Vitamin D and Type 1 Diabetes Risk: A Systematic Review and Meta-Analysis of Genetic Evidence
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion and Exclusion Criteria
- 1.
- Population: human of any gender and age, race and geographical distribution.
- 2.
- Exposure: a biological approach to the selection of genetic variants was used, including variants having a biological link to the exposure. Seven vitamin D related SNPs were selected: CYP2R1 (common variant) rs10741657, CYP2R1 (rare variant/low frequency) rs117913124, DHCR7/NADSYN1 rs12785878, GC rs3755967, CYP24A1 rs17216707, AMDHD1 rs10745742, SEC23A rs8018720. Of these selected SNPs, six are common variants identified based on the results of a recent GWAS for 25(OH)D concentration [21] and one is a low-frequency synonymous coding variant seen with a much larger effect on 25(OH)D concentration [22]. Strong genome-wide associations with 25(OH)D were found in genes located upstream (DHCR7/NADSY1 and both CYP2R1 variants), and two downstream (CYP24A1 and GC) of the 25(OH)D metabolite biochemical pathway. Two genes outside the vitamin D metabolism pathway (AMDHD1 and SEC23A) were also found to be significant variants and hence were included. 25(OH)D related proxies not directly present in the recent GWAS were also included if found in high linkage disequilibrium (r2 > 0.8) using the Ldproxy function in LD link (https://ldlink.nci.nih.gov, accessed on 12 April 2021).
