Combination of Plasma Pharmacochemistry, RNA-Seq, and Molecular Docking Strategies to Reveal the Mechanism of the Alkaloid Fraction of Nelumbinis folium for the Treatment of Hyperlipidemia
Abstract
1. Introduction
2. Results
2.1. Content Determination of 4 Alkaloid Components
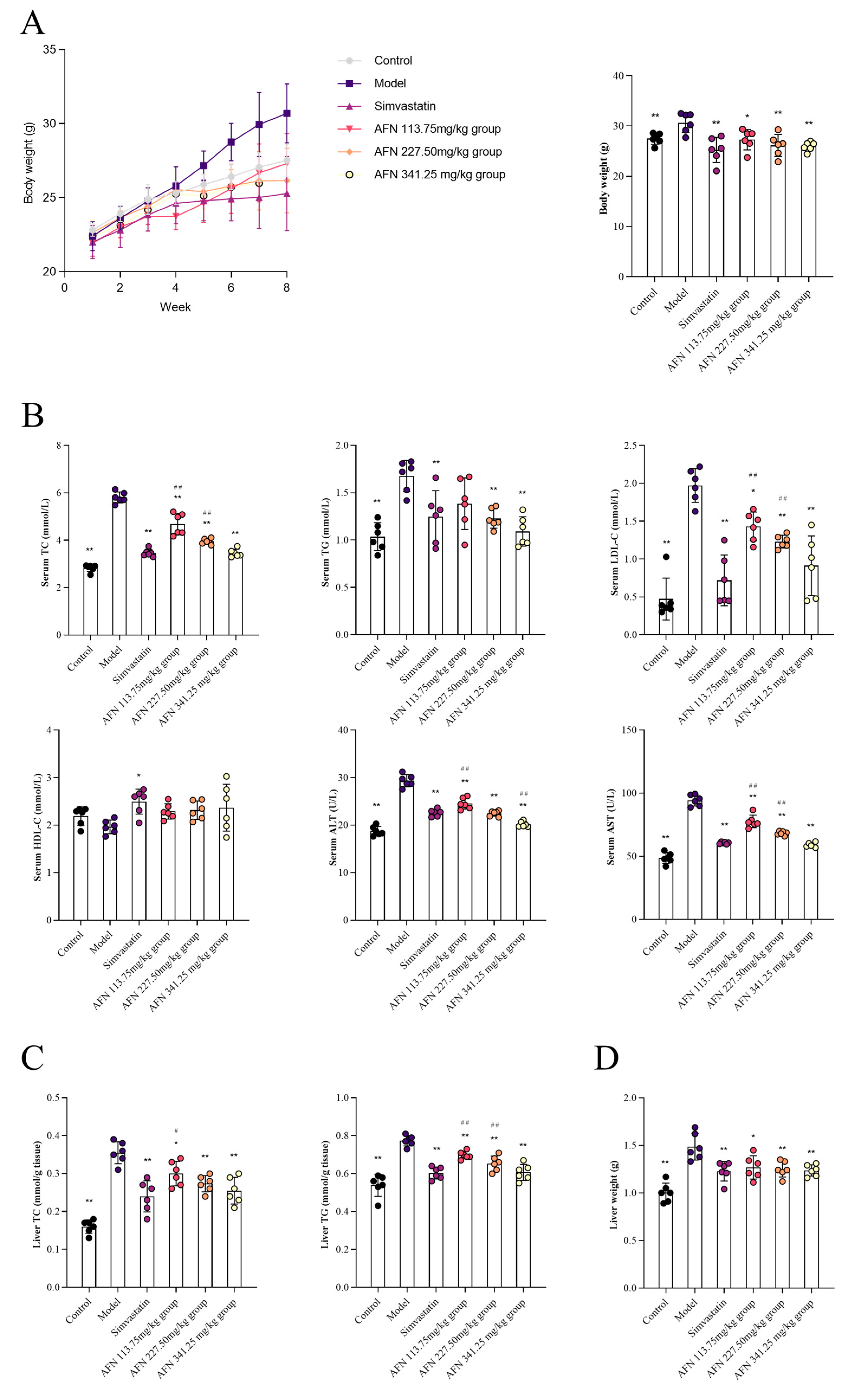
2.2. AFN Reduces Blood Lipids and Ameliorates Liver Damage in Mice
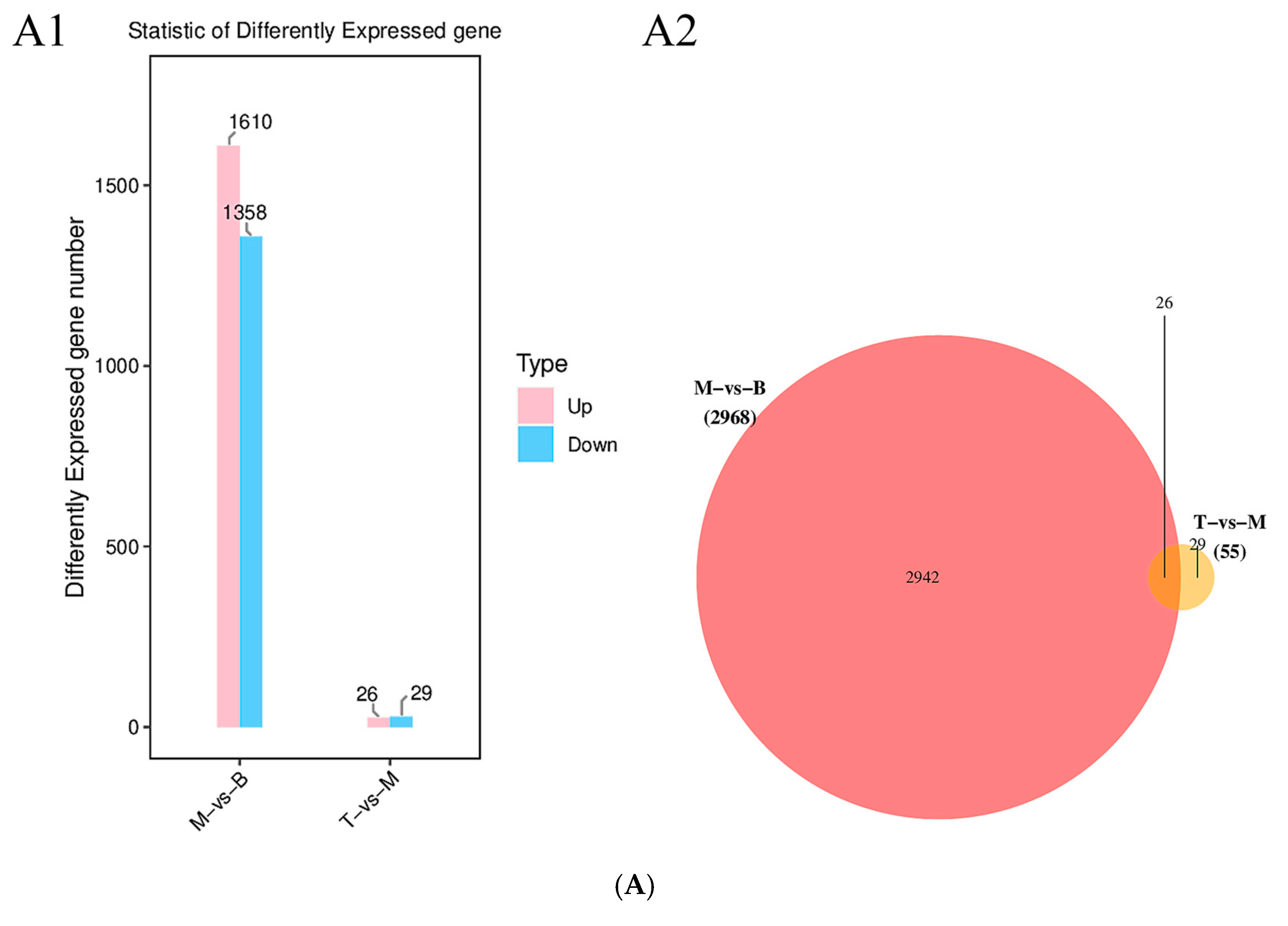
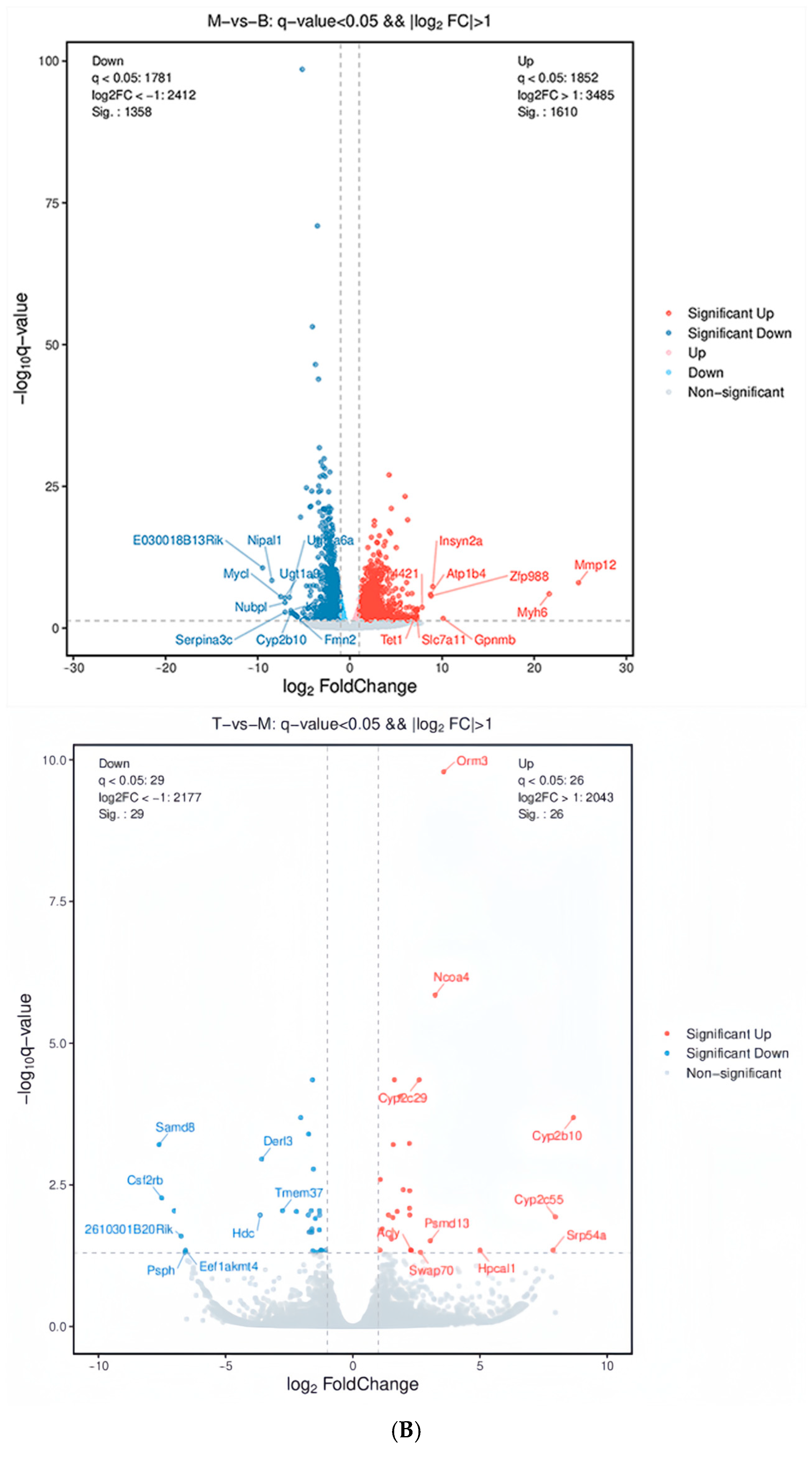
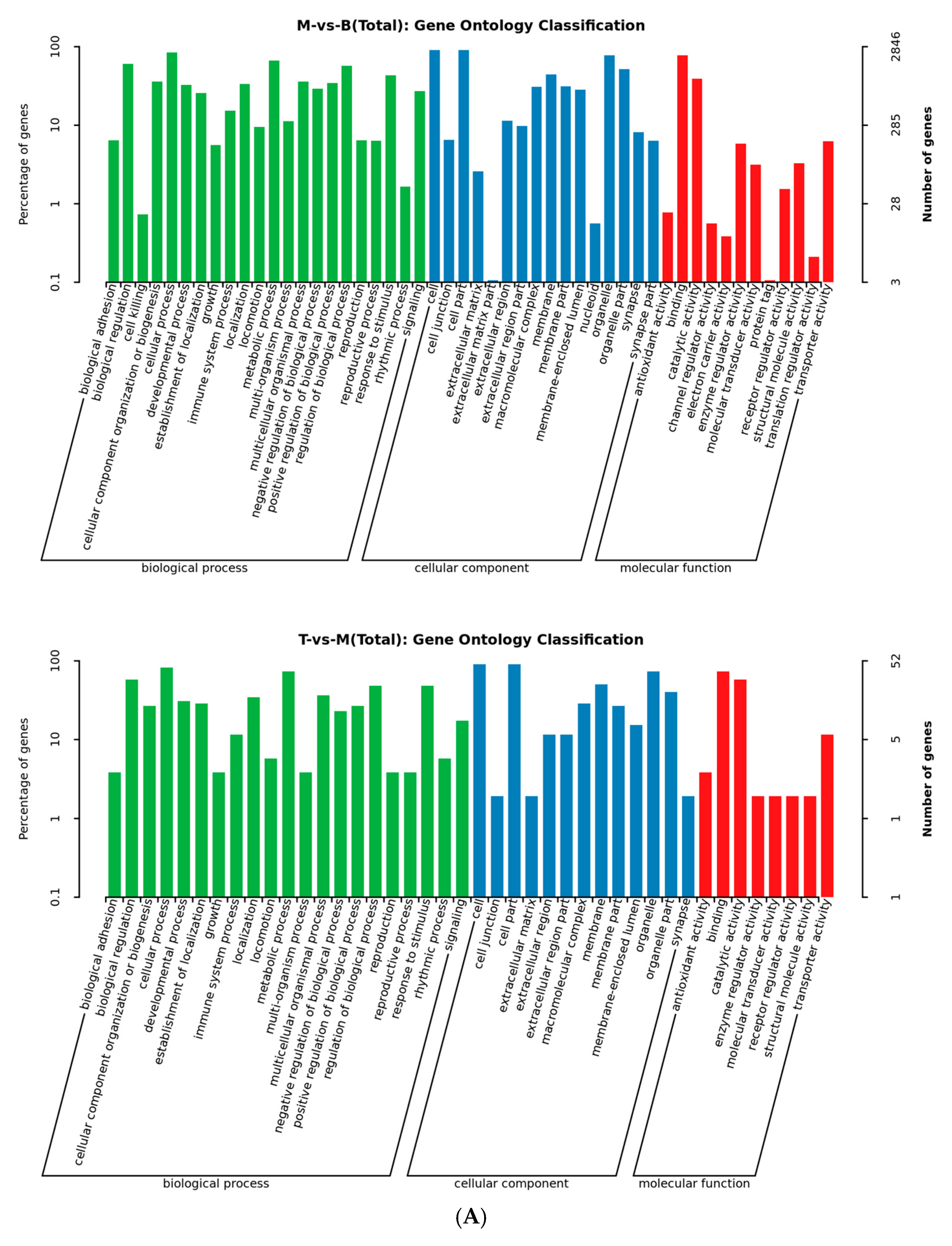
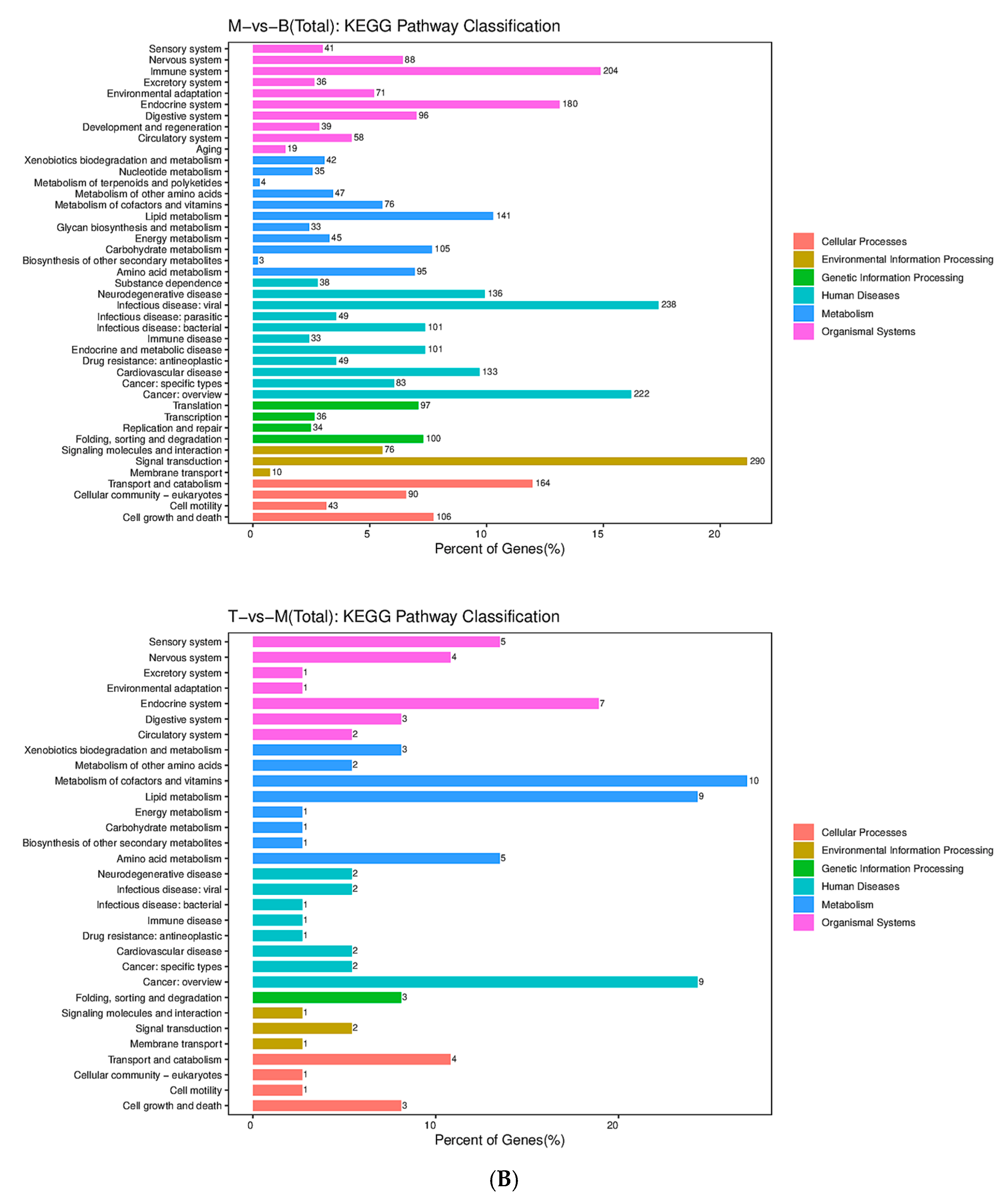
2.3. AFN Regulated Gene Expression in Mice Livers
2.4. Liquid-Mass Spectrometry Technique to Characterize the Components Absorbed into the Plasma at the AFN
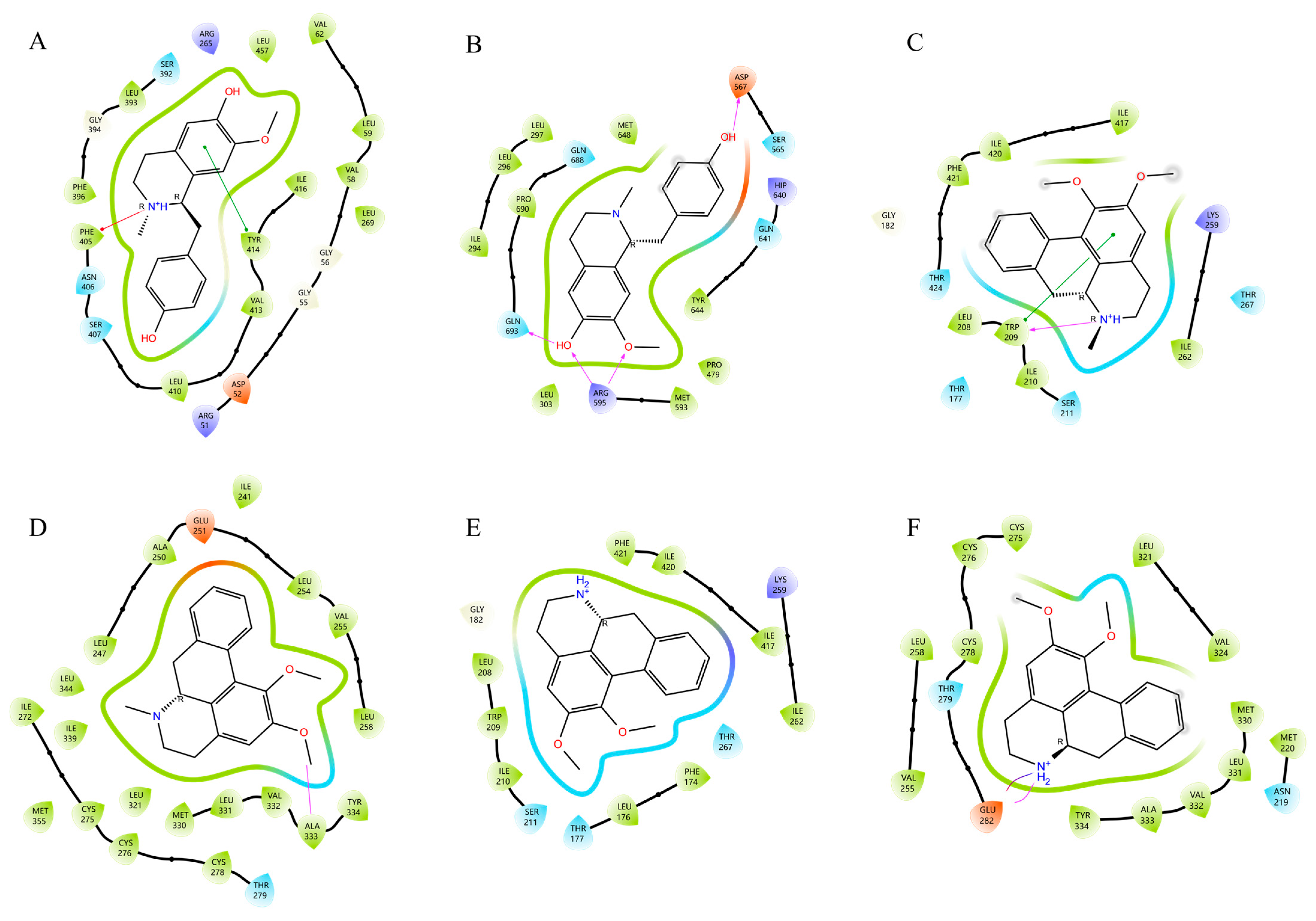
2.5. Ingredient-Target Molecular Docking
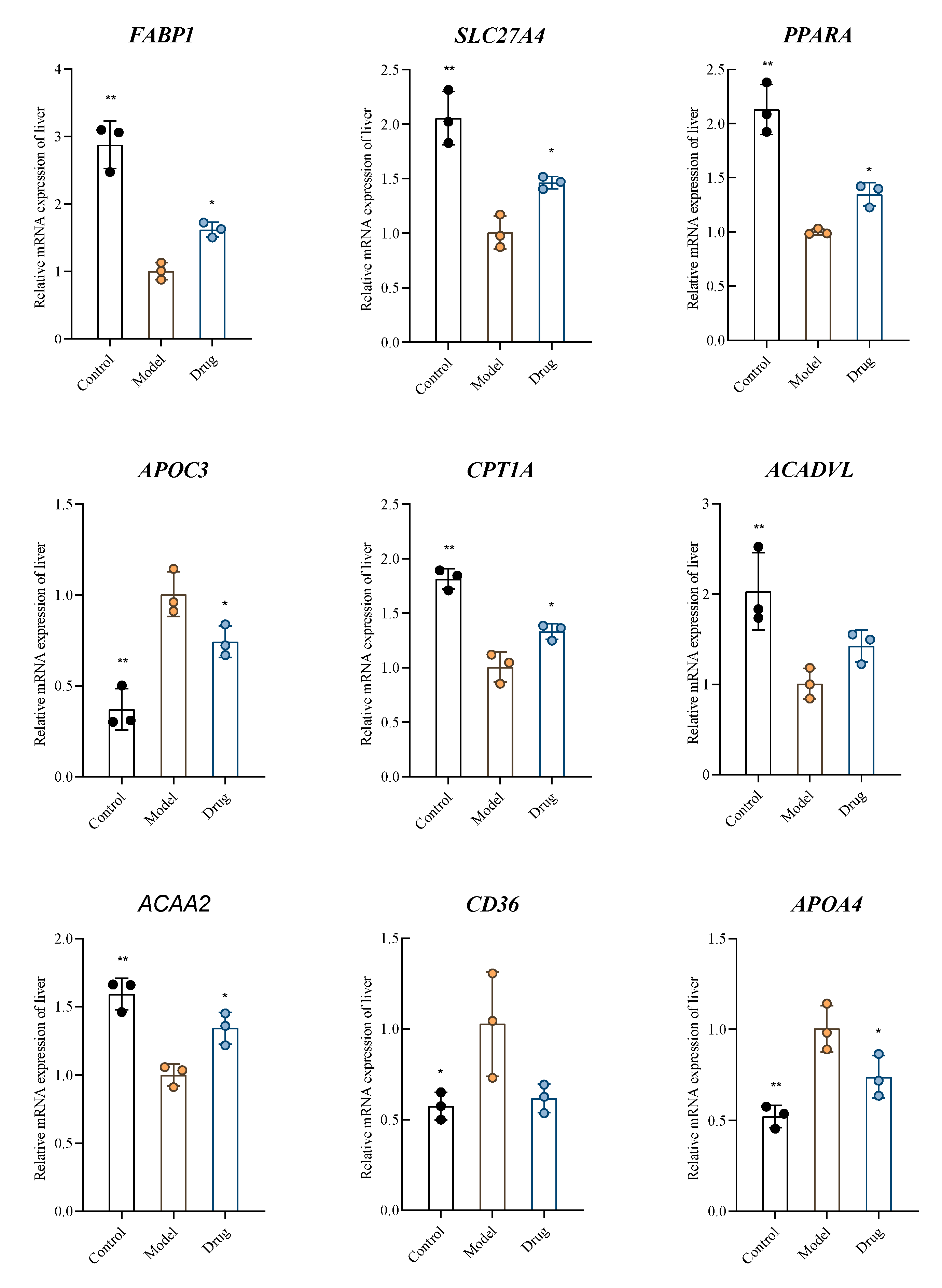
2.6. Verification by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Preparation of AFN
4.2. Determination of AFN by High-Performance Liquid Chromatography
4.3. Hyperlipidemic Mouse Model
4.4. Determination of Serum and Liver Biochemical Indices
4.5. Transcriptomics by RNA-Sequencing
4.6. UPLC-MSn Analysis of AFN Plasma-Absorbed Ingredients
4.7. Computational Analysis
4.8. Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Cui, B.; Yuan, W.K.; Wang, L.D.-L.; Wang, F.R.; Peng, J.; Ma, J.Y.; Chen, X.; Xu, M.Y.; Ke, J.; Tian, Y. Association between cooking patterns and the prevalence of hyperlipidemia in Eastern China. BMC Public Health 2024, 24, 75. [Google Scholar] [CrossRef]
- Karr, S. Epidemiology and Management of Hyperlipidemia. Am. J. Manag. Care 2017, 23, S139–S148. [Google Scholar]
