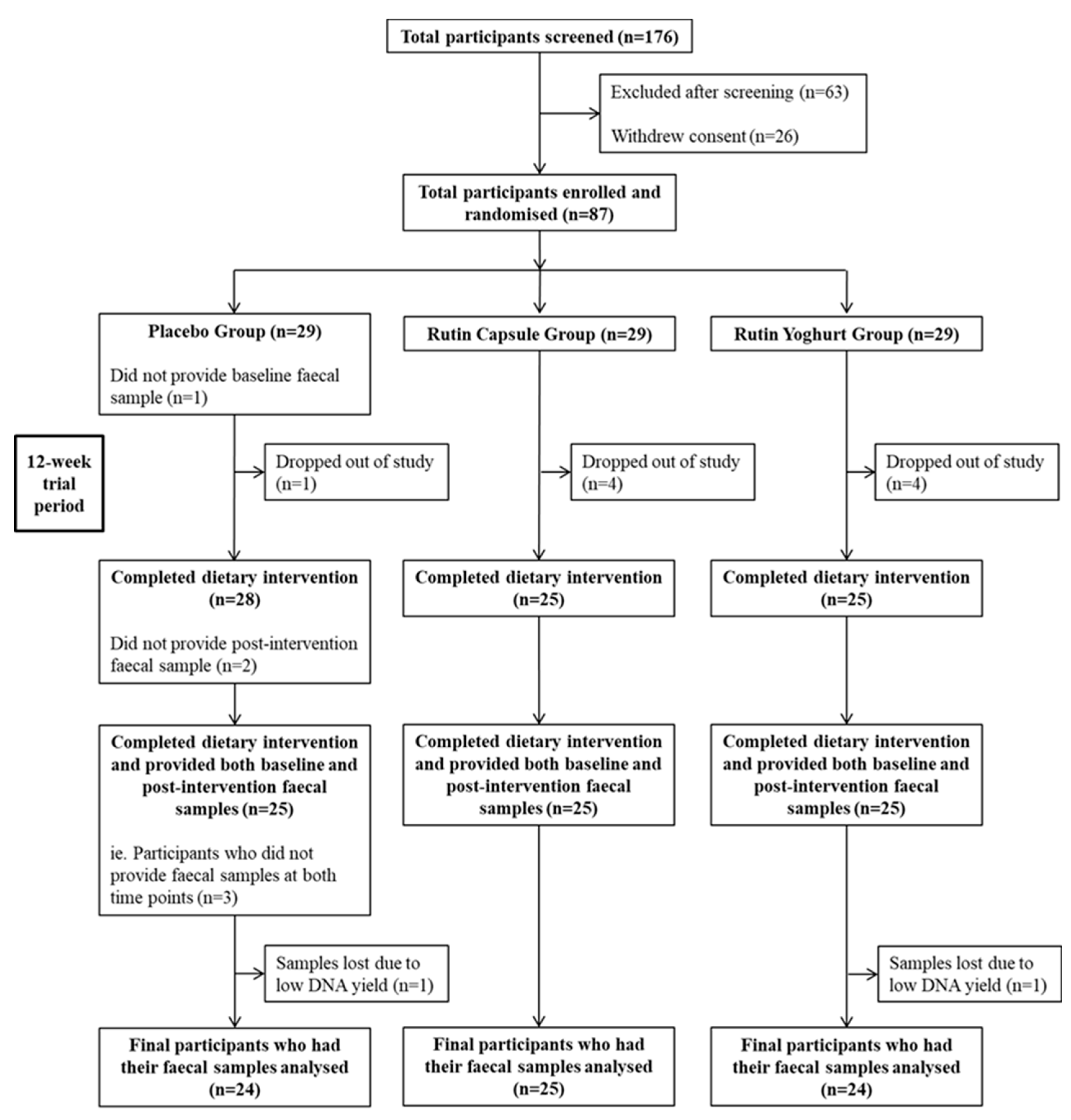
3.1. Participant Baseline Characteristics and Compliance with Treatment
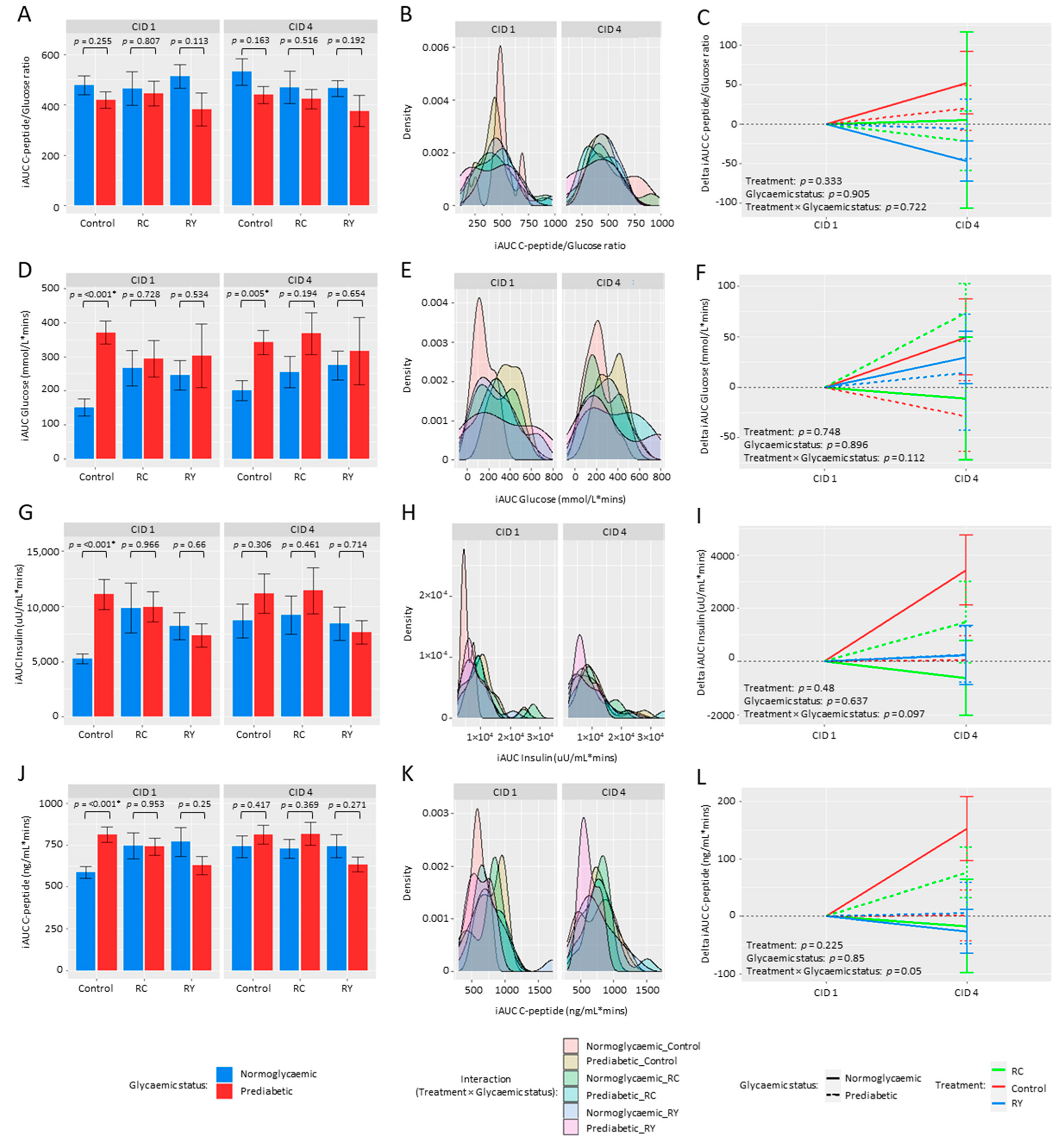
3.2. Effect of Rutin Supplementation on Metabolic Health Parameters
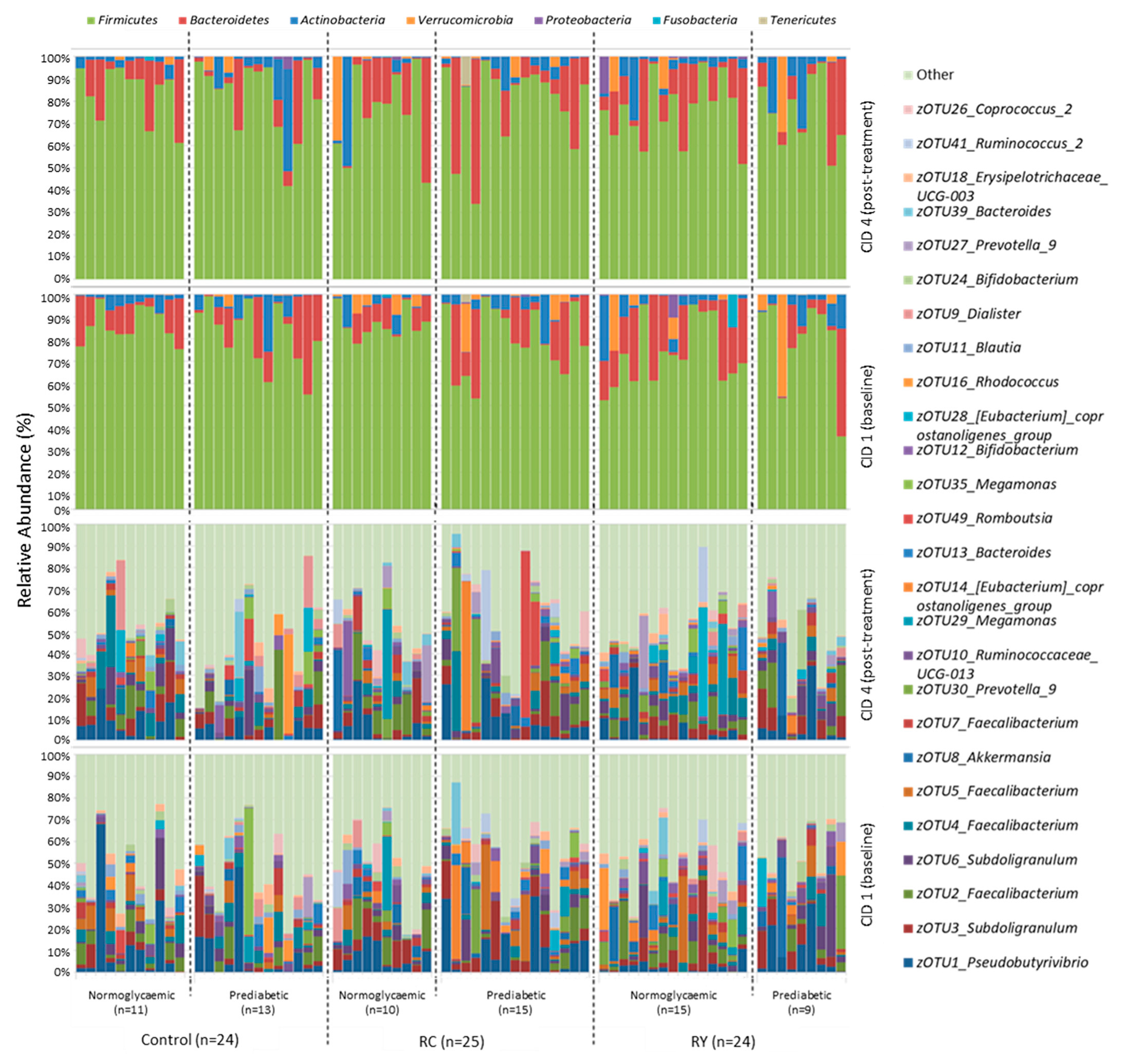
3.3. Effect of Rutin Supplementation on Gut Microbiota Composition
4. Discussion
4.1. Rutin Supplementation and Metabolic Health Parameters
4.2. Rutin Supplementation and the Gut Microbiota
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 2016, 387, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. Diabetes Atlas. 2019. Available online: https://diabetesatlas.org/ (accessed on 31 October 2022).
- Zimmet, P.Z. Diabetes and its drivers: The largest epidemic in human history? Clin. Diabetes Endocrinol. 2017, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- WHO. Diabetes: Fact Sheet. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 31 October 2022).
- Wondmkun, Y.T. Obesity, insulin resistance, and type 2 diabetes: Associations and therapeutic implications. Diabetes Metab. Syndr. Obes. 2020, 13, 3611–3616. [Google Scholar] [CrossRef] [PubMed]
- Eckel, R.H.; Alberti, K.; Grundy, S.M.; Zimmet, P.Z. The metabolic syndrome. Lancet 2010, 375, 181–183. [Google Scholar] [CrossRef]
- Butler, P.C.; Chou, J.; Carter, W.B.; Wang, Y.N.; Bu, B.H.; Chang, D.; Chang, J.K.; Rizza, R.A. Effects of meal ingestion on plasma amylin concentration in NIDDM and nondiabetic humans. Diabetes 1990, 39, 752–756. [Google Scholar] [CrossRef]
- Sanke, T.; Hanabusa, T.; Nakano, Y.; Oki, C.; Okai, K.; Nishimura, S.; Kondo, M.; Nanjo, K. Plasma islet amyloid polypeptide (amylin) levels and their responses to oral glucose in type 2 (non-insulin-dependent) diabetic patients. Diabetologia 1991, 34, 129–132. [Google Scholar] [CrossRef]
- Enoki, S.; Mitsukawa, T.; Takemura, J.; Nakazato, M.; Aburaya, J.; Toshimori, H.; Matsukara, S. Plasma islet amyloid polypeptide levels in obesity, impaired glucose tolerance and non-insulin-dependent diabetes mellitus. Diabetes Res. Clin. Pract. 1992, 15, 97–102. [Google Scholar] [CrossRef]
- Despa, S.; Margulies, K.B.; Chen, L.; Knowlton, A.A.; Havel, P.J.; Taegtmeyer, H.; Bers, D.M.; Despa, F. Hyperamylinemia contributes to cardiac dysfunction in obesity and diabetes: A study in humans and rats. Circ. Res. 2012, 110, 598–608. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, H.; Yu, H.; Cooper, G.J.S. Fas-associated death receptor signaling evoked by human amylin in islet β-cells. Diabetes 2008, 57, 348–356. [Google Scholar] [CrossRef]
- Konarkowska, B.; Aitken, J.F.; Kistler, J.; Zhang, S.; Cooper, G.J. The aggregation potential of human amylin determines its cytotoxicity towards islet beta-cells. FEBS J. 2006, 273, 3614–3624. [Google Scholar] [CrossRef]
- Guardado-Mendoza, R.; Davalli, A.M.; Chavez, A.O.; Hubbard, G.B.; Dick, E.J.; Majluf-Cruz, A.; Tene-Perez, C.E.; Goldschmidt, L.; Hart, J.; Perego, C.; et al. Pancreatic islet amyloidosis, beta-cell apoptosis, and alpha-cell proliferation are determinants of islet remodeling in type-2 diabetic baboons. Proc. Natl. Acad. Sci. USA 2009, 106, 13992–13997. [Google Scholar] [CrossRef]