- 3.
- Outcome: the primary outcome measure, T1D, was defined by the World Health Organization criteria: diabetes symptoms (polyuria, polydipsia and insulin deficiency), accompanied by exogenous insulin usage once T1D had been diagnosed [27]. T1D could be self-reported or doctor-diagnosed when confirming cases.
- 4.
- Study design: peer-reviewed genetic association, cohort, cross-section, or case-control observational studies and Mendelian randomization (MR) studies, as well as clinical trials and unpublished cohort studies.
- 5.
- A sample size of at least 50 cases and 50 controls were mandatory for sufficient data extraction. Where there were multiple publications from the same study population, the most recent highest quality results with the largest sample size were used.
- 6.
- The publication reported genotype distribution in both cases and controls in order to estimate an odds ratio (OR) with a 95% confidence interval (CI).
- 1.
- Conference papers.
- 2.
- Other types of diabetes.
2.3. Study Selection and Data Extraction
2.4. Statistical Analysis
2.5. Risk of Bias and Credibility of the Evidence Assessment
3. Results
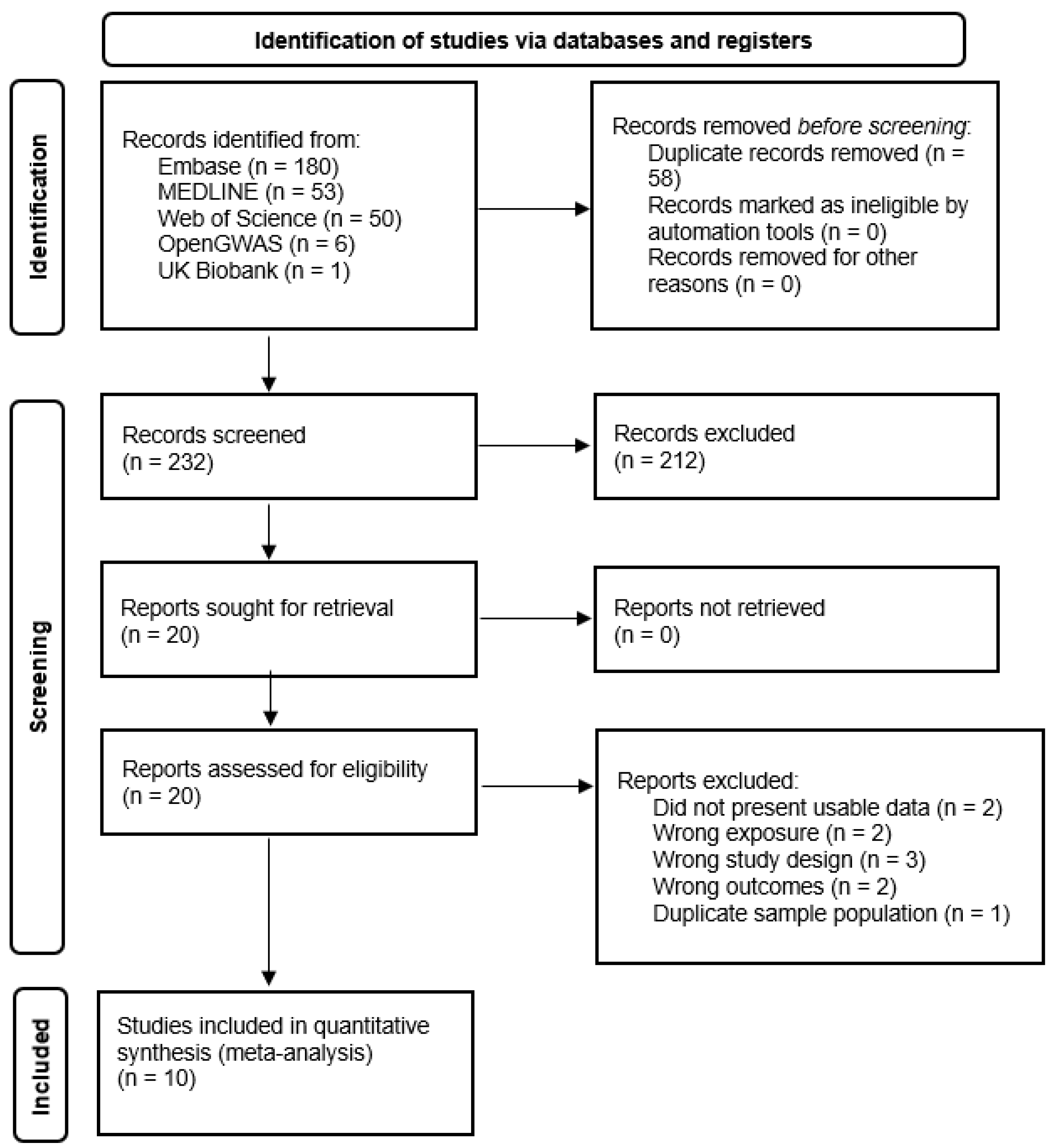
3.1. Study Selection
3.2. Characteristics of Included Studies