- Wu, L.; Wang, X.; Jiang, J.; Chen, Y.; Peng, B.; Jin, W. Mechanism of rhubarb in the treatment of hyperlipidemia: A recent review. Open Med. 2023, 18, 20230812. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Jiang, L.; Li, C.; Sun, Z.; Zhang, Y.; Lin, T.; Jiang, Y.; Liu, B. A triple combination strategy of UHPLC-MSn, hypolipidemic activity and transcriptome sequencing to unveil the hypolipidemic mechanism of Nelumbo nucifera alkaloids. J. Ethnopharmacol. 2022, 282, 114608. [Google Scholar] [CrossRef]
- Guo, F.; Yang, X.; Li, X.; Feng, R.; Guan, C.; Wang, Y.; Li, Y.; Sun, C. Nuciferine prevents hepatic steatosis and injury induced by a high-fat diet in hamsters. PLoS ONE 2013, 8, e63770. [Google Scholar] [CrossRef]
- Pan, P.; Li, Q.; Lin, H.; Dong, L.; Lv, K.; Wang, Z. Exploration of the molecular mechanism of Polygonati Rhizoma in the treatment of hyperlipidemia based on network pharmacology and molecular docking. Medicine 2025, 104, e44087. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, Y.Y.; Jung, J.S.; Hyun, S.H.; Lee, J.; Park, J.-H.; Park, S.K.; Lee, S.H. Cholesterol-modulating effects of Korean red ginseng (KRG) targeting PCSK9 in hyperlipidemia. Sci. Rep. 2025, 15, 30637. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, J.; Wu, X.; Zhang, M.; Zhang, Y.; Chen, P.; Ma, J.; Zhang, S.; Zhang, H.; Li, X.; et al. Mult-omics analysis reveals the lipid-lowering effects of sea buckthorn and milk thistle solid beverage in hyperlipidemic rats. Phytomedicine Int. J. Phytother. Phytopharm. 2025, 144, 156920. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xie, Y.; Huang, L. Advance of Pharmacological Studies on Nuciferine. J. Nanjing Univ. Tradit. Chin. Med. 2021, 37, 619–624. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Wu, T.; Li, X.; Liu, Y.; Wang, H. Analysis of biomarker components of lotus leaf bioactivity effect. J. Food Saf. Qual. 2022, 13, 2105–2112. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, G.; Zhang, Y.; Yang, M.; Chen, J.; Guo, M. Potential hypoglycemic, hypolipidemic, and anti-inflammatory bioactive components in Nelumbo nucifera leaves explored by bioaffinity ultrafiltration with multiple targets. Food Chem. 2022, 375, 131856. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Liu, R.; Wang, D.; Niu, J.; Zhou, Y.; Ji, C. Mechanism of Qinglongyi on Anti-Gastritis Effect Based on Network Pharmacology and Molecular Docking. Chin. Pharm. J. 2023, 58, 599–610. [Google Scholar]
- Xu, M.; Chen, S.; Tan, X.; Ren, F.; Guan, T. Virtual Screening of DNA Topoisomerase IV Inhibitors Based onMolecular Docking. Chin. Pharm. J. 2023, 58, 875–883. [Google Scholar]
- Weisiger, R.A. Cytoplasmic transport of lipids: Role of binding proteins. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1996, 115, 319–331. [Google Scholar] [CrossRef]
- Anderson, C.M.; Stahl, A. SLC27 fatty acid transport proteins. Mol. Asp. Med. 2013, 34, 516–528. [Google Scholar] [CrossRef]
- Buechler, C.; Ritter, M.; Duong, C.Q.; Orso, E.; Kapinsky, M.; Schmitz, G. Adipophilin is a sensitive marker for lipid loading in human blood monocytes. Biochim. Biophys. Acta 2001, 1532, 97–104. [Google Scholar] [CrossRef]
- Luo, T.; Yang, R.; He, W. Neferine ameliorates palmitate-induced lipid by activating AMPK and mediating ACC/CPT1A and SREBP1/FAS pathways in HepG cells. J. Chin. Med. Mater. 2021, 44, 1754–1758. [Google Scholar] [CrossRef]