- Haataja, L.; Gurlo, T.; Huang, C.J.; Butler, P.C. Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis. Endocr. Rev. 2008, 29, 303–316. [Google Scholar] [CrossRef]
- Clark, A.; Saad, M.F.; Nezzer, T.; Uren, C.; Knowler, W.C.; Bennett, P.H.; Turner, R.C. Islet amyloid polypeptide in diabetic and non-diabetic Pima Indians. Diabetologia 1990, 33, 285–289. [Google Scholar] [CrossRef]
- Jaikaran, E.T.; Clark, A. Islet amyloid and type 2 diabetes: From molecular misfolding to islet pathophysiology. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2001, 1537, 179–203. [Google Scholar] [CrossRef]
- Kahn, S.E.; Andrikopoulos, S.; Verchere, C.B. Islet amyloid: A long-recognized but underappreciated pathological feature of type 2 diabetes. Diabetes 1999, 48, 241–253. [Google Scholar] [CrossRef]
- Seeliger, J.; Werkmüller, A.; Winter, R. Macromolecular crowding as a suppressor of human IAPP fibril formation and cytotoxicity. PLoS ONE 2013, 8, e69652. [Google Scholar] [CrossRef] [PubMed]
- Abioye, R.O.; Okagu, O.D.; Udenigwe, C.C. Disaggregation of islet amyloid polypeptide fibrils as a potential anti-fibrillation mechanism of tetrapeptide TNGQ. Int. J. Mol. Sci. 2022, 23, 1972. [Google Scholar] [CrossRef]
- Dall’Asta, M.; Bayle, M.; Neasta, J.; Scazzina, F.; Bruni, R.; Cros, G.; Del Rio, D.; Oiry, C. Protection of pancreatic β-cell function by dietary polyphenols. Phytochem. Rev. 2015, 14, 933–959. [Google Scholar] [CrossRef]
- Aitken, J.F.; Loomes, K.M.; Riba-Garcia, I.; Unwin, R.D.; Prijic, G.; Phillips, A.S.; Phillips, A.R.; Wu, D.; Poppitt, S.D.; Ding, K.; et al. Rutin suppresses human-amylin/hIAPP misfolding and oligomer formation in-vitro, and ameliorates diabetes and its impacts in human-amylin/hIAPP transgenic mice. Biochem. Biophys. Res. Commun. 2017, 482, 625–631. [Google Scholar] [CrossRef]
- Porat, Y.; Abramowitz, A.; Gazit, E. Inhibition of amyloid fibril formation by polyphenols: Structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol. Drug Des. 2006, 67, 27–37. [Google Scholar] [CrossRef]
- Sequeira, I.R.; Poppitt, S.D. Unfolding novel mechanisms of polyphenol flavonoids for better glycaemic control: Targeting pancreatic islet amyloid polypeptide (IAPP). Nutrients 2017, 9, 788. [Google Scholar] [CrossRef] [PubMed]
- Ngoungoure, V.L.N.; Schluesener, J.; Moundipa, P.F.; Schluesener, H. Natural polyphenols binding to amyloid: A broad class of compounds to treat different human amyloid diseases. Mol. Nutr. Food Res. 2015, 59, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Bahadoran, Z.; Mirmiran, P.; Azizi, F. Dietary polyphenols as potential nutraceuticals in management of diabetes: A review. J. Diabetes Metab. Disord. 2013, 12, 43. [Google Scholar] [CrossRef]
- Xiao, J.B.; Högger, P. Dietary polyphenols and type 2 diabetes: Current insights and future perspectives. Curr. Med. Chem. 2015, 22, 23–38. [Google Scholar] [CrossRef]