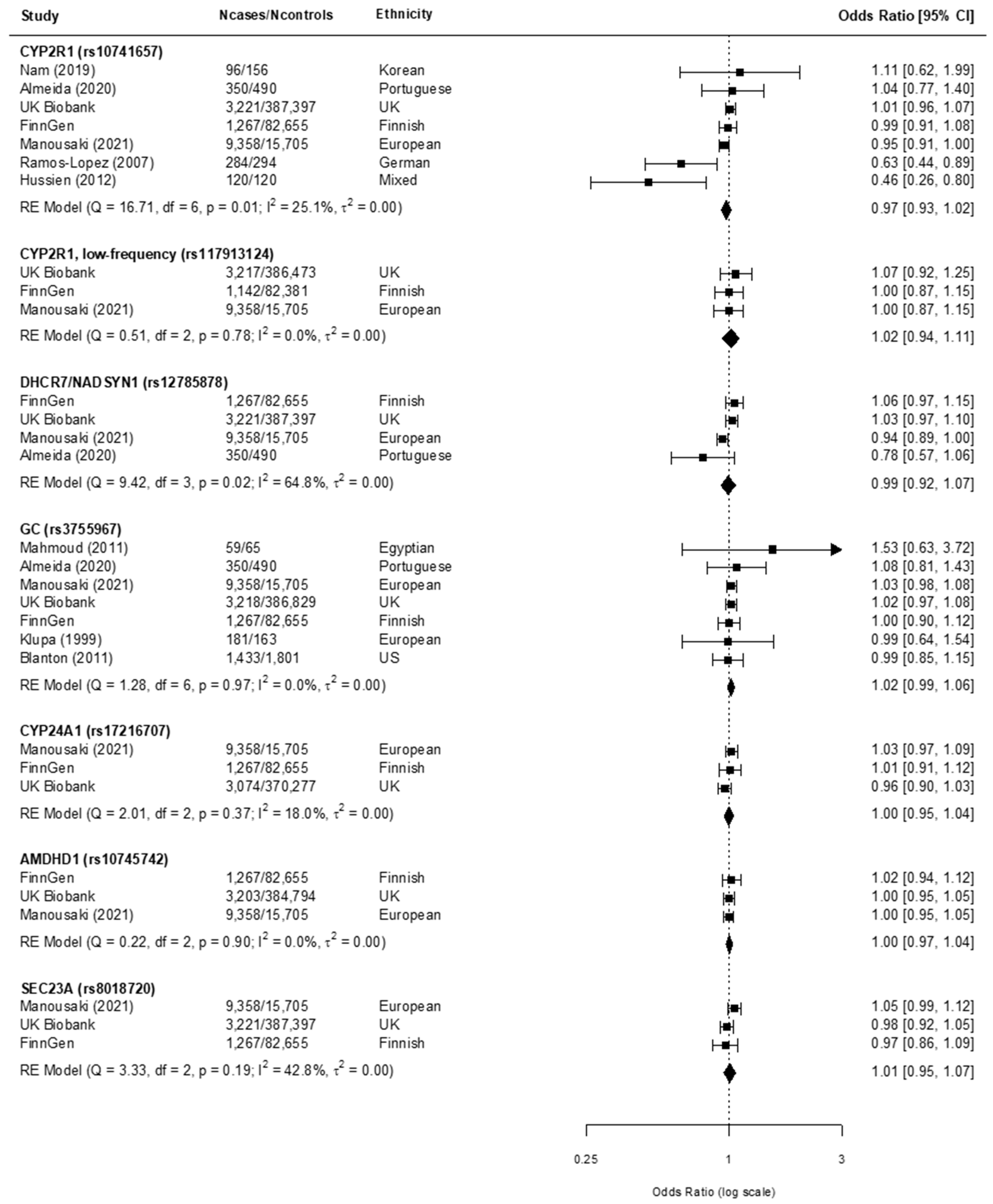
3.3. Findings from the Meta-Analysis
4. Discussion
4.1. Main Findings
4.2. Considerations of Alternative Explanation for Observed Results
4.3. Strengths and Limitations
4.4. Guidelines for Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Paschou, S.A.; Papadopoulou-Marketou, N.; Chrousos, G.P.; Kanaka-Gantenbein, C. On type 1 diabetes mellitus pathogenesis. Endocr. Connect. 2018, 7, R38–R46. [Google Scholar] [CrossRef]
- Cernea, S.; Dobreanu, M.; Raz, I. Prevention of type 1 diabetes: Today and tomorrow. Diabetes/Metab. Res. Rev. 2010, 26, 602–605. [Google Scholar] [CrossRef]
- Huber, A.; Menconi, F.; Corathers, S.; Jacobson, E.M.; Tomer, Y. Joint Genetic Susceptibility to Type 1 Diabetes and Autoimmune Thyroiditis: From Epidemiology to Mechanisms. Endocr. Rev. 2008, 29, 697–725. [Google Scholar] [CrossRef]
- World Health Organization. Global Report on Diabetes; World Health Organization: Geneva, Switzerland, 2016; Available online: http://apps.who.int/iris/bitstream/handle/10665/204871/9789241565257_eng.pdf;jsessionid=CCC2429A03D7EF6D638C05F6F008A3C2?sequence=1 (accessed on 8 July 2020).
- International Diabetes Federation. IDF Diabetes Atlas 8th Edition. 2017. Available online: http://www.diabetesatlas.org/across-the-globe.html (accessed on 10 February 2020).