- Liang, J.; Lin, Y.; Yu, Y.; Wang, Y.; Zhu, J. Cloning and Expression of Goat CPT1A Gene and lts Correlation withIntramuscular Fat Content. Acta Agric. Boreali-Sinica 2019, 34, 231–238. [Google Scholar]
- de Boer, V.C.J.; van Schothorst, E.M.; Dihal, A.A.; van der Woude, H.; Arts, I.C.W.; Rietjens, I.M.C.M.; Hollman, P.C.H.; Keijer, J. Chronic quercetin exposure affects fatty acid catabolism in rat lung. Cell. Mol. Life Sci. CMLS 2006, 63, 2847–2858. [Google Scholar] [CrossRef]
- Goyal, S.; Tanigawa, Y.; Zhang, W.; Chai, J.-F.; Almeida, M.; Sim, X.; Lerner, M.; Chainakul, J.; Ramiu, J.G.; Seraphin, C.; et al. APOC3 genetic variation, serum triglycerides, and risk of coronary artery disease in Asian Indians, Europeans, and other ethnic groups. Lipids Health Dis. 2021, 20, 113. [Google Scholar] [CrossRef]
- Stitziel, N.O.; Kanter, J.E.; Bornfeldt, K.E. Emerging Targets for Cardiovascular Disease Prevention in Diabetes. Trends Mol. Med. 2020, 26, 744–757. [Google Scholar] [CrossRef]
- Simon, T.; Cook, V.R.; Rao, A.; Weinberg, R.B. Impact of murine intestinal apolipoprotein A-IV expression on regional lipid absorption, gene expression, and growth. J. Lipid Res. 2011, 52, 1984–1994. [Google Scholar] [CrossRef]
- VerHague, M.A.; Cheng, D.; Weinberg, R.B.; Shelness, G.S. Apolipoprotein A-IV expression in mouse liver enhances triglyceride secretion and reduces hepatic lipid content by promoting very low density lipoprotein particle expansion. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2501–2508. [Google Scholar] [CrossRef]