- Yang, J.; Lee, H.; Sung, J.; Kim, Y.; Jeong, H.S.; Lee, J. Conversion of rutin to quercetin by acid treatment in relation to biological activities. Prev. Nutr. Food Sci. 2019, 24, 313–320. [Google Scholar] [CrossRef]
- Hollman, P.C.; Bijsman, M.N.; van Gameren, Y.; Cnossen, E.P.; de Vries, J.H.; Katan, M.B. The sugar moiety is a major determinant of the absorption of dietary flavonoid glycosides in man. Free Radic. Res. 1999, 31, 569–573. [Google Scholar] [CrossRef]
- Graefe, E.U.; Wittig, J.; Mueller, S.; Riethling, A.K.; Uehleke, B.; Drewelow, B.; Pforte, H.; Jacobasch, G.; Derendorf, H.; Veit, M. Pharmacokinetics and bioavailability of quercetin glycosides in humans. J. Clin. Pharmacol. 2001, 41, 492–499. [Google Scholar] [CrossRef]
- Manach, C.; Morand, C.; Demigné, C.; Texier, O.; Régérat, F.; Rémésy, C. Bioavailability of rutin and quercetin in rats. FEBS Lett. 1997, 409, 12–16. [Google Scholar] [CrossRef]
- Aitken, J.F.; Loomes, K.M.; Riba-Garcia, I.; Unwin, R.D.; Prijic, G.; Phillips, A.S.; Phillips, A.R.J.; Wu, D.; Poppitt, S.D.; Ding, K.; et al. Quantitative data describing the impact of the flavonol rutin on in-vivo blood-glucose and fluid-intake profiles, and survival of human-amylin transgenic mice. Data Brief 2017, 10, 298–303. [Google Scholar] [CrossRef]
- Boyle, S.P.; Dobson, V.L.; Duthie, S.J.; Hinselwood, D.C.; Kyle, J.A.; Collins, A.R. Bioavailability and efficiency of rutin as an antioxidant: A human supplementation study. Eur. J. Clin. Nutr. 2000, 54, 774–782. [Google Scholar] [CrossRef]
- Sattanathan, K.; Dhanapal, C.K.; Umarani, R.; Manavalan, R. Beneficial health effects of rutin supplementation in patients with diabetes mellitus. J. Appl. Pharm. Sci. 2011, 01, 227–231. [Google Scholar]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Yang, G.; Wei, J.; Liu, P.; Zhang, Q.; Tian, Y.; Hou, G.; Meng, L.; Xin, Y.; Jiang, X. Role of the gut microbiota in type 2 diabetes and related diseases. Metabolism 2021, 117, 154712. [Google Scholar] [CrossRef]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef]
- McNeil, N.I. The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr. 1984, 39, 338–342. [Google Scholar] [CrossRef]
- Arora, T.; Tremaroli, V. Therapeutic potential of butyrate for treatment of type 2 diabetes. Front. Endocrinol. 2021, 12, 761834. [Google Scholar] [CrossRef]
- Wang, X.; He, G.; Peng, Y.; Zhong, W.; Wang, Y.; Zhang, B. Sodium butyrate alleviates adipocyte inflammation by inhibiting NLRP3 pathway. Sci. Rep. 2015, 5, 12676. [Google Scholar] [CrossRef]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Gao, F.; Lv, Y.-W.; Long, J.; Chen, J.-M.; He, J.-M.; Ruan, X.-Z.; Zhu, H.-B. Butyrate improves the metabolic disorder and gut microbiome dysbiosis in mice induced by a high-fat diet. Front. Pharmacol. 2019, 10, 1040. [Google Scholar] [CrossRef]
- Riva, A.; Kolimár, D.; Spittler, A.; Wisgrill, L.; Herbold, C.W.; Abrankó, L.; Berry, D. Conversion of rutin, a prevalent dietary flavonol, by the human gut microbiota. Front. Microbiol. 2020, 11, 585428. [Google Scholar] [CrossRef] [PubMed]