- Pike, J.W.; Christakos, S. Biology and Mechanisms of Action of the Vitamin D Hormone. Endocrinol. Metab. Clin. N. Am. 2017, 46, 815–843. [Google Scholar] [CrossRef]
- Jiang, X.; Kiel, D.P.; Kraft, P. The genetics of vitamin D. Bone 2019, 126, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, J.; DeLuca, H.F. Where is the vitamin D receptor? Arch. Biochem. Biophys. 2012, 523, 123–133. [Google Scholar] [CrossRef]
- Infante, M.; Ricordi, C.; Sanchez, J.; Clare-Salzler, M.J.; Padilla, N.; Fuenmayor, V.; Chavez, C.; Alvarez, A.; Baidal, D.; Alejandro, R.; et al. Influence of Vitamin D on Islet Autoimmunity and Beta-Cell Function in Type 1 Diabetes. Nutrients 2019, 11, 2185. [Google Scholar] [CrossRef] [PubMed]
- Hayes, C.E.; Nashold, F.E.; Spach, K.M.; Pedersen, L.B. The immunological functions of the vitamin D endocrine system. Cell. Mol. Boil. 2003, 49, 277–300. [Google Scholar]
- Hewison, M.; Zehnder, D.; Chakraverty, R.; Adams, J.S. Vitamin D and barrier function: A novel role for extra-renal 1α-hydroxylase. Mol. Cell. Endocrinol. 2004, 215, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, C.; Waer, M.; Laureys, J.; Rutgeerts, O.; Bouillon, R. Prevention of autoimmune diabetes in NOD mice by 1,25 dihydroxyvitamin D3. Diabetol. 1994, 37, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Casteels, K.; Waer, M.; Bouillon, R.; Depovere, J.; Valckx, D.; Laureys, J.; Mathieu, C. 1,25-Dihydroxyvitamin D3 restores sensitivity to cyclophosphamide-induced apoptosis in non-obese diabetic (NOD) mice and protects against diabetes. Clin. Exp. Immunol. 1998, 112, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zella, J.B.; DeLuca, H.F. Vitamin D and autoimmune diabetes. J. Cell. Biochem. 2003, 88, 216–222. [Google Scholar] [CrossRef]
- Delvin, E.; Souberbielle, J.-C.; Viard, J.-P.; Salle, B. Role of vitamin D in acquired immune and autoimmune diseases. Crit. Rev. Clin. Lab. Sci. 2014, 51, 232–247. [Google Scholar] [CrossRef]
- Hyppönen, E.; Läärä, E.; Reunanen, A.; Jarvelin, M.-R.; Virtanen, S. Intake of vitamin D and risk of type 1 diabetes: A birth-cohort study. Lancet 2001, 358, 1500–1503. [Google Scholar] [CrossRef]
- Littorin, B.; Blom, P.; Schölin, A.; Arnqvist, H.J.; Blohmé, G.; Bolinder, J.; Ekbom-Schnell, A.; Eriksson, J.W.; Gudbjörnsdottir, S.; Nyström, L.; et al. Lower levels of plasma 25-hydroxyvitamin D among young adults at diagnosis of autoimmune type 1 diabetes compared with control subjects: Results from the nationwide Diabetes Incidence Study in Sweden (DISS). Diabetologia 2006, 49, 2847–2852. [Google Scholar] [CrossRef]
- Zipitis, C.S.; Akobeng, A.K. Vitamin D supplementation in early childhood and risk of type 1 diabetes: A systematic review and meta-analysis. Arch. Dis. Child. 2008, 93, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Dahlquist, G.; Mustonen, L. Childhood Onset Diabetes—Time Trends and Climatological Factors. Int. J. Epidemiol. 1994, 23, 1234–1241. [Google Scholar] [CrossRef] [PubMed]