- Gomaraschi, M.; Putt, W.E.; Pozzi, S.; Iametti, S.; Barbiroli, A.; Bonomi, F.; Favari, E.; Bernini, F.; Franceschini, G.; Talmud, P.J.; et al. Structure and function of the apoA-IV T347S and Q360H common variants. Biochem. Biophys. Res. Commun. 2010, 393, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Stein, O.; Stein, Y.; Lefevre, M.; Roheim, P.S. The role of apolipoprotein A-IV in reverse cholesterol transport studied with cultured cells and liposomes derived from an ether analog of phosphatidylcholine. Biochim. Biophys. Acta 1986, 878, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
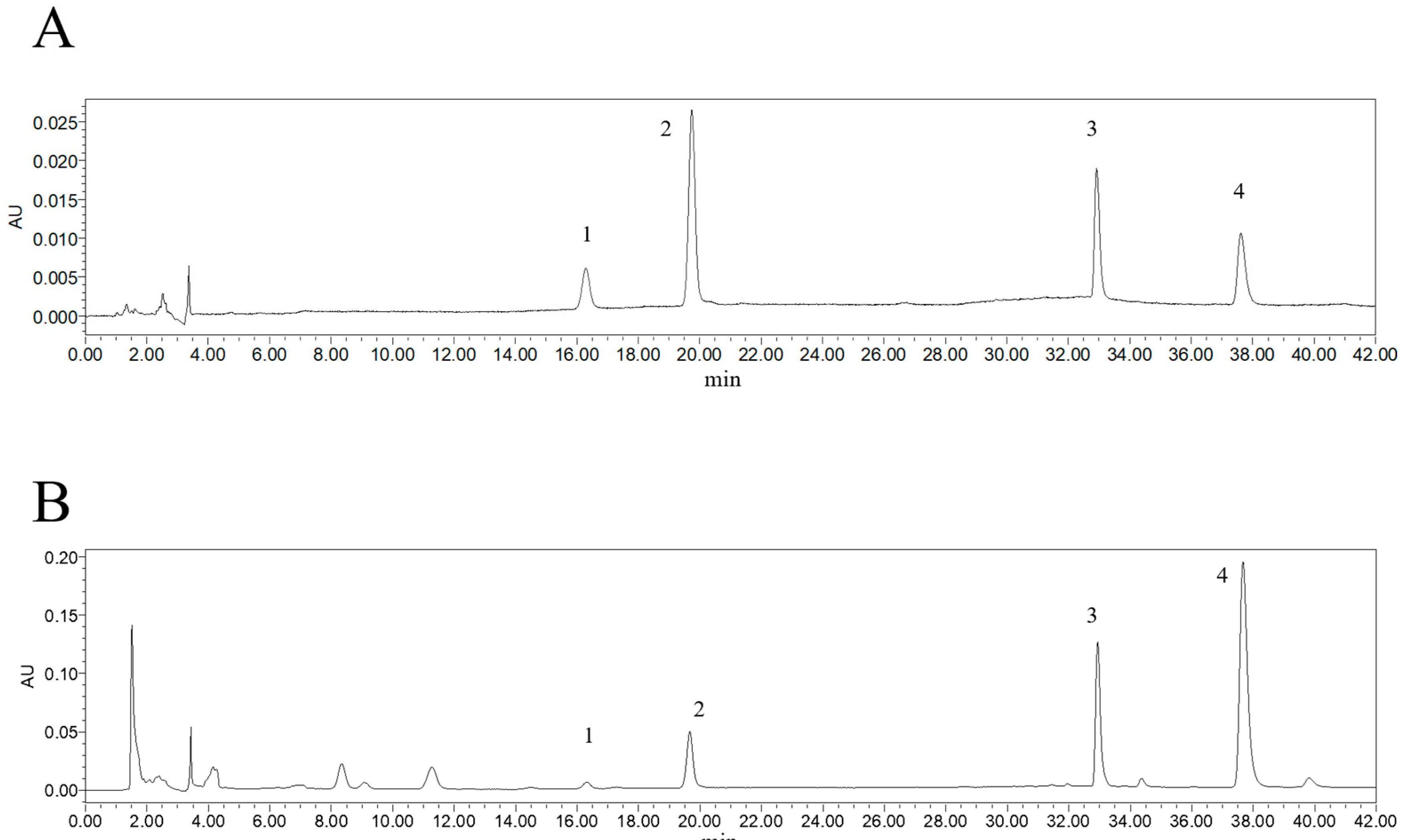
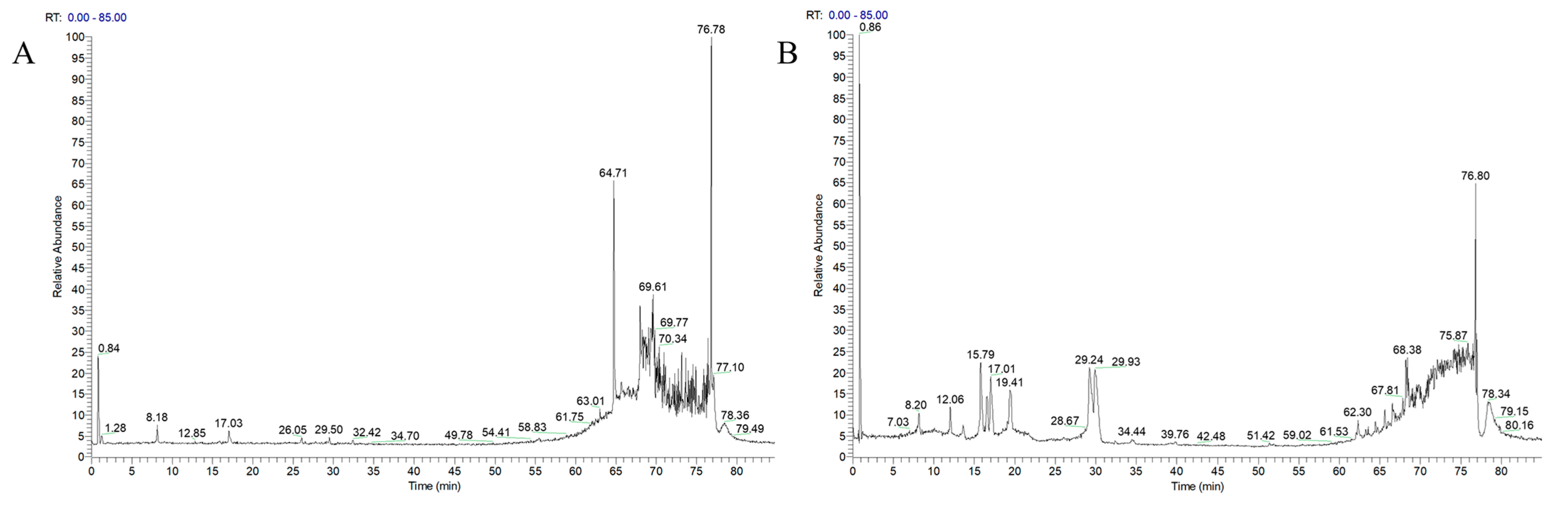
| No. | RT (Min) | Chemical Formula | Theoretical m/z | Measured (m/z) | Mass Error (ppm) | MS/MS (m/z) | Compound |
|---|---|---|---|---|---|---|---|
| 1 | 6.01 | C22H26NO9 | 448.16020 | 448.15952 | −1.535 | 272.12759, 255.10112, 237.09047, 161.05951, 107.04939 | Norcoclaurine-Glucuronide |
| 2 | 7.71 | C24H30NO9 | 476.19150 | 476.19080 | −1.487 | 300.15881, 269.11661, 237.09048, 175.07512, 107.04935 | N-Methylcoclaurine-Glucuronide |
| 3 | 8.08 | C23H28NO9 | 462.17585 | 462.17502 | −1.813 | 286.14325, 269.11673, 175.07516, 107.04937 | N-Norarmepavine desmethyl-Glucuronide |
| 4 | 8.20 | C23H28NO9 | 462.17585 | 462.17499 | −1.878 | 286.14322, 269.11667, 237.09055, 209.09575, 175.07513, 107.04935 | Coclaurine-Glucuronide |
| 5 | 8.73 | C24H30NO9 | 476.19150 | 476.19092 | −1.235 | 300.15894, 283.13239, 252.11403, 189.09076, 107.04939 | Armepavine desmethyl-Glucuronide |
| 6 | 9.54 | C23H26NO8 | 444.16529 | 444.16467 | −1.403 | 268.13269, 251.10629, 236.10664, 219.08018, 191.08528 | Caaverine-Glucuronide |
| 7 | 9.86 | C24H30NO9 | 476.19150 | 476.19095 | −1.172 | 300.15894, 283.13245, 252.11414, 189.09076, 107.04940 | Armepavine desmethyl-Glucuronide |