- Leighton, E.; Sainsbury, C.A.; Jones, G.C. A practical review of C-peptide testing in diabetes. Diabetes Ther. 2017, 8, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Wang, Y.; Dou, T.; Xue, B.; Tan, Y.; Yang, J. Pancreatic fat content is associated with β-cell function and insulin resistance in Chinese type 2 diabetes subjects. Endocr. J. 2019, 66, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Saisho, Y. Postprandial C-peptide to glucose ratio as a marker of β-cell function: Implication for the management of type 2 diabetes. Int. J. Mol. Sci. 2016, 17, 744. [Google Scholar] [CrossRef]
- Lindström, J.; Tuomilehto, J. The diabetes risk score: A practical tool to predict type 2 diabetes risk. Diabetes Care 2003, 26, 725–731. [Google Scholar] [CrossRef]
- Silvestre, M.P.; Jiang, Y.; Volkova, K.; Chisholm, H.; Lee, W.; Poppitt, S.D. Evaluating FINDRISC as a screening tool for type 2 diabetes among overweight adults in the PREVIEW:NZ cohort. Prim. Care Diabetes 2017, 11, 561–569. [Google Scholar] [CrossRef]
- American Diabetes Association. Classification and diagnosis of diabetes: Standards of medical care in diabetes 2018. Diabetes Care 2018, 41, S13–S27. [Google Scholar] [CrossRef]
- Kim, H.S. Blood glucose measurement: Is serum equal to plasma? Diabetes Metab. J. 2016, 40, 365–366. [Google Scholar] [CrossRef]
- Duxbury, M. An enzymatic clinical chemistry laboratory experiment incorporating an introduction to mathematical method comparison techniques. Biochem. Mol. Biol. Educ. 2004, 32, 246–249. [Google Scholar] [CrossRef]
- Malik, V.; Pundir, C.S. Determination of total cholesterol in serum by cholesterol esterase and cholesterol oxidase immobilized and co-immobilized on to arylamine glass. Biotechnol. Appl. Biochem. 2002, 35, 191–197. [Google Scholar] [CrossRef]
- Chauhan, N.; Narang, J.; Pundir, C.S. Covalent immobilization of lipase, glycerol kinase, glycerol-3-phosphate oxidase & horseradish peroxidase onto plasticized polyvinyl chloride (PVC) strip & its application in serum triglyceride determination. Indian J. Med. Res. 2014, 139, 603–609. [Google Scholar]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef]
- Lustig, V.; Papanastasiou-Diamandis, A.; Goldberg, D.M. Evaluation of commercially formulated aspartate aminotransferase and alanine aminotransferase activity determinations by the Scandinavian Committee on Enzymes and IFCC methods as modified for use with automated enzyme analysers. Clin. Biochem. 1988, 21, 283–290. [Google Scholar] [CrossRef]
- Akcakaya, H.; Aroymak, A.; Gokce, S. A quantitative colorimetric method of measuring alkaline phosphatase activity in eukaryotic cell membranes. Cell Biol. Int. 2007, 31, 186–190. [Google Scholar] [CrossRef]
- Szasz, G. A kinetic photometric method for serum gamma-glutamyl transpeptidase. Clin. Chem. 1969, 15, 124–136. [Google Scholar] [CrossRef]
- Rashidinejad, A.; Loveday, S.M.; Jameson, G.B.; Hindmarsh, J.P.; Singh, H. Rutin-casein co-precipitates as potential delivery vehicles for flavonoid rutin. Food Hydrocoll. 2019, 96, 451–462. [Google Scholar] [CrossRef]