- Gregoriou, E.; Mamais, I.; Tzanetakou, I.; Lavranos, G.; Chrysostomou, S. The Effects of Vitamin D Supplementation in Newly Diagnosed Type 1 Diabetes Patients: Systematic Review of Randomized Controlled Trials. Rev. Diabet. Stud. 2017, 14, 260–268. [Google Scholar] [CrossRef]
- Jiang, X.; O’Reilly, P.; Aschard, H.; Hsu, Y.-H.; Richards, J.B.; Dupuis, J.; Ingelsson, E.; Karasik, D.; Pilz, S.; Berry, D.; et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Manousaki, D.; Dudding, T.; Haworth, S.; Hsu, Y.-H.; Liu, C.-T.; Medina-Gomez, C.; Voortman, T.; van der Velde, N.; Melhus, H.; Robinson-Cohen, C.; et al. Low-Frequency Synonymous Coding Variation in CYP2R1 Has Large Effects on Vitamin D Levels and Risk of Multiple Sclerosis. Am. J. Hum. Genet. 2017, 101, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. J. Clin. Epidemiol. 2009, 62, 1006–1012. [Google Scholar] [CrossRef]
- Cooper, J.D.; Smyth, D.J.; Walker, N.M.; Stevens, H.; Burren, O.S.; Wallace, C.; Greissl, C.; Ramos-Lopez, E.; Hyppönen, E.; Dunger, D.B.; et al. Inherited Variation in Vitamin D Genes Is Associated With Predisposition to Autoimmune Disease Type 1 Diabetes. Diabetes 2011, 60, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.T.; Rodrigues, D.; Guimarães, J.; Lemos, M.C. Vitamin D Pathway Genetic Variation and Type 1 Diabetes: A Case–Control Association Study. Genes 2020, 11, 897. [Google Scholar] [CrossRef] [PubMed]
- Moola, S.; Munn, Z.; Sears, K.; Sfetcu, R.; Currie, M.; Lisy, K.; Tufanaru, C.; Qureshi, R.; Mattis, P.; Mu, P. Conducting systematic reviews of association (etiology). Int. J. Evid.-Based Health 2015, 13, 163–169. [Google Scholar] [CrossRef]
- World Health Organization. Diabetes; World Health Organization: Geneva, Switzerland, 2020; Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 8 July 2020).
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials 2015, 45, 139–145. [Google Scholar] [CrossRef] [PubMed]
- CASP. CASP Checklists; CASP: Oxford, UK, 2020; Available online: https://casp-uk.net/ (accessed on 8 July 2020).
- Begg, C.B.; Mazumdar, M. Operating Characteristics of a Rank Correlation Test for Publication Bias. Biometrics 1994, 50, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Frederiksen, B.N.; Kroehl, M.; Fingerlin, T.E.; Wong, R.; Steck, A.K.; Rewers, M.; Norris, J.M. Association between Vitamin D Metabolism Gene Polymorphisms and Risk of Islet Autoimmunity and Progression to Type 1 Diabetes: The Diabetes Autoimmunity Study in the Young (DAISY). J. Clin. Endocrinol. Metab. 2013, 98, E1845–E1851. [Google Scholar] [CrossRef]
- Thorsen, S.U.; Mortensen, H.B.; Carstensen, B.; Fenger, M.; Thuesen, B.; Husemoen, L.; Bergholdt, R.; Brorsson, C.A.; Pociot, F.; Linneberg, A.; et al. No association between type 1 diabetes and genetic variation in vitamin D metabolism genes: A Danish study. Pediatr. Diabetes 2013, 15, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Pani, M.A.; Donner, H.; Herwig, J.; Usadel, K.H.; Badenhoop, K. Vitamin D Binding Protein Alleles and Susceptibility for Type 1 Diabetes in Germans. Autoimmunity 1999, 31, 67–72. [Google Scholar] [CrossRef]