| 8 | 11.04 | C25H32NO9 | 490.20715 | 490.20657 | −1.199 | 314.17447, 283.13239, 252.11391, 189.09074, 107.04939 | Armepavine Glucuronide |
| 9 | 11.33 | C24H30NO9 | 476.19150 | 476.19089 | −1.298 | 300.15881, 283.13232, 251.10623, 175.07518, 107.04935 | N-Methylisococlaurine-Glucuronide |
| 10 | 11.67 | C23H26NO8 | 444.15783 | 444.15726 | −1.374 | 268.13269, 237.09061, 219.08018, 191.08527 | N-Nornuciferine desmethyl-Glucuronide |
| 11 | 11.69 | C23H28NO9 | 462.17585 | 462.17499 | −1.878 | 286.14319, 269.11664, 175.07510, 107.04935 | N-Norarmepavine desmethyl-Glucuronide |
| 12 | 11.81 | C23H28NO9 | 462.17585 | 462.17508 | −1.683 | 286.14328, 269.11670, 237.09074, 178.08604, 107.04937, 143.04901 | Coclaurine-Glucuronide |
| 13 | 12.05 | C24H30NO9 | 476.19150 | 476.19098 | −1.109 | 300.15894, 283.13245, 252.11407, 189.09079, 107.04939 | Armepavine desmethyl-Glucuronide |
| 14 | 12.50 | C23H26NO8 | 444.16529 | 444.16467 | −1.403 | 268.13269, 251.10628, 236.08281, 219.08020, 191.08527 | Caaverine-Glucuronide |
| 15 | 13.19 | C23H28NO9 | 462.17585 | 462.17529 | −1.229 | 286.14334, 269.11679, 237.09071, 209.09579, 178.08606, 143.04903, 107.04940 | Coclaurine-Glucuronide |
| 16 | 13.25 | C23H26NO8 | 444.17846 | 444.17834 | −1.538 | 268.13263, 251.10620, 219.08014, 191.08524 | N-Nornuciferine desmethyl-Glucuronide |
| 17 | 13.47 | C24H28NO8 | 458.18094 | 458.18027 | −1.469 | 282.14844, 251.10628, 219.08020, 191.08528 | O-Nornuciferine-Glucuronide |
| 18 | 16.60 | C17H18NO5S | 348.09001 | 348.08914 | −0.579 | 268.13257, 251.10611, 236.08261, 219.08008, 191.08517 | Caaverine-Sulfate |
| 19 | 17.73 | C18H20NO5S | 362.10566 | 362.10507 | −1.374 | 282.14847, 251.10629, 219.08022, 191.08017 | O-Nornuciferine-Sulfate |