- Singh, D.; Rawat, M.S.M.; Semalty, A.; Semalty, M. Rutin-phospholipid complex: An innovative technique in novel drug delivery system- NDDS. Curr. Drug Deliv. 2012, 9, 305–314. [Google Scholar] [CrossRef]
- Costea, P.I.; Zeller, G.; Sunagawa, S.; Pelletier, E.; Alberti, A.; Levenez, F.; Tramontano, M.; Driessen, M.; Hercog, R.; Jung, F.-E.; et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017, 35, 1069–1076. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Hoggard, M.; Zoing, M.; Biswas, K.; Taylor, M.W.; Douglas, R.G. The sinonasal mycobiota in chronic rhinosinusitis and control patients. Rhinology 2019, 57, 190–199. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016, 81257. [Google Scholar] [CrossRef]
- Edgar, R.C. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 2016, 74161. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, w5–w9. [Google Scholar] [CrossRef]
- Wilkinson, L. SYSTAT; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010; pp. 256–257. [Google Scholar] [CrossRef]
- Utzschneider, K.M.; Prigeon, R.L.; Tong, J.; Gerchman, F.; Carr, D.B.; Zraika, S.; Udayasankar, J.; Montgomery, B.; Mari, A.; Kahn, S.E. Within-subject variability of measures of beta cell function derived from a 2h OGTT: Implications for research studies. Diabetologia 2007, 50, 2516–2525. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Fernandes, A.A.H.; Novelli, E.L.B.; Okoshi, K.; Okoshi, M.P.; Di Muzio, B.P.; Guimarães, J.F.C.; Fernandes Junior, A. Influence of rutin treatment on biochemical alterations in experimental diabetes. Biomed. Pharmacother. 2010, 64, 214–219. [Google Scholar] [CrossRef]
- Niture, N.T.; Ansari, A.A.; Naik, S.R. Anti-hyperglycemic activity of rutin in streptozotocin-induced diabetic rats: An effect mediated through cytokines, antioxidants and lipid biomarkers. Indian J. Exp. Biol. 2014, 52, 720–727. [Google Scholar]
- Stanley Mainzen Prince, P.; Kamalakkannan, N. Rutin improves glucose homeostasis in streptozotocin diabetic tissues by altering glycolytic and gluconeogenic enzymes. J. Biochem. Mol. Toxicol. 2006, 20, 96–102. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Shih, H.-Y.; Chia, Y.-C.; Lee, C.-H.; Ashida, H.; Lai, Y.-K.; Weng, C.-F. Rutin potentiates insulin receptor kinase to enhance insulin-dependent glucose transporter 4 translocation. Mol. Nutr. Food Res. 2014, 58, 1168–1176. [Google Scholar] [CrossRef]
- Ghorbani, A. Mechanisms of antidiabetic effects of flavonoid rutin. Biomed. Pharmacother. 2017, 96, 305–312. [Google Scholar] [CrossRef]
- Ragheb, S.R.; El Wakeel, L.M.; Nasr, M.S.; Sabri, N.A. Impact of rutin and vitamin C combination on oxidative stress and glycemic control in patients with type 2 diabetes. Clin. Nutr. ESPEN 2020, 35, 128–135. [Google Scholar] [CrossRef]
- Saisho, Y. β-cell dysfunction: Its critical role in prevention and management of type 2 diabetes. World J. Diabetes 2015, 6, 109–124. [Google Scholar] [CrossRef]