- Miettinen, M.E.; Smart, M.; Kinnunen, L.; Mathews, C.; Harjutsalo, V.; Surcel, H.-M.; Lamberg-Allardt, C.; Tuomilehto, J.; Hitman, G.A. Maternal VDR variants rather than 25-hydroxyvitamin D concentration during early pregnancy are associated with type 1 diabetes in the offspring. Diabetology 2015, 58, 2278–2283. [Google Scholar] [CrossRef]
- Tapia, G.; Mårild, K.; Dahl, S.R.; Lund-Blix, N.A.; Viken, M.K.; Lie, B.A.; Njølstad, P.R.; Joner, G.; Skrivarhaug, T.; Cohen, A.S.; et al. Maternal and Newborn Vitamin D–Binding Protein, Vitamin D Levels, Vitamin D Receptor Genotype, and Childhood Type 1 Diabetes. Diabetes Care 2019, 42, 553–559. [Google Scholar] [CrossRef]
- Bailey, R.; Cooper, J.D.; Zeitels, L.; Smyth, D.J.; Yang, J.H.M.; Walker, N.M.; Hyppönen, E.; Dunger, D.B.; Ramos-Lopez, E.; Badenhoop, K.; et al. Association of the Vitamin D Metabolism Gene CYP27B1 with Type 1 Diabetes. Diabetes 2007, 56, 2616–2621. [Google Scholar] [CrossRef] [PubMed]
- Norris, J.M.; Lee, H.-S.; Frederiksen, B.; Erlund, I.; Uusitalo, U.; Yang, J.; Lernmark, Å.; Simell, O.; Toppari, J.; Rewers, M.; et al. Plasma 25-Hydroxyvitamin D Concentration and Risk of Islet Autoimmunity. Diabetes 2017, 67, 146–154. [Google Scholar] [CrossRef]
- Mcgovern, A.P.; Hine, J.; De Lusignan, S. The incidence of infection is higher in older people with poor glycemic control. Diabetes 2016, 65, A360–A431. [Google Scholar] [CrossRef][Green Version]
- Ramos-Lopez, E.; Brück, P.; Jansen, T.; Herwig, J.; Badenhoop, K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes/Metab. Res. Rev. 2007, 23, 631–636. [Google Scholar] [CrossRef]
- Hussein, A.G.; Mohamed, R.H.; Alghobashy, A.A. Synergism of CYP2R1 and CYP27B1 polymorphisms and susceptibility to type 1 diabetes in Egyptian children. Cell. Immunol. 2012, 279, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Aakre, K.M.; Watine, J.; Bunting, P.S.; Sandberg, S.; Oosterhuis, W.P. Diabetes mellitus and metabolic syndrome. Clin. Chem. Lab. Med. 2011, 49. [Google Scholar] [CrossRef][Green Version]
- Klupa, T.; Malecki, M.; Hanna, L.; Sieradzka, J.; Frey, J.; Warram, J.H.; Sieradzki, J.; Krolewski, A.S. Amino acid variants of the vitamin D-binding protein and risk of diabetes in white Americans of European origin. Eur. J. Endocrinol. 1999, 141, 490–493. [Google Scholar] [CrossRef]
- Nam, H.; Rhie, Y.; Lee, K. Vitamin D level and gene polymorphisms in Korean children with type 1 diabetes. Pediatr. Diabetes 2019, 20, 750–758. [Google Scholar] [CrossRef]
- Manousaki, D.; Harroud, A.; Mitchell, R.E.; Ross, S.; Forgetta, V.; Timpson, N.J.; Smith, G.D.; Polychronakos, C.; Richards, J.B. Vitamin D levels and risk of type 1 diabetes: A Mendelian randomization study. PLoS Med. 2021, 18, e1003536. [Google Scholar] [CrossRef]
- FinnGen. FinnGen Documentation of R5 Release. FinnGen: Finland. 2021. Available online: https://finngen.gitbook.io/documentation/ (accessed on 21 June 2021).
- UK Biobank. Access Matter: Representativeness of UK Biobank Resource; UK Biobank: Stockport, UK, 2021; Available online: https://www.ukbiobank.ac.uk/ (accessed on 14 August 2021).