| 20 | 17.82 | C18H20NO2 | 282.14885 | 282.14841 | −1.579 | 265.12198, 251.10623, 236.08275, 219.08017, 191.08527 | O-Nornuciferine * |
| 21 | 17.85 | C17H18NO5S | 362.09001 | 362.10495 | −1.143 | 282.14838, 265.28619, 250.12221 | O-Nornuciferine-Sulfate |
| 22 | 21.26 | C17H12NO3 | 278.08116 | 278.08063 | −1.941 | 263.05716, 250.08626, 235.06229 | Desmethyl lysicamine |
| 23 | 21.68 | C17H18NO2 | 268.13320 | 268.13266 | −2.034 | 251.10620, 236.08276, 219.08009, 191.08519 | Caaverine |
| 24 | 25.73 | C18H18NO2 | 280.13320 | 280.13275 | −1.625 | 26310638, 248.08281, 233.05934, 217.78973, 203.88098 | Dehydro-N-Nornuciferine-Glucuronide |
| 25 | 26.30 | C24H28NO8 | 458.18094 | 458.18008 | −1.884 | 282.14838, 251.10623 | N-Nornuciferine-Glucuronide |
| 26 | 29.59 | C17H16NO2 | 266.11755 | 266.12531 | −2.124 | 249.09058, 235.10686, 219.08014 | Anonaine |
| 27 | 29.79 | C18H20NO2 | 282.14885 | 282.14835 | −1.791 | 265.12192, 250.09842, 234.10358 | N-Nornuciferine * |
| 28 | 30.35 | C19H22NO2 | 296.16450 | 296.16400 | −1.707 | 265.12189, 250.09836, 234.10355 | Nuciferine * |
| 29 | 34.58 | C18H14NO3 | 292.09681 | 292.09631 | −1.745 | 277.07294, 248.07016 | Lysicamine |
| 30 | 34.95 | C19H22NO3 | 312.15942 | 312.15869 | −2.339 | 265.12183, 250.09834, 234.10350 | Nuciferoline |
| Primer Name | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| FABP1 | GTGGTCCGCAATGAGTTC | CACCTTCCAGCTTGACGA |
| SLC27A4 | GATTCTCCCTGTTGCTCCTGTA | TATCTCTCCTGACCGTCTTGAT |
| PPARA | GACTCTAAAGATCAGATTCCGC | GTTGAGCTGGTCTAGATCGCA |
| APOC3 | AGAAGGCTTGGGACTCAT | CTCTACCTCTTCAGCTCGG |
| CPT1A | ACCTTGGACCCAAATTGC | ATGTATTCCTCCCACCAGTCA |