- Gerst, F.; Wagner, R.; Oquendo, M.B.; Siegel-Axel, D.; Fritsche, A.; Heni, M.; Staiger, H.; Häring, H.-U.; Ullrich, S. What role do fat cells play in pancreatic tissue? Mol. Metab. 2019, 25, 1–10. [Google Scholar] [CrossRef]
- Mondal, P.; Prasad, A.; Girdhar, K. Interventions to improve β-cell mass and function. Ann. d’Endocrinol. 2017, 78, 469–477. [Google Scholar] [CrossRef]
- Oh, Y.S.; Bae, G.D.; Baek, D.J.; Park, E.-Y.; Jun, H.-S. Fatty acid-induced lipotoxicity in pancreatic beta-cells during development of type 2 diabetes. Front. Endocrinol. 2018, 9, 384. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schafer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Mai, V.; McCrary, Q.M.; Sinha, R.; Glei, M. Associations between dietary habits and body mass index with gut microbiota composition and fecal water genotoxicity: An observational study in African American and Caucasian American volunteers. Nutr. J. 2009, 8, 49. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; Nilaweera, K.; Ross, P.R.; Shanahan, F.; O’Toole, P.W.; Cotter, P.D. The gut microbiota and its relationship to diet and obesity: New insights. Gut Microbes 2012, 3, 186–202. [Google Scholar] [CrossRef]
- Philippot, L.; Andersson, S.G.E.; Battin, T.J.; Prosser, J.I.; Schimel, J.P.; Whitman, W.B.; Hallin, S. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Genet. 2010, 8, 523–529. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef]
- Salamon, D.; Sroka-Oleksiak, A.; Kapusta, P.; Szopa, M.; Mrozińska, S.; Ludwig-Słomczyńska, A.H.; Wołkow, P.P.; Bulanda, M.; Klupa, T.; Małecki, M.T.; et al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on next-generation sequencing of the 16S rRNA gene fragment. Pol. Arch. Intern. Med. 2018, 128, 336–343. [Google Scholar] [CrossRef]
- Remely, M.; Aumueller, E.; Merold, C.; Dworzak, S.; Hippe, B.; Zanner, J.; Pointner, A.; Brath, H.; Haslberger, A.G. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 2014, 537, 85–92. [Google Scholar] [CrossRef]
- Greer, R.L.; Dong, X.; Moraes, A.C.F.; Zielke, R.A.; Fernandes, G.R.; Peremyslova, E.; Vasquez-Perez, S.; Schoenborn, A.A.; Gomes, E.P.; Pereira, A.C.; et al. Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat. Commun. 2016, 7, 13329. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Candela, M.; Biagi, E.; Soverini, M.; Consolandi, C.; Quercia, S.; Severgnini, M.; Peano, C.; Turroni, S.; Rampelli, S.; Pozzilli, P.; et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br. J. Nutr. 2016, 116, 80–93. [Google Scholar] [CrossRef]
- Sedighi, M.; Razavi, S.; Navab-Moghadam, F.; Khamseh, M.E.; Alaei-Shahmiri, F.; Mehrtash, A.; Amirmozafari, N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017, 111, 362–369. [Google Scholar] [CrossRef]
- Wu, X.; Ma, C.; Han, L.; Nawaz, M.; Gao, F.; Zhang, X.; Yu, P.; Zhao, C.; Li, L.; Zhou, A.; et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr. Microbiol. 2010, 61, 69–78. [Google Scholar] [CrossRef]