- Blanton, D.; Han, Z.; Bierschenk, L.; Linga-Reddy, M.P.; Wang, H.; Clare-Salzler, M.; Haller, M.; Schatz, D.; Myhr, C.; She, J.-X.; et al. Reduced Serum Vitamin D-Binding Protein Levels Are Associated With Type 1 Diabetes. Diabetes 2011, 60, 2566–2570. [Google Scholar] [CrossRef]
- Terrin, N.; Schmid, C.; Lau, J. In an empirical evaluation of the funnel plot, researchers could not visually identify publication bias. J. Clin. Epidemiol. 2005, 58, 894–901. [Google Scholar] [CrossRef]
- Jones, P.; Lucock, M.; Chaplin, G.; Jablonski, N.G.; Veysey, M.; Scarlett, C.; Beckett, E. Distribution of variants in multiple vitamin D-related loci (DHCR7/NADSYN1, GC, CYP2R1, CYP11A1, CYP24A1, VDR, RXRα and RXRγ) vary between European, East-Asian and Sub-Saharan African-ancestry populations. Genes Nutr. 2020, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kuan, V.; Martineau, A.R.; Griffiths, C.J.; Hyppönen, E.; Walton, R. DHCR7 mutations linked to higher vitamin D status allowed early human migration to Northern latitudes. BMC Evol. Biol. 2013, 13, 1–10. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Liu, J.; Wu, W.; Ouyang, H.; Zhang, Q.; Wang, Y.; Liu, L.; Yang, R.; Liu, X.; et al. Polymorphisms in the vitamin D receptor gene and type 1 diabetes mellitus risk: An update by meta-analysis. Mol. Cell. Endocrinol. 2012, 355, 135–142. [Google Scholar] [CrossRef]
- Bouillon, R. Genetic and Racial Differences in the Vitamin D Endocrine System. Endocrinol. Metab. Clin. N. Am. 2017, 46, 1119–1135. [Google Scholar] [CrossRef]
- Berry, D.; Hypponen, E. Determinants of vitamin D status: Focus on genetic variations. Curr. Opin. Nephrol. Hypertens. 2011, 20, 331–336. [Google Scholar] [CrossRef]
- Raulio, S.; Erlund, I.; Männistö, S.; Sarlio-Lähteenkorva, S.; Sundvall, J.; Tapanainen, H.; Vartiainen, E.; Virtanen, S. Successful nutrition policy: Improvement of vitamin D intake and status in Finnish adults over the last decade. Eur. J. Public Health 2016, 27, 268–273. [Google Scholar] [CrossRef]
- Mäkinen, M.; Simell, V.; Mykkänen, J.; Ilonen, J.; Veijola, R.; Hyöty, H.; Knip, M.; Simell, O.; Toppari, J.; Hermann, R. An Increase in Serum 25-Hydroxyvitamin D Concentrations Preceded a Plateau in Type 1 Diabetes Incidence in Finnish Children. J. Clin. Endocrinol. Metab. 2014, 99, E2353–E2356. [Google Scholar] [CrossRef]
- Fry, A.; Littlejohns, T.J.; Sudlow, C.; Doherty, N.; Adamska, L.; Sprosen, T.; Collins, R.; Allen, N.E. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants with Those of the General Population. Am. J. Epidemiol. 2017, 186, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
| Study Details | Participant Characteristics | Polymorphism Details | Findings | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author; Year | Country | Study Design | Ethnicity | n Cases/n Controls | Mean Age of Cases/Controls (Year) | Mean Age of Onset in Cases (Years) | T1D Diagnostic Criteria | Genotyping | Adjusted Factors | Gene | Variant | EA a | Relevant Key Findings |
| Manousaki et al., 2021 [45] | Canada, United Kingdom, United States | Cohort | European | 9358/15,705 | NI | NI | Multiple criteria | PCR-RFLP | Age, sex, season of 25OHD measurement, genotype batch, genotype array, assessment centre (proxy for latitude) | CYP2R1 CYP2R1 (low frequency) DHCR7/NADSYN1 GC CYP24A1 AMDHDI SEC23A | rs10741657 rs117913124 rs12785878 rs3755967 b rs17216707 rs10745742 rs8018720 | A G T C c T T G | No association of individual SNPs with T1D. |
| Almeida et al., 2020 [25] | Portugal | Case-control | Caucasian Portuguese | 350/490 | 29.0/32.2 | 17.2 | Classic clinical presentation d | PCR-RFLP | Age at bleed, sex, BMI, month of bleed, geographical region | CYP2R1 DHCR7/NADSYN1 GC | rs10741657 rs12785878 rs3755967 b | A T C c | No association of individual SNP with T1D. |