| ACADVL | TCTGCCCAGCGACTTTAT | TGGTGGAAGCATCAGAGGA |
| ACAA2 | TCTGGTTTCCAGTCCATCG | TCTCTGTTCCTCCACACAAG |
| CD36 | GTCCTTACACATACAGAGTTCG | CTCTGTTCCAACAGACAGTGA |
| APOA4 | GGTGGAGCCAACTCAAGAA | GGCCTCTTGGACTTTAGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Huang, R.; Lin, T.; Yang, L.; Zhou, C.; Li, Y.; Liu, B.; Dong, S.; Jiang, Y. Combination of Plasma Pharmacochemistry, RNA-Seq, and Molecular Docking Strategies to Reveal the Mechanism of the Alkaloid Fraction of Nelumbinis folium for the Treatment of Hyperlipidemia. Molecules 2025, 30, 3727. https://doi.org/10.3390/molecules30183727
Cai Y, Huang R, Lin T, Yang L, Zhou C, Li Y, Liu B, Dong S, Jiang Y. Combination of Plasma Pharmacochemistry, RNA-Seq, and Molecular Docking Strategies to Reveal the Mechanism of the Alkaloid Fraction of Nelumbinis folium for the Treatment of Hyperlipidemia. Molecules. 2025; 30(18):3727. https://doi.org/10.3390/molecules30183727
Chicago/Turabian StyleCai, Yuan, Rong Huang, Tianfeng Lin, Leyi Yang, Chang Zhou, Yumiao Li, Bin Liu, Shifen Dong, and Yanyan Jiang. 2025. "Combination of Plasma Pharmacochemistry, RNA-Seq, and Molecular Docking Strategies to Reveal the Mechanism of the Alkaloid Fraction of Nelumbinis folium for the Treatment of Hyperlipidemia" Molecules 30, no. 18: 3727. https://doi.org/10.3390/molecules30183727
APA StyleCai, Y., Huang, R., Lin, T., Yang, L., Zhou, C., Li, Y., Liu, B., Dong, S., & Jiang, Y. (2025). Combination of Plasma Pharmacochemistry, RNA-Seq, and Molecular Docking Strategies to Reveal the Mechanism of the Alkaloid Fraction of Nelumbinis folium for the Treatment of Hyperlipidemia. Molecules, 30(18), 3727. https://doi.org/10.3390/molecules30183727