- Barengolts, E.; Green, S.J.; Eisenberg, Y.; Akbar, A.; Reddivari, B.; Layden, B.T.; Dugas, L.; Chlipala, G. Gut microbiota varies by opioid use, circulating leptin and oxytocin in African American men with diabetes and high burden of chronic disease. PLoS ONE 2018, 13, e0194171. [Google Scholar] [CrossRef]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Kedenko, I.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Adachi, K.; Sugiyama, T.; Shimozato, A.; Ebi, M.; Ogasawara, N.; Funaki, Y.; Goto, C.; Sasaki, M.; Kasugai, K. Association of intestinal microbiota with metabolic markers and dietary habits in patients with type 2 diabetes. Digestion 2016, 94, 66–72. [Google Scholar] [CrossRef]
- Allin, K.H.; Tremaroli, V.; Caesar, R.; Jensen, B.A.H.; Damgaard, M.T.F.; Bahl, M.I.; Licht, T.R.; Hansen, T.H.; Nielsen, T.; Dantoft, T.M.; et al. Aberrant intestinal microbiota in individuals with prediabetes. Diabetologia 2018, 61, 810–820. [Google Scholar] [CrossRef]
- Egshatyan, L.; Kashtanova, D.; Popenko, A.; Tkacheva, O.; Tyakht, A.; Alexeev, D.; Karamnova, N.; Kostryukova, E.; Babenko, V.; Vakhitova, M.; et al. Gut microbiota and diet in patients with different glucose tolerance. Endocr. Connect. 2016, 5, 1–9. [Google Scholar] [CrossRef]
- Inoue, R.; Ohue-Kitano, R.; Tsukahara, T.; Tanaka, M.; Masuda, S.; Inoue, T.; Yamakage, H.; Kusakabe, T.; Hasegawa, K.; Shimatsu, A.; et al. Prediction of functional profiles of gut microbiota from 16S rRNA metagenomic data provides a more robust evaluation of gut dysbiosis occurring in Japanese type 2 diabetic patients. J. Clin. Biochem. Nutr. 2017, 61, 217–221. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed]
| All (n = 87) | Normoglycaemic (n = 47) | Prediabetic (n = 40) | p-Value | |
|---|---|---|---|---|
| Sex (M:F) | 39:48 | 17:30 | 22:18 | 0.088 |
| Ethnicity (C:A) | 23:64 | 11:36 | 12:28 | 0.626 |
| Age (years) | 44.3 (21–64) | 42.7 (22–64) | 46.2 (21–64) | 0.182 |
| Body weight (kg) | 79.5 (54.3–124.2) | 77.9 (54.3–105.3) | 81.3 (56.1–124.2) | 0.293 |
| Height (m) | 1.7 (1.4–1.9) | 1.7 (1.4–1.9) | 1.7 (1.5–1.9) | 0.386 |
| Body mass index, BMI (kg/m2) | 27.6 (22.1–37.8) | 27.4 (22.1–37.8) | 27.9 (22.3–35.8) | 0.537 |
| Waist circumference (cm) | 93.9 (73–122) | 92.2 (73–112) | 95.8 (81–122) | 0.102 |
| Hip circumference (cm) | 104.3 (83.5–127) | 104.5 (88–127) | 104.1 (83.5–125) | 0.8 |
| Systolic blood pressure (mmHg) | 120.4 (91–167) | 119.4 (91–157) | 121.5 (91–167) | 0.562 |
| Diastolic blood pressure (mmHg) | 65.3 (47–101) | 64.6 (50–101) | 66.1 (47–89.7) | 0.526 |
| Body Composition | ||||
| Total body fat (%) | 36.4 (19–52.1) | 37.4 (19–52.1) | 35.2 (21.9–50.2) | 0.188 |
| Abdominal fat (%) | 42.6 (18.4–60.6) | 43.1 (18.4–60.5) | 42.1 (19.9–60.6) | 0.617 |
| Visceral fat (%) | 42.9 (2.3–90.6) | 39.4 (2.3–72.9) | 47.2 (7.6–90.6) | 0.014 * |
| Subcutaneous fat (%) | 57.1 (9.4–97.7) | 60.6 (27.1–97.7) | 52.8 (9.4–92.4) | 0.014 * |
| Glycaemic, Liver Function, Lipid biomarkers | ||||
| Fasting plasma glucose, FPG (mmol/L) | 5.5 (4.5–6.7) | 5.1 (4.5–5.5) | 5.9 (5.6–6.7) | <0.001 * |