| Nam et al., 2019 [44] | Korea | Case-control | Korean | 96/156 | 14.7/14.0 | NI | Classic clinical presentation d | PCR | 25OHD and 1α,25(OH)2D levels. (25OHD measurement obtained in same season) | CYP2R1 | rs10741657 | A | No association of individual SNP with T1D. |
| Hussein et al., 2012 [41] | Egypt | Matched case-control | Egyptian | 120/120 | 11.7/11.1 | NI | WHO and ADA | PCR-RFLP | Nil | CYP2R1 | rs10741657 | A | An association of GG genotype of CYP2R1 polymorphism (coded by 25(OH)D decreasing alleles) with risk of T1D in Egyptian children [OR = 2.6, 95% CI = 1.1–6.1, p = 0.03]. A synergistic effect of multiple risk alleles between GG genotype of CYP2R1 and CC genotype of CYP27B1 and T1D risk found. |
| Mahmoud et al., 2011 [42] | Egypt | Matched case-control | Egyptian | 59/65 | 13/>24 | 7.5 | WHO | PCR-RFLP | Nil | GC | rs3755967 b | C c | No association between VDBP polymorphisms with T1D. |
| Blanton et al., 2011 [48] | United States | Case-control | American | 1705/2033 | NI | 12.9 | Classic clinical presentation d | TaqMan PCR Assays | Sex, onset of T1D, HLA risk | GC | rs3755967 b | C c | No association between VDBP polymorphisms with T1D detected. An association of the phenotype of lower VDBP levels with T1D. |
| Ramos-Lopez et al., 2007 [40] | Germany | Case-control | German | 284/294 | NI | 11.5 | WHO | PCR-RFLP | 25(OH)D3 levels | CYP2R1 | rs10741657 | A | An association of the ‘G’ allele of CYP2R1 common variant polymorphisms (coded by 25(OH)D decreasing alleles) with T1D risk. |
| Klupa et al., 1999 [43] | United States | Case-control | European | 181/163 | 36.2/52.55 | 10.9 | WHO | PCR | Nil; sensitivity confirmed via stratification by obesity and age at examination | GC | rs3755967 b | C c | No association of individual SNP with T1D. |
| FinnGen [46] | Finland | Cohort | Finnish | 1143–1267/82,381–82,655 | NI | NI | Strict definition (Minimal/absent insulin production by pancreas) | Illumina and Affymetrix Chip Arrays | Sex, age, 10 PCs, genotyping batch | CYP2R1 CYP2R1 (low frequency) DHCR7/NADSYN1 GC CYP24A1 AMDHDI SEC23A | rs10741657 rs117913124 b rs12785878 rs3755967 rs17216707 rs10745742 rs8018720 | A G c T C T T G | NI |
| UK Biobank [47] | United Kingdom | Cohort | Caucasian British | 3074–3221/370,277–387,397 | NI | NI | WHO | UK Biobank Axiom Array | Age, sex, birth location, assessment centre, SNP array, pc1-pc40, account for relatedness | CYP2R1 CYP2R1 (low frequency) DHCR7/NADSYN1 GC CYP24A1 AMDHDI SEC23A | rs10741657 rs117913124 rs12785878 rs3755967 rs17216707 rs10745742 rs8018720 | A G T C T T G | NI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Najjar, L.; Sutherland, J.; Zhou, A.; Hyppönen, E. Vitamin D and Type 1 Diabetes Risk: A Systematic Review and Meta-Analysis of Genetic Evidence. Nutrients 2021, 13, 4260. https://doi.org/10.3390/nu13124260
Najjar L, Sutherland J, Zhou A, Hyppönen E. Vitamin D and Type 1 Diabetes Risk: A Systematic Review and Meta-Analysis of Genetic Evidence. Nutrients. 2021; 13(12):4260. https://doi.org/10.3390/nu13124260
Chicago/Turabian StyleNajjar, Liana, Joshua Sutherland, Ang Zhou, and Elina Hyppönen. 2021. "Vitamin D and Type 1 Diabetes Risk: A Systematic Review and Meta-Analysis of Genetic Evidence" Nutrients 13, no. 12: 4260. https://doi.org/10.3390/nu13124260
APA StyleNajjar, L., Sutherland, J., Zhou, A., & Hyppönen, E. (2021). Vitamin D and Type 1 Diabetes Risk: A Systematic Review and Meta-Analysis of Genetic Evidence. Nutrients, 13(12), 4260. https://doi.org/10.3390/nu13124260