| Fasting insulin (uU/mL) | 12 (2.3–42.6) | 10.6 (2.3–42.6) | 13.6 (4.69–32.4) | 0.03 * |
| Fasting C-peptide (ng/mL) | 2.3 (0.8–4.9) | 2.1 (0.8–4.9) | 2.6 (1.4–4.3) | 0.016 * |
| Fasting C-peptide (ng/mL)/FPG (mg/dL) ratio (×100) | 2.3 (1–5.2) | 2.3 (1–5.2) | 2.4 (1.3–3.9) | 0.552 |
| Alanine aminotransferase, ALT (U/L) | 14.1 (2.7–51.9) | 12.8 (2.7–48.6) | 15.6 (4.1–51.9) | 0.197 |
| Aspartate aminotransferase, AST (U/L) | 23.4 (11.9–111.2) | 23.8 (12.9–97.5) | 23.1 (11.9–111.2) | 0.834 |
| Alkaline phosphatase, ALP (U/L) | 65.4 (35–120) | 63.4 (35–110) | 67.8 (36–120) | 0.279 |
| Gamma-glutamyl transferase, GGT (U/L) | 26.4 (5–162) | 24.9 (5–162) | 28.1 (8–108) | 0.581 |
| Total cholesterol (mmol/L) | 4.9 (2.3–7.6) | 5.0 (3.4–7.6) | 4.7 (2.3–6.5) | 0.093 |
| HDL-C (mmol/L) | 1.2 (0.7–2) | 1.3 (0.7–2) | 1.2 (0.7–1.9) | 0.327 |
| LDL-C (mmol/L) | 3 (0.9–5.9) | 3.1 (1.9–5.9) | 2.9 (0.9–4.3) | 0.124 |
| Triglyceride (mmol/L) | 1.6 (0.5–7.6) | 1.5 (0.5–7.6) | 1.6 (0.6–5.3) | 0.767 |
| Variable | Treatment Group | zOTUs Identified | Spearman Coefficient | p-Value | |
|---|---|---|---|---|---|
| (FDR-adj.) | |||||
| Normoglycaemic cohort | Δ Bacterial richness | Control | Otu32_Bacteroidetes_Bacteroides | 0.890 | 0.039 * |
| RY | Otu79_Bacteroidetes_Bacteroides_Bacteroides_sp_’Smarlab_BioMol-2301151′ | 0.810 | 0.041 * | ||
| Δ Shannon diversity | Control | Otu154_Firmicutes_[Eubacterium]_hallii_groups:butyrate-producing_bacterium_SL6/1/1 | 0.914 | 0.013 * | |
| Δ FI | Control | Otu67_Firmicutes_Lachnoclostridium:[Ruminococcus]_torques | −0.936 | 0.004 * | |
| Prediabetic cohort | Δ FPG | Control | Otu34_Firmicutes_Roseburia_Roseburia_inulinivorans | −0.861 | 0.025 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathrani, A.; Yip, W.; Sequeira-Bisson, I.R.; Barnett, D.; Stevenson, O.; Taylor, M.W.; Poppitt, S.D. Effect of a 12-Week Polyphenol Rutin Intervention on Markers of Pancreatic β-Cell Function and Gut Microbiota in Adults with Overweight without Diabetes. Nutrients 2023, 15, 3360. https://doi.org/10.3390/nu15153360
Mathrani A, Yip W, Sequeira-Bisson IR, Barnett D, Stevenson O, Taylor MW, Poppitt SD. Effect of a 12-Week Polyphenol Rutin Intervention on Markers of Pancreatic β-Cell Function and Gut Microbiota in Adults with Overweight without Diabetes. Nutrients. 2023; 15(15):3360. https://doi.org/10.3390/nu15153360
Chicago/Turabian StyleMathrani, Akarsh, Wilson Yip, Ivana R. Sequeira-Bisson, Daniel Barnett, Oliver Stevenson, Michael W. Taylor, and Sally D. Poppitt. 2023. "Effect of a 12-Week Polyphenol Rutin Intervention on Markers of Pancreatic β-Cell Function and Gut Microbiota in Adults with Overweight without Diabetes" Nutrients 15, no. 15: 3360. https://doi.org/10.3390/nu15153360
APA StyleMathrani, A., Yip, W., Sequeira-Bisson, I. R., Barnett, D., Stevenson, O., Taylor, M. W., & Poppitt, S. D. (2023). Effect of a 12-Week Polyphenol Rutin Intervention on Markers of Pancreatic β-Cell Function and Gut Microbiota in Adults with Overweight without Diabetes. Nutrients, 15(15), 3360. https://doi.org/10.3390/nu15153360






